- Notifications
You must be signed in to change notification settings - Fork5
Visualising single-cell trajectories, including comparisons between two models 📈
License
Unknown, MIT licenses found
Licenses found
dynverse/dynplot
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation
ℹ️ Tutorials ℹ️ Referencedocumentation
Visualise a single-cell trajectory as a graph or dendrogram, as adimensionality reduction or heatmap of the expression data, or acomparison between two trajectories as a pairwise scatterplot ordimensionality reduction projection.
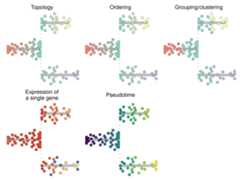
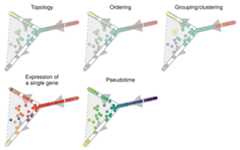
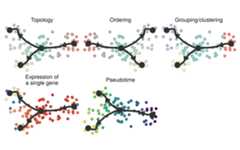
Here’s a summary of the different plotting functions for visualisingsingle-cell trajectories.
library(tidyverse)library(dyno)# get trajectorydata(example_bifurcating)trajectory<-example_bifurcating %>% add_root()# gather some prior informationgrouping<-trajectory$prior_information$groups_idgroups<- tibble(group_id=trajectory$milestone_ids,color=dynplot:::milestone_palette_list$auto(length(group_id)))features_oi<- apply(as.matrix(trajectory$counts),2,sd) %>% sort() %>% names() %>% tail(10)feature_oi<-features_oi[[10]]
patchwork::wrap_plots( plot_dendro(trajectory)+ labs(title="Topology"), plot_dendro(trajectory,"milestone")+ labs(title="Ordering"), plot_dendro(trajectory,grouping=grouping,groups=groups)+ labs(title="Grouping/clustering"), plot_dendro(trajectory,feature_oi=feature_oi)+ labs(title="Expression of\na single gene"), plot_dendro(trajectory,"pseudotime")+ labs(title="Pseudotime"),byrow=TRUE,ncol=3)& theme(legend.position="none")
patchwork::wrap_plots( plot_onedim(trajectory)+ labs(title="Topology"), plot_onedim(trajectory,"milestone")+ labs(title="Ordering"), plot_onedim(trajectory,grouping=grouping,groups=groups)+ labs(title="Grouping/clustering"), plot_onedim(trajectory,feature_oi=feature_oi)+ labs(title="Expression of\na single gene"), plot_onedim(trajectory,"pseudotime")+ labs(title="Pseudotime"),byrow=TRUE,ncol=2)& theme(legend.position="none")
patchwork::wrap_plots( plot_graph(trajectory)+ labs(title="Topology"), plot_graph(trajectory,"milestone")+ labs(title="Ordering"), plot_graph(trajectory,grouping=grouping,groups=groups)+ labs(title="Grouping/clustering"), plot_graph(trajectory,feature_oi=feature_oi)+ labs(title="Expression of\na single gene"), plot_graph(trajectory,"pseudotime")+ labs(title="Pseudotime"),byrow=TRUE,ncol=3)& theme(legend.position="none")
patchwork::wrap_plots( plot_dimred(trajectory)+ labs(title="Topology"), plot_dimred(trajectory,"milestone")+ labs(title="Ordering"), plot_dimred(trajectory,grouping=grouping,groups=groups)+ labs(title="Grouping/clustering"), plot_dimred(trajectory,feature_oi=feature_oi)+ labs(title="Expression of\na single gene"), plot_dimred(trajectory,"pseudotime")+ labs(title="Pseudotime"),byrow=TRUE,ncol=3)& theme(legend.position="none")
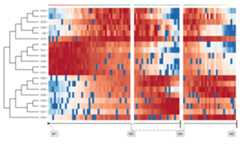
In addition, you can also plot the expression of genes along thetrajectory as a heatmap.
plot_heatmap(trajectory,grouping=trajectory$prior_information$grouping_assignment)
You can compare multiple trajectories (for the same cells) by creating ascatterplot between the two trajectories.
prediction<- infer_trajectory(trajectory, ti_comp1())#> v0.9.9.01: Pulling from dynverse/ti_comp1#> 844c33c7e6ea: Pulling fs layer#> 4f11e4e30170: Pulling fs layer#> a9724dff2655: Pulling fs layer#> f5f3a048c9c3: Pulling fs layer#> a53c4db932de: Pulling fs layer#> bd5da474b8ba: Pulling fs layer#> be9b916e18e7: Pulling fs layer#> b8b469f67972: Pulling fs layer#> e33132a1a81b: Pulling fs layer#> 32bd550d2fc1: Pulling fs layer#> a828ddf00b38: Pulling fs layer#> d97023e2f782: Pulling fs layer#> 84d8340a282e: Pulling fs layer#> be9b916e18e7: Waiting#> e33132a1a81b: Waiting#> 32bd550d2fc1: Waiting#> a828ddf00b38: Waiting#> a53c4db932de: Waiting#> b8b469f67972: Waiting#> f5f3a048c9c3: Waiting#> bd5da474b8ba: Waiting#> d97023e2f782: Waiting#> 844c33c7e6ea: Verifying Checksum#> 844c33c7e6ea: Download complete#> 844c33c7e6ea: Pull complete#> f5f3a048c9c3: Verifying Checksum#> f5f3a048c9c3: Download complete#> a53c4db932de: Verifying Checksum#> a53c4db932de: Download complete#> bd5da474b8ba: Verifying Checksum#> bd5da474b8ba: Download complete#> be9b916e18e7: Download complete#> 4f11e4e30170: Verifying Checksum#> 4f11e4e30170: Download complete#> 4f11e4e30170: Pull complete#> e33132a1a81b: Verifying Checksum#> e33132a1a81b: Download complete#> a9724dff2655: Verifying Checksum#> a9724dff2655: Download complete#> 32bd550d2fc1: Verifying Checksum#> 32bd550d2fc1: Download complete#> a9724dff2655: Pull complete#> f5f3a048c9c3: Pull complete#> a53c4db932de: Pull complete#> bd5da474b8ba: Pull complete#> be9b916e18e7: Pull complete#> a828ddf00b38: Verifying Checksum#> a828ddf00b38: Download complete#> 84d8340a282e: Verifying Checksum#> 84d8340a282e: Download complete#> d97023e2f782: Download complete#> b8b469f67972: Verifying Checksum#> b8b469f67972: Download complete#> b8b469f67972: Pull complete#> e33132a1a81b: Pull complete#> 32bd550d2fc1: Pull complete#> a828ddf00b38: Pull complete#> d97023e2f782: Pull complete#> 84d8340a282e: Pull complete#> Digest: sha256:012b3bfef2250767bcd3f3b389ebff82d4a2dc23d39b1bdadf7076e23b833680#> Status: Downloaded newer image for dynverse/ti_comp1:v0.9.9.01#> docker.io/dynverse/ti_comp1:v0.9.9.01trajectory$id<-"Bifurcating"prediction$id<-"Linear"plot_linearised_comparison(trajectory,prediction)
Check outnews(package = "dynwrap") orNEWS.md for a fulllist of changes.
BUG FIX
project_waypoints_coloured(): Fix refactoring issue “Mustsupply a symbol or a string as argument” (#54).BUG FIX
project_waypoints_coloured(): Fix wrong results whenprojecting waypoint segments (#54 bis).
Initial release on CRAN.
MINOR CHANGE: Add
arrowparameter to all plot functions.BUG FIX: Apply fixes for new versions of tibble, tidyr, and ggraph.
BUG FIX
optimise_order(): Fix problem where GA::ga() wouldn’t runon milestone networks with 1 or 4 edges.BUG FIX
linearise_cells(): Fix ordering issue whenequal_cell_widthisTRUE.MINOR CHANGE: Clean imports and supposed undefined variables.
MINOR CHANGE
plot_dendro(): Allow plotting of disconnected graphs(#32). This assumes thatdynwrap::add_root(traj, root_milestone_id = c(...))has beencalled properly.DOCUMENTATION: Extend documentation on usage of parameters and theexpected output values of functions.
About
Visualising single-cell trajectories, including comparisons between two models 📈
Topics
Resources
License
Unknown, MIT licenses found
Licenses found
Uh oh!
There was an error while loading.Please reload this page.
Stars
Watchers
Forks
Packages0
Uh oh!
There was an error while loading.Please reload this page.
Contributors4
Uh oh!
There was an error while loading.Please reload this page.