- FASTA Sequence
- PDBx/mmCIF Format
- PDBx/mmCIF Format (gz)
- BinaryCIF Format (gz)
- Legacy PDB Format
- Legacy PDB Format (gz)
- PDBML/XML Format (gz)
- NMR Restraints (Text)
- NMR Restraints (Text - gz)
- NMR Restraints v2 (Text)
- NMR Restraints v2 (Text - gz)
- Validation Full (PDF - gz)
- Validation (XML - gz)
- Validation (CIF - gz)
- Biological Assembly 1 (CIF - gz)
- Biological Assembly 1 (PDB - gz)
2KOE|pdb_00002koe
Human cannabinoid receptor 1 - helix 7/8 peptide
- PDB DOI: https://doi.org/10.2210/pdb2KOE/pdb
- BMRB: 16504
- Classification: MEMBRANE PROTEIN, SIGNALING PROTEIN
- Organism(s): Homo sapiens
- Mutation(s): Yes
- Deposited: 2009-09-18 Released: 2009-10-06
- Deposition Author(s): Deshmukh, L.,Vinogradova, O.,Makriyannis, A.,Tiburu, E.,Tyukhtenko, S.,Janero, D.
Experimental Data Snapshot
- Method: SOLUTION NMR
- Conformers Calculated: 90
- Conformers Submitted: 18
- Selection Criteria: structures with the lowest energy
wwPDB Validation 3D Report Full Report
NMR solution structure of human cannabinoid receptor-1 helix 7/8 peptide: candidate electrostatic interactions and microdomain formation.
Tyukhtenko, S., Tiburu, E.K., Deshmukh, L., Vinogradova, O., Janero, D.R., Makriyannis, A.(2009) Biochem Biophys Res Commun 390: 441-446
- PubMed: 19766594 Search on PubMedSearch on PubMed Central
- DOI: https://doi.org/10.1016/j.bbrc.2009.09.053
- Primary Citation of Related Structures:
2KOE - PubMed Abstract:
We report the NMR solution structure of a synthetic 40-mer (T(377)-E(416)) that encompasses human cannabinoid receptor-1 (hCB1) transmembrane helix 7 (TMH7) and helix 8 (H8) [hCB1(TMH7/H8)] in 30% trifluoroethanol/H(2)O. Structural features include, from the peptide's amino terminus, a hydrophobic alpha-helix (TMH7); a loop-like, 11 residue segment featuring a pronounced Pro-kink within the conserved NPxxY motif; a short amphipathic alpha-helix (H8) orthogonal to TMH7 with cationic and hydrophobic amino-acid clusters; and an unstructured C-terminal end. The hCB1(TMH7/H8) NMR solution structure suggests multiple electrostatic amino-acid interactions, including an intrahelical H8 salt bridge and a hydrogen-bond network involving the peptide's loop-like region. Potential cation-pi and cation-phenolic OH interactions between Y(397) in the TMH7 NPxxY motif and R(405) in H8 are identified as candidate structural forces promoting interhelical microdomain formation. This microdomain may function as a flexible molecular hinge during ligand-induced hCB1 conformer transitions.
- Department of Chemistry and Chemical Biology, Northeastern University, Center for Drug Discovery, Boston, MA 02115-5000, USA.
Organizational Affiliation:
Explore in 3D: Structure |Sequence Annotations |Validation Report
Global Symmetry: Asymmetric - C1
Global Stoichiometry: Monomer - A1
Find Similar Assemblies
Biological assembly 1 assigned by authors.
Macromolecule Content
- Total Structure Weight: 4.52 kDa
- Atom Count: 317
- Modeled Residue Count: 40
- Deposited Residue Count: 40
- Unique protein chains: 1
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| human cannabinoid receptor 1 - helix 7/8 peptide | 40 | Homo sapiens | Mutation(s): 2 | 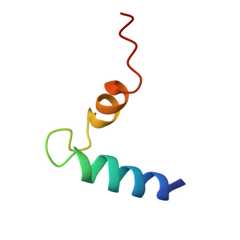 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P21554 (Homo sapiens) Explore P21554 Go to UniProtKB: P21554 | |||||
PHAROS: P21554 GTEx: ENSG00000118432 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P21554 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Experimental Data
- Method: SOLUTION NMR
- Conformers Calculated: 90
- Conformers Submitted: 18
- Selection Criteria: structures with the lowest energy
Deposition Data
- Released Date: 2009-10-06 Deposition Author(s): Deshmukh, L.,Vinogradova, O.,Makriyannis, A.,Tiburu, E.,Tyukhtenko, S.,Janero, D.
Revision History (Full details and data files)
- Version 1.0: 2009-10-06
Type: Initial release - Version 1.1: 2011-07-13
Changes: Version format compliance - Version 1.2: 2021-10-13
Changes: Data collection, Database references, Derived calculations - Version 1.3: 2024-05-22
Changes: Data collection
- RCSB PDB is a member of
- RCSB Partners
- Nucleic Acid Knowledgebase
RCSB PDB Core Operations are funded by theU.S. National Science Foundation (DBI-2321666), theUS Department of Energy (DE-SC0019749), and theNational Cancer Institute,National Institute of Allergy and Infectious Diseases, andNational Institute of General Medical Sciences of theNational Institutes of Health under grant R01GM157729.















