Introduction
The organisms of the fungal lineage include mushrooms, rusts, smuts, puffballs, truffles, morels, molds, and yeasts, as well as many less well-known organisms (Alexopoulos et al., 1996). More than 70,000 species of fungi have been described; however, some estimates of total numbers suggest that 1.5 million species may exist (Hawksworth, 1991; Hawksworth et al., 1995).
As the sister group of animals and part of the eukaryotic crown group that radiated about a billion years ago, the fungi constitute an independent group equal in rank to that of plants and animals. They share with animals the ability to export hydrolytic enzymes that break down biopolymers, which can be absorbed for nutrition. Rather than requiring a stomach to accomplish digestion, fungi live in their own food supply and simply grow into new food as the local environment becomes nutrient depleted.
Most biologists have seen dense filamentous fungal colonies growing on rich nutrient agar plates, but in nature the filaments can be much longer and the colonies less dense. When one of the filaments contacts a food supply, the entire colony mobilizes and reallocates resources to exploit the new food. Should all food become depleted, sporulation is triggered. Although the fungal filaments and spores are microscopic, the colony can be very large with individuals of some species rivaling the mass of the largest animals or plants.
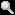
Click on an image to view larger version & data in a new window

Figure 1: Hyphae of a wood-decaying fungus found growing on the underside of a fallen log. The metabolically active hyphae have secreted droplets on their surfaces. Copyright © M. Blackwell 1996.
Prior to mating in sexual reproduction, individual fungi communicate with other individuals chemically via pheromones. In every phylum at least one pheromone has been characterized, and they range from sesquiterpines and derivatives of the carotenoid pathway in chytridiomycetes and zygomycetes to oligopeptides in ascomycetes and basidiomycetes.
Within their varied natural habitats fungi usually are the primary decomposer organisms present. Many species are free-living saprobes (users of carbon fixed by other organisms) in woody substrates, soils, leaf litter, dead animals, and animal exudates. The large cavities eaten out of living trees by wood-decaying fungi provide nest holes for a variety of animals, and extinction of the ivory billed woodpecker was due in large part to loss, through human activity, of nesting trees in bottom land hardwoods. In some low nitrogen environments several independent groups of fungi have adaptations such as nooses and sticky knobs with which to trap and degrade nematodes and other small animals. A number of references on fungal ecology are available (Carroll and Wicklow, 1992; Cooke and Whipps, 1993; Dix and Webster, 1995).
However, many other fungi are biotrophs, and in this role a number of successful groups form symbiotic associations with plants (including algae), animals (especially arthropods), and prokaryotes. Examples are lichens, mycorrhizae, and leaf and stem endophytes. Although lichens may seem infrequent in polluted cities, they can form the dominant vegetation in nordic environments, and there is a better than 80% chance that any plant you find is mycorrhizal. Leaf and stem endophytes are a more recent discovery, and some of these fungi can protect the plants they inhabit from herbivory and even influence flowering and other aspects of plant reproductive biology. Fungi are our most important plant pathogens, and include rusts, smuts, and many ascomycetes such as the agents of Dutch elm disease and chestnut blight. Among the other well known associations are fungal parasites of animals. Humans, for example, may succumb to diseases caused byPneumocystis(a type of pneumonia that affects individuals with supressed immune systems),Coccidioides(valley fever),Ajellomyces(blastomycosis and histoplasmosis), andCryptococcus(cryptococcosis) (Kwon-Chung and Bennett, 1992).
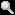
Click on an image to view larger version & data in a new window

Figure 2: The fluffy white hyphae of the mycorrhizal fungusRhizopogon rubescens has enveloped the smaller roots of a Virginia pine seedling. Note that some of the mycelium extends out into the surrounding environment. Copyright © J. B. Anderson 1996.
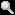
Click on an image to view larger version & data in a new window

Figure 3:Entomophthora, "destroyer of insects", is the agent of a fungual infection that kills flies. After their death the fungal growth erupts through the fly cuticle, and dispersal by forcible spore discharge is a source of inoculum for infection of new flies. Copyright © G. L. Barron 1996.
Fungal spores may be actively or passively released for dispersal by several effective methods. The air we breathe is filled with spores of species that are air dispersed. These usually are species that produce large numbers of spores, and examples include many species pathogenic on agricultural crops and trees. Other species are adapted for dispersal within or on the surfaces of animals (particularly arthropods). Some fungi are rain splash or flowing water dispersed. In a few cases the forcible release of spores is sufficient to serve as the dispersal method as well. The function of some spores is not primarily for dispersal, but to allow the organisms to survive as resistant cells during periods when the conditions of the environment are not conducive to growth.
Fungi are vital for their ecosystem functions, some of which we have reviewed in the previous paragraphs. In addition a number of fungi are used in the processing and flavoring of foods (baker's and brewer's yeasts, Penicillia in cheese-making) and in production of antibiotics and organic acids. Other fungi produce secondary metabolites such as aflatoxins that may be potent toxins and carcinogens in food of birds, fish, humans, and other mammals.
A few species are studied as model organisms that can be used to gain knowledge of basic processes such as genetics, physiology, biochemistry, and molecular biology with results that are applicable to many organisms (Taylor et al., 1993). Some of the fungi that have been intensively studied in this way includeSaccharomyces cereviseae,Neurospora crassa, andUstilago maydis.
Most phyla appear to be terrestrial in origin, although all major groups have invaded marine and freshwater habitats. An exception to this generality is the flagellum-bearing phyla Chytridiomycota, Blastocladiomycota, and Neocallimastigomycota (collectively referred to as chytrids), which probably had an aquatic origin. Extant chytrid species also occur in terrestrial environments as plant pathogenic fungi, soil fungi, and even as anaerobic inhabitants of the guts of herbivores such as cows (all Neocallimastigomycota).
Discussion of Phylogenetic Relationships
The kingdom Fungi is a diverse clade of heterotrophic organisms that shares some characters with animals such as chitinous structures, storage of glycogen, and mitochondrial codon UGA encoding tryptophan. Both animals and fungi have spores or gametes with a single smooth, posteriorly inserted flagellum, but only species of the basal chytrid phyla have retained this primitive character (Barr, 1992; Cavalier-Smith, 1987, 1995). Fungi, animals, and other heterotrophic protist-like organisms such as choanoflagellates and Mesomycetozoea are now considered part of the larger group termed opisthokonts (Cavalier-Smith, 1987) in reference to the posterior flagellum.
The branch uniting the fungi and animals is well-supported based on a number of molecular phylogenetic datasets, including the nuclear small subunit ribosomal RNA gene (Wainwright et al., 1993; Bruns et al. 1993), unique and shared sequence insertions in proteins such as elongation factor 1α (Baldauf and Palmer, 1993), entire mitochondrial genomes (Lang et al., 2002), and concatenated protein-coding genes (Steenkamp et al., 2006).
Prior classification systems of Fungi based primarily on morphology are in need of updating to more accurately reflect phylogenetic relationships as determined by molecular systematics. Molecular characters have been essential for phylogenetic analysis in cases when morphological characters are convergent, reduced, or missing among the taxa considered. This is especially true of species that never reproduce sexually, because characters of sexual reproduction traditionally have been the basis for classification of Fungi. Use of molecular characters allows asexual fungi to be placed among their closest relatives.
Previous classifications placed early-diverging fungal groups (non-Ascomycota or Basidiomycota) into two phyla: Chytridiomycota and Zygomycota. Numerous phylogenetic studies now suggest that neither is monophyletic, and the latest classification scheme includes six phyla and an additional four unplaced subphyla (Hibbett et al., 2007). At present, because of the ancient divergence times between the fungal phyla, the exact phylogenetic relationships are ambiguous. Chytrids appear to be a paraphyletic group at the base of the fungal phylogeny and merely fungal lineages which have retained the character of flagellated spores. Three phyla of flagellated fungi are proposed (Blastocladiomycota, Chytridiomycota, and Neocallimastigomycota; Hibbett et al., 2007) and two chytrid generaOlpidium andRozella, are of uncertain phylogenetic position (James et al., 2006a, 2006b). These genera are interesting because they are both highly reduced endoparasites (living inside the host cell) whose entire thallus consists of only a spherical body absorbing nutrients from the host material that surrounds it.Rozella appears in an isolated position in the fungal phylogeny as the very earliest lineage to diverge from the rest of the fungi (James et al., 2006a, 2006b). In contrast,Olpidium brassicae appears to have diverged after the majority of chytrids and is more closely related to some zygomycete fungi (James et al., 2006a, 2006b).
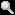
Click on an image to view larger version & data in a new window

Figure 7: The endoparasitic chytridRozella allomycis inside the hyphae of another chytridAllomyces. Thick spiny spores of the parasite are seen inside some cells while zoospores are produced in other cells. ©Timothy Y. James
Fungi with non-septate or irregularly septate hyphae and thick-walled spores were traditionally placed in the phylum Zygomycota. However, evidence for a monophyletic Zygomycota is lacking (Seif et al., 2005), and the deconstruction of the Zygomycota into four unordered subphyla (Entomophthoromycotina, Kickxellomycotina, Mucoromycotina, Zoopagomycotina) has been proposed (Hibbett et al., 2007). The separation of the superficially similar arbuscular mycorrhizal fungi (that lack septa in hyphae but also lack zygospores) into the phylum Glomeromycota has been previously proposed (Schüßler et al., 2001). Whether this phylum is more closely related to the Ascomycota and Basidiomycota lineage or to other zygomycete lineages is controversial (Redecker et al., 2006).
Evidence from shared morphological characters such as regularly septate hyphae and a dikaryotic stage (two separate and different nuclei in a single hyphal segment) in the life cycle usually has been interpreted as support for a close relationship between Basidiomycota and Ascomycota. Numerous phylogenetic studies such as SSU rDNA (Berbee and Taylor, 1992), RNA polymerase genes (Liu et al., 2006), and mitochondrial genome sequencing (Seif et al., 2005) provide strong support for this relationship. A subkingdom termed Dikarya is proposed (Hibbett et al., 2007), creating a division between a highly speciose subkingdom (Dikarya) and the remaining early diverging lineages whose relationships are not precisely known.
Fungal classification is far from static, and even which organisms are actually members of Fungi is changing. For example, the group trichomycetes describes gut inhabitants of arthropods that share similarities with zygomycetes. Molecular phylogenetic studies have demonstrated that two of the four orders of trichomycetes are actually members of the Mesomycetozoea protist group (Benny and O’Donnell, 2000; Cafaro, 2005). Other organisms that were previously considered to be Fungi because of their heterotrophic, mold-like growth forms are now classified as stramenopiles (Oomycota, Hyphochytriomycota, and Labyrinthulomycota) or slime molds (Myxomycota, Plasmodiomycota, Dictyosteliomycota, Acrasiomycota) (Bhattacharya et al., 1992; Leipe et al., 1994; Van der Auwera et al., 1995). More interesting for mycologists are the findings that some species previously considered protozoa are actually Fungi. For example, the speciesHyaloraphidium curvatum was assumed to be a green alga that had adopted a heterotrophic lifecycle concomitantly with losing its chloroplast. It is now known to be a chytrid fungus related to Monoblephariomycetes but lacking a flagellated stage (Ustinova et al., 2000). Other examples include the parasitic organisms presumed to be protozoa, such as the cockroach parasiteNepridiophaga (Wylezich et al., 2004) and theDaphnia parasitePolycarum (Johnson et al., 2006) recently demonstrated to be members of the fungal kingdom based on SSU rDNA phylogenies.
The most revolutionary addition to the fungal lineage has occurred with phylogenetic evidence indicating the protist group microsporidia is closely related to Fungi–possibly derived from zygomycetes (Keeling, 2003) or sister to the genusRozella on the earliest branch in the fungal kingdom (James et al., 2006a). Microsporidia are highly specialized intracellular parasites (primarily of animals) that lack mitochondria but have chitin and trehalose in their spores (similar to Fungi). All molecular studies have shown that microsporidia evolve at an extremely accelerated rate of evolution, making their placement in the Tree of Life difficult. The relationship with fungi is supported by many single and multiple gene phylogenies (e.g., Liu et al., 2006), but an exact placement within the fungi has not received strong support (Keeling and Fast, 2002).
More recently the nuclearid amoebae have been demonstrated to be a sister group to the Fungi with strong support (Steenkamp et al., 2006). This finding is significant becauseNuclearia lacks a cell wall and has phagotrophic nutrition in which the food source (such as a bacterium or algal cell) is engulfed wholly, unlike fungi and microsporidia which utilize absorptive nutrition. Further sampling of basal fungal lineages will be needed to determine whether aNuclearia-like organism was the cenancestor (most recent common ancestor) of Fungi.
References
Alexopoulos, C. J., C. W. Mims, and M. Blackwell. 1996. Introductory Mycology (4th Ed.). John Wiley and Sons, New York, USA. 868p.
Baldauf, S. L., and J. D. Palmer. 1993. Animals and fungi are each other's closest relatives: congruent evidence form multiple proteins. Proceedings of the National Academy of Sciences (USA) 90:11558-11562.
Barr, D. J. S. 1992. Evolution and kingdoms of organisms from the perspective of a mycologist. Mycologia 84:1-11.
Benny G. L. and K. O'Donnell. 2000. Amoebidium parasiticum is a protozoan, not a Trichomycete. Mycologia 92: 1133-1137.
Berbee, M. L., and J. W. Taylor. 1992. Two ascomycete classes based on fruiting-body characters and ribosomal DNA sequence. Molecular Biology and Evolution 9:278-284.
Berbee, M. L., and J. W. Taylor. 1993. Dating the evolutionary radiations of the true fungi. Canadian Journal of Botany 71:1114-1127.
Bhattacharya, D., L. Medlin, P. O. Wainright, E. V. Ariztia, C. Bibeau, S. K. Stickel, and M. L. Sogin. 1992. Algae containing chlorophylls a +c are paraphyletic: molecular evolutionary analysis of the Chromophyta. Evolution 46:801-1817.
Bruns, T. D., T. J. White, and J. W. Taylor. 1991. Fungal molecular systematics. Annual Review of Ecology and Systematics 22:525-564.
Bruns, T. D., R. Vilgalys, S. M. Barns, D. Gonzalez, D. S. Hibbett, D. J. Lane, L. Simon, S. Stickel, T. M. Szaro, W. G. Weisburg, and M. L. Sogin. 1993. Evolutionary relationships within the fungi: analysis of nuclear small subunit rRNA sequences. Molecular Phylogenetics and Evolution 1:231 241.
Cafaro, M. J. 2005. Eccrinales (Trichomycetes) are not fungi, but a clade of protists at the early divergence of animals and fungi. Molecular Phylogenetics and Evolution. 35: 21-34.
Carroll, G.C., and D.T. Wicklow. 1992. The Fungal Community: Its Organization and Role in the Ecosystem. Marcel Deker, Inc., New York.
Cavalier-Smith, T. 1987. The origin of Fungi and Pseudofungi. Pp. 339-353. In: Evolutionary Biology of the Fungi. Eds. A. D. M. Rayner, C. M. Brasier, and D. Moore. Cambridge University Press, Cambridge, United Kingdom.
Cooke, R.C., and J.M. Whipps. 1993. Ecophysiology of Fungi. Blackwell Scientific Pub., London, U.K.
Dix, N.J., and J.W. Webster. 1995. Fungal Ecology. Capman and Hall. London, U.K.
Griffin, D. 1993. Fungal Physiology (2nd Ed.). Wiley-Liss. New York.
Hasegawa, M., T. Hashimoto, J. Adachi, N. Iwabe, and T. Miyata. 1993. Early branchings in the evolution of eukaryotes: ancient divergence of Entamoeba that lacks mitochondria revealed by protein sequence data. Journal of Molecular Evolution 36:380-388.
Hass, H., T. N. Taylor, and W. Remy. 1994. Fungi from the Lower Devonian Rhynie Chert - mycoparasitism. American Journal of Botany 81:29-37.
Hawksworth, D. L. 1991. The fungal dimension of biodiversity: magnitude, significance, and conservation. Mycological Research 95:641-655.
Hawksworth, D. L., P. M. Kirk, B. C. Sutton, and D. N. Pegler. 1995. Ainsworth and Bisby's Dictionary of the Fungi (8th Ed.). CAB International, Wallingford, United Kingdom. 616p.
Hibbett, D. S., M. Binder, J. F. Bischoff, M. Blackwell, P. F. Cannon, O. E. Eriksson, S. Huhndorf, T. James, P. M. Kirk, R. Lücking, T. Lumbsch, F. Lutzoni, P. B. Matheny, D. J. Mclaughlin, M. J. Powell, S. Redhead, C. L. Schoch, J. W. Spatafora, J. A. Stalpers, R. Vilgalys, M. C. Aime, A. Aptroot, R. Bauer, D. Begerow, G. L. Benny, L. A. Castlebury, P. W. Crous, Y.-C. Dai, W. Gams, D. M. Geiser, G. W. Griffith, C. Gueidan, D. L. Hawksworth, G. Hestmark, K. Hosaka, R. A. Humber, K. Hyde, J. E. Ironside, U. Kõljalg, C. P. Kurtzman, K.-H. Larsson, R. Lichtwardt, J. Longcore, J. Miądlikowska, A. Miller, J.-M. Moncalvo, S. Mozley-Standridge, F. Oberwinkler, E. Parmasto, V. Reeb, J. D. Rogers, C. Roux, L. Ryvarden, J. P. Sampaio, A. Schüßler, J. Sugiyama, R. G. Thorn, L. Tibell, W. A. Untereiner, C. Walker, Z. Wang, A. Weir, M. Weiß, M. M. White, K. Winka, Y.-J. Yao, and N. Zhang. 2007. A higher-level phylogenetic classification of the Fungi. Mycological Research 111: 509-547.
Isaac, S., J. C. Frankland, R. Watling, and A. J. S. Whalley. 1993. Aspects of Tropical Mycology. Cambridge University Press, Cambridge, U.K.
James, T. Y., F. Kauff, C. Schoch, P. B. Matheny, V. Hofstetter, C. Cox, G. Celio, C. Gueidan, E. Fraker, J. Miadlikowska, H. T. Lumbsch, A. Rauhut, V. Reeb, A. E. Arnold, A. Amtoft, J. E. Stajich, K. Hosaka, G.-H. Sung, D. Johnson, B. ORourke, M. Crockett, M. Binder, J. M. Curtis, J. C. Slot, Z. Wang, A. W. Wilson, A. Schüßler, J. E. Longcore, K. ODonnell, S. Mozley-Standridge, D. Porter, P. M. Letcher, M. J. Powell, J. W. Taylor, M. M. White, G. W. Griffith, D. R. Davies, R. A. Humber, J. B. Morton, J. Sugiyama, A. Y. Rossman, J. D. Rogers, D. H. Pfister, D. Hewitt, K. Hansen, S. Hambleton, R. A. Shoemaker, J. Kohlmeyer, B. Volkmann-Kohlmeyer, R. A. Spotts, M. Serdani, P. W. Crous, K. W. Hughes, K. Matsuura, E. Langer, G. Langer, W. A. Untereiner, R. Lücking, B. Büdel, D. M. Geiser, A. Aptroot, P. Diederich, I. Schmitt, M. Schultz, R. Yahr, D. Hibbett, F Lutzoni, D. McLaughlin, J. Spatafora, and R. Vilgalys. 2006a. Reconstructing the early evolution of the fungi using a six gene phylogeny. Nature 443:818-822.
James, T. Y., P. M. Letcher, J. E. Longcore, S. E. Mozley-Standridge, D. Porter, M. J. Powell, G. W. Griffith, and R. Vilgalys. 2006b. A molecular phylogeny of the flagellated Fungi (Chytridiomycota) and a proposal for a new phylum (Blastocladiomycota). Mycologia 98: 860-871.
Johnson, P. T. J., J. E. Longcore, D. E. Stanton, R. B. Carnegie, J. D. Shields, and E. R. Preu. 2006. Chytrid infections of Daphnia pulicaria: development, ecology, pathology and phylogeny of Polycaryum laeve. Freshwater Biology 51: 634-648.
Keeling, P. J. 2003. Congruent evidence from alpha-tubulin and beta-tubulin gene phylogenies for a zygomycete origin of microsporidia. Fungal Genetics and Biology 38: 298-309.
Keeling, P. J., and N. M. Fast. 2002. Microsporidia: biology and evolution of highly reduced intracellular parasites. Annual Review of Microbiology 56: 93-116.
Kwon-Chung, K.J., and J.E. Bennett. 1992. Medical Mycology. Lea and Febiger, Philadelphia.
Lang, B. F., C. O'Kelly, T. Nerad, M. W. Gray, and G. Burger. 2002. Current Biology 12: 1773-1778.
Leipe, D. D., P. O. Wainright, J. H. Gunderson, D. Porter, D. J. Patterson, F. Valois, S. Himmerich, and M. L. Sogin. 1994. The straminopiles from a molecular perspective: 16S-like rRNA sequences from Labyrinthula minuta and Cafeteria roenbergensis. Phycologia 33:369-377.
Liu, Y., M. C. Hodson, and B. D. Hall. 2006. Loss of the flagellum happened only once in the fungal lineage: phylogenetic structure of kingdom Fungi inferred from RNA polymerase II subunit genes. BMC Evolutionary Biology 6:74.
Matthews, P. (Ed.). 1994. Guinness Book of Records. Bantum Books, New York. 819p.
Morehouse, E. A., T. Y. James, A. R. D. Ganley, R. Vilgalys, L. Berger, P. J. Murphy, and J. E. Longcore. 2003. Multilocus sequence typing suggests the chytrid pathogen of amphibians is a recently emerged clone. Molecular Ecology 12:395-403.
Morgan, J. A. T., V. T. Vredenburg, L. J. Rachowicz, R. A. Knapp, M. J. Stice, T. Tunstall, R. E. Bingham, J. M. Parker, J. E. Longcore, C. Moritz, C. J. Briggs, and J. W. Taylor. 2007. Population genetics of the frog killing fungus Batrachochytrium dendrobatidis. Proceedings of the National Academy of Sciences 104:13845-13850.
Nagahama, T., H. Sato, M. Shimazu, and J. Sugiyama. 1995. Phylogenetic divergence of the entomophthoralean fungi: evidence from nuclear 18S ribosomal RNA gene sequences. Mycologia 87:203-209.
Redecker, D., and P. Raab. 2006. Phylogeny of the Glomeromycota (arbuscular mycorrhizal fungi): recent developments and new gene markers. Mycologia 98: 885-895.
Remy, W., T. N. Taylor, and H. Hass. 1994a. Early Devonian fungi - a blastocladalean fungus with sexual reproduction. American Journal of Botany 81:690-702.
Remy, W., T. N. Taylor, H. Hass, and H. Kerp. 1994b. Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proceedings of the National Academy of Sciences (USA) 91:11841-11843.
Rodrigo, A. G., P. R. Bergquist, and P. I. Bergquist. 1994. Inadequate support for an evolutionary link between the metazoa and the fungi. Systematic Biology 43:578-584.
Schüßler, A., D. Schwarzott, and C. Walker. 2001. A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycological Research. 105: 1413-1421.
Seif, E., J. Leigh, Y. Liu, I. Roewer, L. Forget, and B. F. Lang. 2005. Comparative mitochondrial genomics in zygomycetes: bacteria-like RNase P RNAs, mobile elements and a close source of the group I intron invasion in angiosperms. Nucleic Acids Research 33:734-744.
Sherwood-Pike, M. A., and J. Gray. 1985. Silurian fungal remains: probable records of the class Ascomycota. Lethaia 18:1-20.
Sidow, A., and W. K. Thomas. 1994. A molecular evolutionary framework for eukaryotic model organisms. Current Biology 4:596-603.
Steenkamp, E. T., J. Wright, and S. L. Baldauf. 2006. The protistan origins of animals and fungi. Mol. Biol. Evol. 23:93-106.
Taylor, J. W., B. Bowman, M. L. Berbee, and T. J. White. 1993. Fungal model organisms: phylogenetics of Saccharomyces, Aspergillus and Neurospora.. Systematic Biology 42:440-457.
Taylor, T. N., W. Remy, H. Hass 1994a. Allomyces in the Devonian. Nature 367:601-601.
Taylor, T. N., J. Galtier, B. J. Axsmith. 1994b. Fungi from the Lower Carboniferous of central France. Review of Palaeobotany and Palynology 83:253-260.
Taylor, T. N., W. Remy, H. Hass, H. Kerp. 1995a. Fossil arbuscular mycorrhizae from the Early Devonian. Mycologia 87:560-573.
Ustinova, I., L. Krienitz and V. A. R. Huss. 2000. Hyaloraphidium curvatum is not a green alga, but a lower fungus; Amobedium parasiticum is not a fungus, but a member of the DRIPs. Protist 151: 253-262.
Van der Auwera, G., R. De Baere, Y. Van de Peer, P. De Rijk, I. Van den Broeck, and R. De Wachter. 1995. The phylogeny of Hyphochytriomycota as deduced from ribosomal RNA sequences of Hyphochytrium catenoides. Molecular Biology and Evolution 12:671-678.
Wainright, P. O., G. Hinkle, M. L. Sogin, and S. K. Stickel. 1993. Monophyletic origins of the Metazoa: an evolutionary link with fungi. Science 260:340-342.
Wylezich, C., R. Radek, and M. Schlegel. 2004. Phylogenetic analysis of the 18S rRNA identifies the parasitic protist Nephridiophaga blattellae (Nephridiophagidae) as a representative of the Zygomycota (Fungi). Denisia 13: 435-442.
 Formed in 2009, the Archive Team (not to be confused with the archive.org Archive-It Team) is a rogue archivist collective dedicated to saving copies of rapidly dying or deleted websites for the sake of history and digital heritage. The group is 100% composed of volunteers and interested parties, and has expanded into a large amount of related projects for saving online and digital history.
Formed in 2009, the Archive Team (not to be confused with the archive.org Archive-It Team) is a rogue archivist collective dedicated to saving copies of rapidly dying or deleted websites for the sake of history and digital heritage. The group is 100% composed of volunteers and interested parties, and has expanded into a large amount of related projects for saving online and digital history.






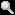



















 This media file is licensed under the
This media file is licensed under the


 Go to quick linksGo to quick searchGo to navigation for this section of the ToL siteGo to detailed links for the ToL site
Go to quick linksGo to quick searchGo to navigation for this section of the ToL siteGo to detailed links for the ToL site