More About PyGAD¶
Multi-Objective Optimization¶
InPyGAD3.2.0,the library supports multi-objective optimization using thenon-dominated sorting genetic algorithm II (NSGA-II). The code isexactly similar to the regular code used for single-objectiveoptimization except for 1 difference. It is the return value of thefitness function.
In single-objective optimization, the fitness function returns a singlenumeric value. In this example, the variablefitness is expected tobe a numeric value.
deffitness_func(ga_instance,solution,solution_idx):...returnfitness
But in multi-objective optimization, the fitness function returns any ofthese data types:
listtuplenumpy.ndarray
deffitness_func(ga_instance,solution,solution_idx):...return[fitness1,fitness2,...,fitnessN]
Whenever the fitness function returns an iterable of these data types,then the problem is considered multi-objective. This holds even if thereis a single element in the returned iterable.
Other than the fitness function, everything else could be the same inboth single and multi-objective problems.
But it is recommended to use one of these 2 parent selection operatorsto solve multi-objective problems:
nsga2: This selects the parents based on non-dominated sortingand crowding distance.tournament_nsga2: This selects the parents using tournamentselection which uses non-dominated sorting and crowding distance torank the solutions.
This is a multi-objective optimization example that optimizes these 2linear functions:
y1=f(w1:w6)=w1x1+w2x2+w3x3+w4x4+w5x5+6wx6y2=f(w1:w6)=w1x7+w2x8+w3x9+w4x10+w5x11+6wx12
Where:
(x1,x2,x3,x4,x5,x6)=(4,-2,3.5,5,-11,-4.7)andy=50(x7,x8,x9,x10,x11,x12)=(-2,0.7,-9,1.4,3,5)andy=30
The 2 functions use the same parameters (weights)w1 tow6.
The goal is to use PyGAD to find the optimal values for such weightsthat satisfy the 2 functionsy1 andy2.
importpygadimportnumpy"""Given these 2 functions: y1 = f(w1:w6) = w1x1 + w2x2 + w3x3 + w4x4 + w5x5 + 6wx6 y2 = f(w1:w6) = w1x7 + w2x8 + w3x9 + w4x10 + w5x11 + 6wx12 where (x1,x2,x3,x4,x5,x6)=(4,-2,3.5,5,-11,-4.7) and y=50 and (x7,x8,x9,x10,x11,x12)=(-2,0.7,-9,1.4,3,5) and y=30What are the best values for the 6 weights (w1 to w6)? We are going to use the genetic algorithm to optimize these 2 functions.This is a multi-objective optimization problem.PyGAD considers the problem as multi-objective if the fitness function returns: 1) List. 2) Or tuple. 3) Or numpy.ndarray."""function_inputs1=[4,-2,3.5,5,-11,-4.7]# Function 1 inputs.function_inputs2=[-2,0.7,-9,1.4,3,5]# Function 2 inputs.desired_output1=50# Function 1 output.desired_output2=30# Function 2 output.deffitness_func(ga_instance,solution,solution_idx):output1=numpy.sum(solution*function_inputs1)output2=numpy.sum(solution*function_inputs2)fitness1=1.0/(numpy.abs(output1-desired_output1)+0.000001)fitness2=1.0/(numpy.abs(output2-desired_output2)+0.000001)return[fitness1,fitness2]num_generations=100num_parents_mating=10sol_per_pop=20num_genes=len(function_inputs1)ga_instance=pygad.GA(num_generations=num_generations,num_parents_mating=num_parents_mating,sol_per_pop=sol_per_pop,num_genes=num_genes,fitness_func=fitness_func,parent_selection_type='nsga2')ga_instance.run()ga_instance.plot_fitness(label=['Obj 1','Obj 2'])solution,solution_fitness,solution_idx=ga_instance.best_solution(ga_instance.last_generation_fitness)print(f"Parameters of the best solution :{solution}")print(f"Fitness value of the best solution ={solution_fitness}")prediction=numpy.sum(numpy.array(function_inputs1)*solution)print(f"Predicted output 1 based on the best solution :{prediction}")prediction=numpy.sum(numpy.array(function_inputs2)*solution)print(f"Predicted output 2 based on the best solution :{prediction}")
This is the result of the print statements. The predicted outputs areclose to the desired outputs.
Parametersofthebestsolution:[0.79676439-2.98823386-4.126776625.70539445-2.02797016-1.07243922]Fitnessvalueofthebestsolution=[1.68090829349.8591915]Predictedoutput1basedonthebestsolution:50.59491545442283Predictedoutput2basedonthebestsolution:29.99714270722312
This is the figure created by theplot_fitness() method. The fitnessof the first objective has the green color. The blue color is used forthe second objective fitness.
Limit the Gene Value Range using thegene_space Parameter¶
InPyGAD2.11.0,thegene_space parameter supported a new feature to allowcustomizing the range of accepted values for each gene. Let’s take aquick review of thegene_space parameter to build over it.
Thegene_space parameter allows the user to feed the space of valuesof each gene. This way the accepted values for each gene is retracted tothe user-defined values. Assume there is a problem that has 3 geneswhere each gene has different set of values as follows:
Gene 1:
[0.4,12,-5,21.2]Gene 2:
[-2,0.3]Gene 3:
[1.2,63.2,7.4]
Then, thegene_space for this problem is as given below. Note thatthe order is very important.
gene_space=[[0.4,12,-5,21.2],[-2,0.3],[1.2,63.2,7.4]]
In case all genes share the same set of values, then simply feed asingle list to thegene_space parameter as follows. In this case,all genes can only take values from this list of 6 values.
gene_space=[33,7,0.5,95.6.3,0.74]
The previous example restricts the gene values to just a set of fixednumber of discrete values. In case you want to use a range of discretevalues to the gene, then you can use therange() function. Forexample,range(1,7) means the set of allowed values for the geneare1,2,3,4,5,and6. You can also use thenumpy.arange() ornumpy.linspace() functions for the same purpose.
The previous discussion only works with a range of discrete values notcontinuous values. InPyGAD2.11.0,thegene_space parameter can be assigned a dictionary that allowsthe gene to have values from a continuous range.
Assuming you want to restrict the gene within this half-open range [1 to5) where 1 is included and 5 is not. Then simply create a dictionarywith 2 items where the keys of the 2 items are:
'low': The minimum value in the range which is 1 in the example.'high': The maximum value in the range which is 5 in the example.
The dictionary will look like that:
{'low':1,'high':5}
It is not acceptable to add more than 2 items in the dictionary or useother keys than'low' and'high'.
For a 3-gene problem, the next code creates a dictionary for each geneto restrict its values in a continuous range. For the first gene, it cantake any floating-point value from the range that starts from 1(inclusive) and ends at 5 (exclusive).
gene_space=[{'low':1,'high':5},{'low':0.3,'high':1.4},{'low':-0.2,'high':4.5}]
More about thegene_space Parameter¶
Thegene_space parameter customizes the space of values of eachgene.
Assuming that all genes have the same global space which include thevalues 0.3, 5.2, -4, and 8, then those values can be assigned to thegene_space parameter as a list, tuple, or range. Here is a listassigned to this parameter. By doing that, then the gene values arerestricted to those assigned to thegene_space parameter.
gene_space=[0.3,5.2,-4,8]
If some genes have different spaces, thengene_space should accept anested list or tuple. In this case, the elements could be:
Number (of
int,float, orNumPydata types): A singlevalue to be assigned to the gene. This means this gene will have thesame value across all generations.list,tuple,numpy.ndarray, or any range likerange,numpy.arange(), ornumpy.linspace: It holds the space foreach individual gene. But this space is usually discrete. That isthere is a set of finite values to select from.dict: To sample a value for a gene from a continuous range. Thedictionary must have 2 mandatory keys which are"low"and"high"in addition to an optional key which is"step". Arandom value is returned between the values assigned to the itemswith"low"and"high"keys. If the"step"exists, thenthis works as the previous options (i.e. discrete set of values).None: A gene with its space set toNoneis initializedrandomly from the range specified by the 2 parametersinit_range_lowandinit_range_high. For mutation, its valueis mutated based on a random value from the range specified by the 2parametersrandom_mutation_min_valandrandom_mutation_max_val. If all elements in thegene_spaceparameter areNone, the parameter will not have any effect.
Assuming that a chromosome has 2 genes and each gene has a differentvalue space. Then thegene_space could be assigned a nestedlist/tuple where each element determines the space of a gene.
According to the next code, the space of the first gene is[0.4,-5]which has 2 values and the space for the second gene is[0.5,-3.2,8.8,-9] which has 4 values.
gene_space=[[0.4,-5],[0.5,-3.2,8.2,-9]]
For a 2 gene chromosome, if the first gene space is restricted to thediscrete values from 0 to 4 and the second gene is restricted to thevalues from 10 to 19, then it could be specified according to the nextcode.
gene_space=[range(5),range(10,20)]
Thegene_space can also be assigned to a single range, as givenbelow, where the values of all genes are sampled from the same range.
gene_space=numpy.arange(15)
Thegene_space can be assigned a dictionary to sample a value from acontinuous range.
gene_space={"low":4,"high":30}
A step also can be assigned to the dictionary. This works as if a rangeis used.
gene_space={"low":4,"high":30,"step":2.5}
Setting a
dictlike{"low":0,"high":10}in thegene_spacemeans that random values from the continuous range [0,10) are sampled. Note that0is included but10is notincluded while sampling. Thus, the maximum value that could bereturned is less than10like9.9999. But if the user decidedto round the genes using, for example,[float,2], then thisvalue will become 10. So, the user should be careful to the inputs.
If aNone is assigned to only a single gene, then its value will berandomly generated initially using theinit_range_low andinit_range_high parameters in thepygad.GA class’s constructor.During mutation, the value are sampled from the range defined by the 2parametersrandom_mutation_min_val andrandom_mutation_max_val.This is an example where the second gene is given aNone value.
gene_space=[range(5),None,numpy.linspace(10,20,300)]
If the user did not assign the initial population to theinitial_population parameter, the initial population is createdrandomly based on thegene_space parameter. Moreover, the mutationis applied based on this parameter.
How Mutation Works with thegene_space Parameter?¶
Mutation changes based on whether thegene_space has a continuousrange or discrete set of values.
If a gene has itsstatic/discrete space defined in thegene_space parameter, then mutation works by replacing the genevalue by a value randomly selected from the gene space. This happens forbothint andfloat data types.
For example, the followinggene_space has the static space[1,2,3] defined for the first gene. So, this gene can only have avalue out of these 3 values.
Genespace:[[1,2,3],None]Solution:[1,5]
For a solution like[1,5], then mutation happens for the first geneby simply replacing its current value by a randomly selected value(other than its current value if possible). So, the value 1 will bereplaced by either 2 or 3.
For the second gene, its space is set toNone. So, traditionalmutation happens for this gene by:
Generating a random value from the range defined by the
random_mutation_min_valandrandom_mutation_max_valparameters.Adding this random value to the current gene’s value.
If its current value is 5 and the random value is-0.5, then the newvalue is 4.5. If the gene type is integer, then the value will berounded.
On the other hand, if a gene has acontinuous space defined in thegene_space parameter, then mutation occurs by adding a random valueto the current gene value.
For example, the followinggene_space has the continuous spacedefined by the dictionary{'low':1,'high':5}. This applies to allgenes. So, mutation is applied to one or more selected genes by adding arandom value to the current gene value.
Genespace:{'low':1,'high':5}Solution:[1.5,3.4]
Assumingrandom_mutation_min_val=-1 andrandom_mutation_max_val=1, then a random value such as0.3 canbe added to the gene(s) participating in mutation. If only the firstgene is mutated, then its new value changes from1.5 to1.5+0.3=1.8. Note that PyGAD verifies that the new value is withinthe range. In the worst scenarios, the value will be set to eitherboundary of the continuous range. For example, if the gene value is 1.5and the random value is -0.55, then the new value is 0.95 which smallerthan the lower boundary 1. Thus, the gene value will be rounded to 1.
If the dictionary has a step like the example below, then it isconsidered a discrete range and mutation occurs by randomly selecting avalue from the set of values. In other words, no random value is addedto the gene value.
Genespace:{'low':1,'high':5,'step':0.5}
Gene Constraint¶
InPyGAD3.5.0,a new parameter calledgene_constraint is added to the constructorof thepygad.GA class. An instance attribute of the same name iscreated for any instance of thepygad.GA class.
Thegene_constraint parameter allows the users to define constraintsto be enforced (as much as possible) when selecting a value for a gene.For example, this constraint is enforced when applying mutation to makesure the new gene value after mutation meets the gene constraint.
The default value of this parameter isNone which means no geneshave constraints. It can be assigned a list but the length of this listmust be equal to the number of genes as specified by thenum_geneparameter.
When assigned a list, the allowed values for each element are:
None: No constraint for the gene.callable: A callable/function that accepts 2 parameters:The solution where the gene exists.
A list or NumPy array of candidate values for the gene.
It is the user’s responsibility to build such callables to filter thepassed list of values and return a new list with the values that meetsthe gene constraint. If no value meets the constraint, return an emptylist or NumPy array.
For example, if the gene must be smaller than 5, then use this callable:
lambdasolution,values:[valforvalinvaluesifval<5]
The first parameter is the solution where the target gene exists. It ispassed just in case you would like to compare the gene value with othergenes. The second parameter is the list of candidate values for thegene. The objective of the lambda function is to filter the values andreturn only the valid values that are less than 5.
A lambda function is used in this case but we can use a regularfunction:
defconstraint_func(solution,values):return[valforvalinvaluesifval<5]
Assumingnum_genes is 2, then here is a valid value for thegene_constraint parameter.
importpygaddeffitness_func(...):...returnfitnessga_instance=pygad.GA(num_genes=2,sample_size=200,...gene_constraint=[lambdasolution,values:[valforvalinvaluesifval<5],lambdasolution,values:[valforvalinvaluesifval>[solution[0]]])
The first lambda function filters the values for the first gene by onlyconsidering the gene values that are less than 5. If the passed valuesis[-5,2,6,13,3,4,0], then the returned filtered values willbe[-5,2,3,4,0].
The constraint for the second gene makes sure the selected value islarger than the value of the first gene. Assuming the values for the 2parameters are:
solution=[1,4]values=[17,2,-1,0.5,-2.1,1.4]
Then the value of the first gene in the passed solution is1. Byfiltering the passed values using the callable corresponding to thesecond gene, then the returned values will be[17,2,1.4] becausethese are the only values that are larger than the first gene value of1.
Sometimes it is normal for PyGAD to fail to find a gene value thatsatisfies the constraint. For example, if the possible gene values areonly[20,30,40] and the gene constraint restricts the values to begreater than 50, then it is impossible to meet the constraint.
For some other cases, the constraint can be met but with some changes.For example, increasing the range from which a value is sampled. If thegene_space is used and assignedrange(10), then the geneconstraint can be met by usingrange(50) so that we can find valuesgreater than 50.
Even if the the gene space is already assignedrange(1000), it mightstill not find values meeting the constraints This is because PyGADsamples a number of values equal to thesample_size parameter whichdefaults to100.
Out of the range of1000 numbers, all the 100 values might not besatisfying the constraint. This issue could be solved by simplyassigning a larger value for thesample_size parameter.
PyGAD does not yet handle thedependencies among the genes in the
gene_constraintparameter.This is an example where gene 0 depends on gene 1. To efficientlyenforce the constraints, the constraint for gene 1 must be enforcedfirst (if not
None) then the constraint for gene 0.gene_constraint=[lambdasolution,values:[valforvalinvaluesifval<solution[1]],lambdasolution,values:[valforvalinvaluesifval>10]]PyGAD applies constraints sequentially, starting from the first geneto the last. To ensure correct behavior when genes depend on eachother, structure your GA problem so that if gene X depends on gene Y,then gene Y appears earlier in the chromosome (solution) than gene X.As a result, its gene constraint will be earlier in the list.
Full Example¶
For a full example, please check the`examples/example_gene_constraint.pyscript <https://github.com/ahmedfgad/GeneticAlgorithmPython/blob/master/examples/example_gene_constraint.py>`__.
sample_size Parameter¶
InPyGAD3.5.0,a new parameter calledsample_size. It is used in some situationswhere PyGAD seeks a single value for a gene out of a range. Two of theimportant use cases are:
Find a unique value for the gene. This is when the
allow_duplicate_genesparameter is set toFalseto reject theduplicate gene values within the same solution.Find a value that satisfies the
gene_constraintparameter.
Given that we are sampling values from a continuous range as defined bythe 2 attributes:
random_mutation_min_val=0random_mutation_max_val=100
PyGAD samples a fixed number of values out of this continuous range. Thenumber of values in the sample is defined by thesample_sizeparameter which defaults to100.
If the objective is to find a unique value or enforce the geneconstraint, then the 100 values are filtered to keep only the valuesthat keep the gene unique or meet the constraint.
Sometimes 100 values is not enough and PyGAD sometimes fails to find agood value. In this case, it is highly recommended to increase thesample_size parameter. This is to create a larger sample to increasethe chance of finding a value that meets our objectives.
Stop at Any Generation¶
InPyGAD2.4.0,it is possible to stop the genetic algorithm after any generation. Allyou need to do it to return the string"stop" in the callbackfunctionon_generation. When this callback function is implementedand assigned to theon_generation parameter in the constructor ofthepygad.GA class, then the algorithm immediately stops aftercompleting its current generation. Let’s discuss an example.
Assume that the user wants to stop algorithm either after the 100generations or if a condition is met. The user may assign a value of 100to thenum_generations parameter of thepygad.GA classconstructor.
The condition that stops the algorithm is written in a callback functionlike the one in the next code. If the fitness value of the best solutionexceeds 70, then the string"stop" is returned.
deffunc_generation(ga_instance):ifga_instance.best_solution()[1]>=70:return"stop"
Stop Criteria¶
InPyGAD2.15.0,a new parameter namedstop_criteria is added to the constructor ofthepygad.GA class. It helps to stop the evolution based on somecriteria. It can be assigned to one or more criterion.
Each criterion is passed asstr that consists of 2 parts:
Stop word.
Number.
It takes this form:
"word_num"The current 2 supported words arereach andsaturate.
Thereach word stops therun() method if the fitness value isequal to or greater than a given fitness value. An example forreachis"reach_40" which stops the evolution if the fitness is >= 40.
saturate stops the evolution if the fitness saturates for a givennumber of consecutive generations. An example forsaturate is"saturate_7" which means stop therun() method if the fitnessdoes not change for 7 consecutive generations.
Here is an example that stops the evolution if either the fitness valuereached127.4 or if the fitness saturates for15 generations.
importpygadimportnumpyequation_inputs=[4,-2,3.5,8,9,4]desired_output=44deffitness_func(ga_instance,solution,solution_idx):output=numpy.sum(solution*equation_inputs)fitness=1.0/(numpy.abs(output-desired_output)+0.000001)returnfitnessga_instance=pygad.GA(num_generations=200,sol_per_pop=10,num_parents_mating=4,num_genes=len(equation_inputs),fitness_func=fitness_func,stop_criteria=["reach_127.4","saturate_15"])ga_instance.run()print(f"Number of generations passed is{ga_instance.generations_completed}")
Multi-Objective Stop Criteria¶
When multi-objective is used, then there are 2 options to use thestop_criteria parameter with thereach keyword:
Pass a single value to use along the
reachkeyword to use acrossall the objectives.Pass multiple values along the
reachkeyword. But the number ofvalues must equal the number of objectives.
For thesaturate keyword, it is independent to the number ofobjectives.
Suppose there are 3 objectives, this is a working example. It stops whenthe fitness value of the 3 objectives reach or exceed 10, 20, and 30,respectively.
stop_criteria='reach_10_20_30'
More than one criterion can be used together. In this case, pass thestop_criteria parameter as an iterable. This is an example. It stopswhen either of these 2 conditions hold:
The fitness values of the 3 objectives reach or exceed 10, 20, and30, respectively.
The fitness values of the 3 objectives reach or exceed 90, -5.7, and10, respectively.
stop_criteria=['reach_10_20_30','reach_90_-5.7_10']
Elitism Selection¶
InPyGAD2.18.0,a new parameter calledkeep_elitism is supported. It accepts aninteger to define the number of elitism (i.e. best solutions) to keep inthe next generation. This parameter defaults to1 which means onlythe best solution is kept in the next generation.
In the next example, thekeep_elitism parameter in the constructorof thepygad.GA class is set to 2. Thus, the best 2 solutions ineach generation are kept in the next generation.
importnumpyimportpygadfunction_inputs=[4,-2,3.5,5,-11,-4.7]desired_output=44deffitness_func(ga_instance,solution,solution_idx):output=numpy.sum(solution*function_inputs)fitness=1.0/numpy.abs(output-desired_output)returnfitnessga_instance=pygad.GA(num_generations=2,num_parents_mating=3,fitness_func=fitness_func,num_genes=6,sol_per_pop=5,keep_elitism=2)ga_instance.run()
The value passed to thekeep_elitism parameter must satisfy 2conditions:
It must be
>=0.It must be
<=sol_per_pop. That is its value cannot exceed thenumber of solutions in the current population.
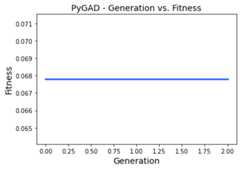
In the previous example, if thekeep_elitism parameter is set equalto the value passed to thesol_per_pop parameter, which is 5, thenthere will be no evolution at all as in the next figure. This is becauseall the 5 solutions are used as elitism in the next generation and nooffspring will be created.
...ga_instance=pygad.GA(...,sol_per_pop=5,keep_elitism=5)ga_instance.run()
Note that if thekeep_elitism parameter is effective (i.e. isassigned a positive integer, not zero), then thekeep_parentsparameter will have no effect. Because the default value of thekeep_elitism parameter is 1, then thekeep_parents parameter hasno effect by default. Thekeep_parents parameter is only effectivewhenkeep_elitism=0.
Random Seed¶
InPyGAD2.18.0,a new parameter calledrandom_seed is supported. Its value is usedas a seed for the random function generators.
PyGAD uses random functions in these 2 libraries:
NumPy
random
Therandom_seed parameter defaults toNone which means no seedis used. As a result, different random numbers are generated for eachrun of PyGAD.
If this parameter is assigned a proper seed, then the results will bereproducible. In the next example, the integer 2 is used as a randomseed.
importnumpyimportpygadfunction_inputs=[4,-2,3.5,5,-11,-4.7]desired_output=44deffitness_func(ga_instance,solution,solution_idx):output=numpy.sum(solution*function_inputs)fitness=1.0/numpy.abs(output-desired_output)returnfitnessga_instance=pygad.GA(num_generations=2,num_parents_mating=3,fitness_func=fitness_func,sol_per_pop=5,num_genes=6,random_seed=2)ga_instance.run()best_solution,best_solution_fitness,best_match_idx=ga_instance.best_solution()print(best_solution)print(best_solution_fitness)
This is the best solution found and its fitness value.
[2.77249188-4.065706620.04196872-3.47770796-0.57502138-3.22775267]0.04872203136549972
After running the code again, it will find the same result.
[2.77249188-4.065706620.04196872-3.47770796-0.57502138-3.22775267]0.04872203136549972
Continue without Losing Progress¶
InPyGAD2.18.0,and thanks forFelix Bernhard foropeningthis GitHubissue,the values of these 4 instance attributes are no longer reset after eachcall to therun() method.
self.best_solutionsself.best_solutions_fitnessself.solutionsself.solutions_fitness
This helps the user to continue where the last run stopped withoutlosing the values of these 4 attributes.
Now, the user can save the model by calling thesave() method.
importpygaddeffitness_func(ga_instance,solution,solution_idx):...returnfitnessga_instance=pygad.GA(...)ga_instance.run()ga_instance.plot_fitness()ga_instance.save("pygad_GA")
Then the saved model is loaded by calling theload() function. Aftercalling therun() method over the loaded instance, then the datafrom the previous 4 attributes are not reset but extended with the newdata.
importpygaddeffitness_func(ga_instance,solution,solution_idx):...returnfitnessloaded_ga_instance=pygad.load("pygad_GA")loaded_ga_instance.run()loaded_ga_instance.plot_fitness()
The plot created by theplot_fitness() method will show the datacollected from both the runs.
Note that the 2 attributes (self.best_solutions andself.best_solutions_fitness) only work if thesave_best_solutions parameter is set toTrue. Also, the 2attributes (self.solutions andself.solutions_fitness) only workif thesave_solutions parameter isTrue.
Change Population Size during Runtime¶
Starting fromPyGAD3.3.0,the population size can changed during runtime. In other words, thenumber of solutions/chromosomes and number of genes can be changed.
The user has to carefully arrange the list ofparameters andinstanceattributes that have to be changed to keep the GA consistent before andafter changing the population size. Generally, change everything thatwould be used during the GA evolution.
CAUTION: If the user failed to change a parameter or an instanceattributes necessary to keep the GA running after the population sizechanged, errors will arise.
These are examples of the parameters that the user should decide whetherto change. The user should check thelist ofparametersand decide what to change.
population: The population. Itmust be changed.num_offspring: The number of offspring to produce out of thecrossover and mutation operations. Change this parameter if thenumber of offspring have to be changed to be consistent with the newpopulation size.num_parents_mating: The number of solutions to select as parents.Change this parameter if the number of parents have to be changed tobe consistent with the new population size.fitness_func: If the way of calculating the fitness changes afterthe new population size, then the fitness function have to bechanged.sol_per_pop: The number of solutions per population. It is notcritical to change it but it is recommended to keep this numberconsistent with the number of solutions in thepopulationparameter.
These are examples of the instance attributes that might be changed. Theuser should check thelist of instanceattributesand decide what to change.
All the
last_generation_*parameterslast_generation_fitness: A 1D NumPy array of fitness values ofthe population.last_generation_parentsandlast_generation_parents_indices: Two NumPy arrays: 2D arrayrepresenting the parents and 1D array of the parents indices.last_generation_elitismandlast_generation_elitism_indices: Must be changed ifkeep_elitism!=0. The default value ofkeep_elitismis 1.Two NumPy arrays: 2D array representing the elitism and 1D arrayof the elitism indices.
pop_size: The population size.
Prevent Duplicates in Gene Values¶
InPyGAD2.13.0,a new bool parameter calledallow_duplicate_genes is supported tocontrol whether duplicates are supported in the chromosome or not. Inother words, whether 2 or more genes might have the same exact value.
Ifallow_duplicate_genes=True (which is the default case), genes mayhave the same value. Ifallow_duplicate_genes=False, then no 2 geneswill have the same value given that there are enough unique values forthe genes.
The next code gives an example to use theallow_duplicate_genesparameter. A callback generation function is implemented to print thepopulation after each generation.
importpygaddeffitness_func(ga_instance,solution,solution_idx):return0defon_generation(ga):print("Generation",ga.generations_completed)print(ga.population)ga_instance=pygad.GA(num_generations=5,sol_per_pop=5,num_genes=4,mutation_num_genes=3,random_mutation_min_val=-5,random_mutation_max_val=5,num_parents_mating=2,fitness_func=fitness_func,gene_type=int,on_generation=on_generation,sample_size=200,allow_duplicate_genes=False)ga_instance.run()
Here are the population after the 5 generations. Note how there are noduplicate values.
Generation1[[2-2-33][0123][5-363][-31-24][-10-23]]Generation2[[-10-23][-31-24][0-3-26][-30-23][1-424]]Generation3[[1-424][-30-23][40-21][-40-2-3][-4203]]Generation4[[-4203][-40-2-3][-254-3][-12-44][-420-3]]Generation5[[-420-3][-12-44][34-40][-102-2][-42-11]]
Theallow_duplicate_genes parameter is configured with use with thegene_space parameter. Here is an example where each of the 4 geneshas the same space of values that consists of 4 values (1, 2, 3, and 4).
importpygaddeffitness_func(ga_instance,solution,solution_idx):return0defon_generation(ga):print("Generation",ga.generations_completed)print(ga.population)ga_instance=pygad.GA(num_generations=1,sol_per_pop=5,num_genes=4,num_parents_mating=2,fitness_func=fitness_func,gene_type=int,gene_space=[[1,2,3,4],[1,2,3,4],[1,2,3,4],[1,2,3,4]],on_generation=on_generation,sample_size=200,allow_duplicate_genes=False)ga_instance.run()
Even that all the genes share the same space of values, no 2 genesduplicate their values as provided by the next output.
Generation1[[2314][2314][2413][2314][1324]]Generation2[[1324][2314][1324][2341][1342]]Generation3[[1342][2341][1342][3142][3241]]Generation4[[3241][3142][3241][1243][1342]]Generation5[[1342][1243][2143][1243][1243]]
You should care of giving enough values for the genes so that PyGAD isable to find alternatives for the gene value in case it duplicates withanother gene.
If PyGAD failed to find a unique gene while there is still room to finda unique value, one possible option is to set thesample_sizeparameter to a larger value. Check thesample_sizeParametersection for more information.
Limitation¶
There might be 2 duplicate genes where changing either of the 2duplicating genes will not solve the problem. For example, ifgene_space=[[3,0,1],[4,1,2],[0,2],[3,2,0]] and thesolution is[3200], then the values of the last 2 genesduplicate. There are no possible changes in the last 2 genes to solvethe problem.
This problem can be solved by randomly changing one of thenon-duplicating genes that may make a room for a unique value in one the2 duplicating genes. For example, by changing the second gene from 2 to4, then any of the last 2 genes can take the value 2 and solve theduplicates. The resultant gene is then[3420]. But this option isnot yet supported in PyGAD.
Solve Duplicates using a Third Gene¶
Whenallow_duplicate_genes=False and a user-definedgene_spaceis used, it sometimes happen that there is no room to solve theduplicates between the 2 genes by simply replacing the value of one geneby another gene. InPyGAD3.1.0,the duplicates are solved by looking for a third gene that will help insolving the duplicates. The following examples explain how it works.
Example 1:
Let’s assume that this gene space is used and there is a solution with 2duplicate genes with the same value 4.
Genespace:[[2,3],[3,4],[4,5],[5,6]]Solution:[3,4,4,5]
By checking the gene space, the second gene can have the values[3,4] and the third gene can have the values[4,5]. To solvethe duplicates, we have the value of any of these 2 genes.
If the value of the second gene changes from 4 to 3, then it will beduplicate with the first gene. If we are to change the value of thethird gene from 4 to 5, then it will duplicate with the fourth gene. Asa conclusion, trying to just selecting a different gene value for eitherthe second or third genes will introduce new duplicating genes.
When there are 2 duplicate genes but there is no way to solve theirduplicates, then the solution is to change a third gene that makes aroom to solve the duplicates between the 2 genes.
In our example, duplicates between the second and third genes can besolved by, for example,:
Changing the first gene from 3 to 2 then changing the second gene from4 to 3.
Or changing the fourth gene from 5 to 6 then changing the third genefrom 4 to 5.
Generally, this is how to solve such duplicates:
For any duplicate geneGENE1, select another value.
Check which other geneGENEX has duplicate with this new value.
Find ifGENEX can have another value that will not cause any moreduplicates. If so, go to step 7.
If all the other values ofGENEX will cause duplicates, then tryanother geneGENEY.
Repeat steps 3 and 4 until exploring all the genes.
If there is no possibility to solve the duplicates, then there is notway to solve the duplicates and we have to keep the duplicate value.
If a value for a geneGENEM is found that will not cause moreduplicates, then use this value for the geneGENEM.
Replace the value of the geneGENE1 by the old value of the geneGENEM. This solves the duplicates.
This is an example to solve the duplicate for the solution[3,4,4,5]:
Let’s use the second gene with value 4. Because the space of thisgene is
[3,4], then the only other value we can select is 3.The first gene also have the value 3.
The first gene has another value 2 that will not cause moreduplicates in the solution. Then go to step 7.
Skip.
Skip.
Skip.
The value of the first gene 3 will be replaced by the new value 2.The new solution is [2, 4, 4, 5].
Replace the value of the second gene 4 by the old value of the firstgene which is 3. The new solution is [2, 3, 4, 5]. The duplicate issolved.
Example 2:
Genespace:[[0,1],[1,2],[2,3],[3,4]]Solution:[1,2,2,3]
The quick summary is:
Change the value of the first gene from 1 to 0. The solution becomes[0, 2, 2, 3].
Change the value of the second gene from 2 to 1. The solution becomes[0, 1, 2, 3]. The duplicate is solved.
More about thegene_type Parameter¶
Thegene_type parameter allows the user to control the data type forall genes at once or each individual gene. InPyGAD2.15.0,thegene_type parameter also supports customizing the precision forfloat data types. As a result, thegene_type parameter helps to:
Select a data type for all genes with or without precision.
Select a data type for each individual gene with or withoutprecision.
Let’s discuss things by examples.
Data Type for All Genes without Precision¶
The data type for all genes can be specified by assigning the numericdata type directly to thegene_type parameter. This is an example tomake all genes ofint data types.
gene_type=int
Given that the supported numeric data types of PyGAD include Python’sint andfloat in addition to all numeric types ofNumPy,then any of these types can be assigned to thegene_type parameter.
If no precision is specified for afloat data type, then thecomplete floating-point number is kept.
The next code uses anint data type for all genes where the genes inthe initial and final population are only integers.
importpygadimportnumpyequation_inputs=[4,-2,3.5,8,-2]desired_output=2671.1234deffitness_func(ga_instance,solution,solution_idx):output=numpy.sum(solution*equation_inputs)fitness=1.0/(numpy.abs(output-desired_output)+0.000001)returnfitnessga_instance=pygad.GA(num_generations=10,sol_per_pop=5,num_parents_mating=2,num_genes=len(equation_inputs),fitness_func=fitness_func,gene_type=int)print("Initial Population")print(ga_instance.initial_population)ga_instance.run()print("Final Population")print(ga_instance.population)
InitialPopulation[[1-120-3][0-20-3-1][0-1-120][-23-233][002-2-2]]FinalPopulation[[1-1220][1-1220][1-1220][1-1220][1-1220]]
Data Type for All Genes with Precision¶
A precision can only be specified for afloat data type and cannotbe specified for integers. Here is an example to use a precision of 3for thefloat data type. In this case, all genes are of typefloat and their maximum precision is 3.
gene_type=[float,3]
The next code uses prints the initial and final population where thegenes are of typefloat with precision 3.
importpygadimportnumpyequation_inputs=[4,-2,3.5,8,-2]desired_output=2671.1234deffitness_func(ga_instance,solution,solution_idx):output=numpy.sum(solution*equation_inputs)fitness=1.0/(numpy.abs(output-desired_output)+0.000001)returnfitnessga_instance=pygad.GA(num_generations=10,sol_per_pop=5,num_parents_mating=2,num_genes=len(equation_inputs),fitness_func=fitness_func,gene_type=[float,3])print("Initial Population")print(ga_instance.initial_population)ga_instance.run()print("Final Population")print(ga_instance.population)
InitialPopulation[[-2.417-0.4873.6232.457-2.362][-1.2310.079-1.631.629-2.637][0.692-2.0980.7050.914-3.633][2.637-1.339-1.107-0.781-3.896][-1.4951.378-1.0263.5222.379]]FinalPopulation[[1.714-1.0243.6233.185-2.362][0.692-1.0243.6233.185-2.362][0.692-1.0243.6233.375-2.362][0.692-1.0244.0413.185-2.362][1.714-0.6443.6233.185-2.362]]
Data Type for each Individual Gene without Precision¶
InPyGAD2.14.0,thegene_type parameter allows customizing the gene type for eachindividual gene. This is by using alist/tuple/numpy.ndarraywith number of elements equal to the number of genes. For each element,a type is specified for the corresponding gene.
This is an example for a 5-gene problem where different types areassigned to the genes.
gene_type=[int,float,numpy.float16,numpy.int8,float]
This is a complete code that prints the initial and final population fora custom-gene data type.
importpygadimportnumpyequation_inputs=[4,-2,3.5,8,-2]desired_output=2671.1234deffitness_func(ga_instance,solution,solution_idx):output=numpy.sum(solution*equation_inputs)fitness=1.0/(numpy.abs(output-desired_output)+0.000001)returnfitnessga_instance=pygad.GA(num_generations=10,sol_per_pop=5,num_parents_mating=2,num_genes=len(equation_inputs),fitness_func=fitness_func,gene_type=[int,float,numpy.float16,numpy.int8,float])print("Initial Population")print(ga_instance.initial_population)ga_instance.run()print("Final Population")print(ga_instance.population)
InitialPopulation[[00.86155223600268280.7021484375-23.5301821368185866][-32.648189378595294-3.8300781251-0.9586271572917742][33.77298275701107141.2529296875-31.395741994211889][01.04906871780532821.51953125-20.7243617940450235][0-0.6550158436937226-2.861328125-21.8212734549263097]]FinalPopulation[[33.77298275701107142.05500.7243617940450235][33.77298275701107141.4580-0.14638754050305036][33.77298275701107141.45800.0869406120516778][33.77298275701107141.45800.7243617940450235][33.77298275701107141.4580-0.14638754050305036]]
Data Type for each Individual Gene with Precision¶
The precision can also be specified for thefloat data types as inthe next line where the second gene precision is 2 and last geneprecision is 1.
gene_type=[int,[float,2],numpy.float16,numpy.int8,[float,1]]
This is a complete example where the initial and final populations areprinted where the genes comply with the data types and precisionsspecified.
importpygadimportnumpyequation_inputs=[4,-2,3.5,8,-2]desired_output=2671.1234deffitness_func(ga_instance,solution,solution_idx):output=numpy.sum(solution*equation_inputs)fitness=1.0/(numpy.abs(output-desired_output)+0.000001)returnfitnessga_instance=pygad.GA(num_generations=10,sol_per_pop=5,num_parents_mating=2,num_genes=len(equation_inputs),fitness_func=fitness_func,gene_type=[int,[float,2],numpy.float16,numpy.int8,[float,1]])print("Initial Population")print(ga_instance.initial_population)ga_instance.run()print("Final Population")print(ga_instance.population)
InitialPopulation[[-2-1.221.716796875-10.2][-1-1.58-3.0917968750-1.3][33.35-0.1074218751-3.3][-2-3.58-1.77929687500.6][2-3.732.652343753-0.5]]FinalPopulation[[2-4.223.473-1.3][2-3.733.473-1.3][2-4.223.472-1.3][2-4.583.473-1.3][2-3.733.473-1.3]]
Parallel Processing in PyGAD¶
Starting fromPyGAD2.17.0,parallel processing becomes supported. This section explains how to useparallel processing in PyGAD.
According to thePyGADlifecycle,parallel processing can be parallelized in only 2 operations:
Population fitness calculation.
Mutation.
The reason is that the calculations in these 2 operations areindependent (i.e. each solution/chromosome is handled independently fromthe others) and can be distributed across different processes orthreads.
For the mutation operation, it does not do intensive calculations on theCPU. Its calculations are simple like flipping the values of some genesfrom 0 to 1 or adding a random value to some genes. So, it does not takemuch CPU processing time. Experiments proved that parallelizing themutation operation across the solutions increases the time instead ofreducing it. This is because running multiple processes or threads addsoverhead to manage them. Thus, parallel processing cannot be applied onthe mutation operation.
For the population fitness calculation, parallel processing can helpmake a difference and reduce the processing time. But this isconditional on the type of calculations done in the fitness function. Ifthe fitness function makes intensive calculations and takes muchprocessing time from the CPU, then it is probably that parallelprocessing will help to cut down the overall time.
This section explains how parallel processing works in PyGAD and how touse parallel processing in PyGAD
How to Use Parallel Processing in PyGAD¶
Starting fromPyGAD2.17.0,a new parameter calledparallel_processing added to the constructorof thepygad.GA class.
importpygad...ga_instance=pygad.GA(...,parallel_processing=...)...
This parameter allows the user to do the following:
Enable parallel processing.
Select whether processes or threads are used.
Specify the number of processes or threads to be used.
These are 3 possible values for theparallel_processing parameter:
None: (Default) It means no parallel processing is used.A positive integer referring to the number of threads to be used(i.e. threads, not processes, are used.
list/tuple: If a list or a tuple of exactly 2 elements isassigned, then:The first element can be either
'process'or'thread'tospecify whether processes or threads are used, respectively.The second element can be:
A positive integer to select the maximum number of processes orthreads to be used
0to indicate that 0 processes or threads are used. Itmeans no parallel processing. This is identical to settingparallel_processing=None.Noneto use the default value as calculated by theconcurrent.futuresmodule.
These are examples of the values assigned to theparallel_processingparameter:
parallel_processing=4: Because the parameter is assigned apositive integer, this means parallel processing is activated where 4threads are used.parallel_processing=["thread",5]: Use parallel processing with 5threads. This is identical toparallel_processing=5.parallel_processing=["process",8]: Use parallel processing with 8processes.parallel_processing=["process",0]: As the second element is giventhe value 0, this means do not use parallel processing. This isidentical toparallel_processing=None.
Examples¶
The examples will help you know the difference between using processesand threads. Moreover, it will give an idea when parallel processingwould make a difference and reduce the time. These are dummy exampleswhere the fitness function is made to always return 0.
The first example uses 10 genes, 5 solutions in the population whereonly 3 solutions mate, and 9999 generations. The fitness function uses afor loop with 100 iterations just to have some calculations. In theconstructor of thepygad.GA class,parallel_processing=Nonemeans no parallel processing is used.
importpygadimporttimedeffitness_func(ga_instance,solution,solution_idx):for_inrange(99):passreturn0ga_instance=pygad.GA(num_generations=9999,num_parents_mating=3,sol_per_pop=5,num_genes=10,fitness_func=fitness_func,suppress_warnings=True,parallel_processing=None)if__name__=='__main__':t1=time.time()ga_instance.run()t2=time.time()print("Time is",t2-t1)
When parallel processing is not used, the time it takes to run thegenetic algorithm is1.5 seconds.
In the comparison, let’s do a second experiment where parallelprocessing is used with 5 threads. In this case, it take5 seconds.
...ga_instance=pygad.GA(...,parallel_processing=5)...
For the third experiment, processes instead of threads are used. Also,only 99 generations are used instead of 9999. The time it takes is99 seconds.
...ga_instance=pygad.GA(num_generations=99,...,parallel_processing=["process",5])...
This is the summary of the 3 experiments:
No parallel processing & 9999 generations: 1.5 seconds.
Parallel processing with 5 threads & 9999 generations: 5 seconds
Parallel processing with 5 processes & 99 generations: 99 seconds
Because the fitness function does not need much CPU time, the normalprocessing takes the least time. Running processes for this simpleproblem takes 99 compared to only 5 seconds for threads because managingprocesses is much heavier than managing threads. Thus, most of the CPUtime is for swapping the processes instead of executing the code.
In the second example, the loop makes 99999999 iterations and only 5generations are used. With no parallelization, it takes 22 seconds.
importpygadimporttimedeffitness_func(ga_instance,solution,solution_idx):for_inrange(99999999):passreturn0ga_instance=pygad.GA(num_generations=5,num_parents_mating=3,sol_per_pop=5,num_genes=10,fitness_func=fitness_func,suppress_warnings=True,parallel_processing=None)if__name__=='__main__':t1=time.time()ga_instance.run()t2=time.time()print("Time is",t2-t1)
It takes 15 seconds when 10 processes are used.
...ga_instance=pygad.GA(...,parallel_processing=["process",10])...
This is compared to 20 seconds when 10 threads are used.
...ga_instance=pygad.GA(...,parallel_processing=["thread",10])...
Based on the second example, using parallel processing with 10 processestakes the least time because there is much CPU work done. Generally,processes are preferred over threads when most of the work in on theCPU. Threads are preferred over processes in some situations like doinginput/output operations.
Before releasingPyGAD2.17.0,LászlóFazekaswrote an article to parallelize the fitness function with PyGAD. Checkit:How Genetic Algorithms Can Compete with Gradient Descent andBackprop.
Print Lifecycle Summary¶
InPyGAD2.19.0,a new method calledsummary() is supported. It prints a Keras-likesummary of the PyGAD lifecycle showing the steps, callback functions,parameters, etc.
This method accepts the following parameters:
line_length=70: An integer representing the length of the singleline in characters.fill_character="": A character to fill the lines.line_character="-": A character for creating a line separator.line_character2="=": A secondary character to create a lineseparator.columns_equal_len=False: The table rows are split into equal-sizedcolumns or split subjective to the width needed.print_step_parameters=True: Whether to print extra parametersabout each step inside the step. Ifprint_step_parameters=Falseandprint_parameters_summary=True, then the parameters of eachstep are printed at the end of the table.print_parameters_summary=True: Whether to print parameters summaryat the end of the table. Ifprint_step_parameters=False, then theparameters of each step are printed at the end of the table too.
This is a quick example to create a PyGAD example.
importpygadimportnumpyfunction_inputs=[4,-2,3.5,5,-11,-4.7]desired_output=44defgenetic_fitness(solution,solution_idx):output=numpy.sum(solution*function_inputs)fitness=1.0/(numpy.abs(output-desired_output)+0.000001)returnfitnessdefon_gen(ga):passdefon_crossover_callback(a,b):passga_instance=pygad.GA(num_generations=100,num_parents_mating=10,sol_per_pop=20,num_genes=len(function_inputs),on_crossover=on_crossover_callback,on_generation=on_gen,parallel_processing=2,stop_criteria="reach_10",fitness_batch_size=4,crossover_probability=0.4,fitness_func=genetic_fitness)
Then call thesummary() method to print the summary with the defaultparameters. Note that entries for the crossover and generation callbackfunction are created because their callback functions are implementedthrough theon_crossover_callback() andon_gen(), respectively.
ga_instance.summary()
----------------------------------------------------------------------PyGADLifecycle======================================================================StepHandlerOutputShape======================================================================FitnessFunctiongenetic_fitness()(1)Fitnessbatchsize:4----------------------------------------------------------------------ParentSelectionsteady_state_selection()(10,6)NumberofParents:10----------------------------------------------------------------------Crossoversingle_point_crossover()(10,6)Crossoverprobability:0.4----------------------------------------------------------------------OnCrossoveron_crossover_callback()None----------------------------------------------------------------------Mutationrandom_mutation()(10,6)MutationGenes:1RandomMutationRange:(-1.0,1.0)MutationbyReplacement:FalseAllowDuplicatedGenes:True----------------------------------------------------------------------OnGenerationon_gen()NoneStopCriteria:[['reach',10.0]]----------------------------------------------------------------------======================================================================PopulationSize:(20,6)NumberofGenerations:100InitialPopulationRange:(-4,4)KeepElitism:1GeneDType:[<class'float'>,None]ParallelProcessing:['thread',2]SaveBestSolutions:FalseSaveSolutions:False======================================================================
We can set theprint_step_parameters andprint_parameters_summary parameters toFalse to not print theparameters.
ga_instance.summary(print_step_parameters=False,print_parameters_summary=False)
----------------------------------------------------------------------PyGADLifecycle======================================================================StepHandlerOutputShape======================================================================FitnessFunctiongenetic_fitness()(1)----------------------------------------------------------------------ParentSelectionsteady_state_selection()(10,6)----------------------------------------------------------------------Crossoversingle_point_crossover()(10,6)----------------------------------------------------------------------OnCrossoveron_crossover_callback()None----------------------------------------------------------------------Mutationrandom_mutation()(10,6)----------------------------------------------------------------------OnGenerationon_gen()None----------------------------------------------------------------------======================================================================
Logging Outputs¶
InPyGAD3.0.0,theprint() statement is no longer used and the outputs are printedusing theloggingmodule. A a new parameter calledlogger is supported to accept theuser-defined logger.
importlogginglogger=...ga_instance=pygad.GA(...,logger=logger,...)
The default value for this parameter isNone. If there is no loggerpassed (i.e.logger=None), then a default logger is created to logthe messages to the console exactly like how theprint() statementworks.
Some advantages of using the thelogging moduleinstead of theprint() statement are:
The user has more control over the printed messages specially ifthere is a project that uses multiple modules where each moduleprints its messages. A logger can organize the outputs.
Using the proper
Handler, the user can log the output messages tofiles and not only restricted to printing it to the console. So, itis much easier to record the outputs.The format of the printed messages can be changed by customizing the
Formatterassigned to the Logger.
This section gives some quick examples to use thelogging module andthen gives an example to use the logger with PyGAD.
Logging to the Console¶
This is an example to create a logger to log the messages to theconsole.
importlogging# Create a loggerlogger=logging.getLogger(__name__)# Set the logger level to debug so that all the messages are printed.logger.setLevel(logging.DEBUG)# Create a stream handler to log the messages to the console.stream_handler=logging.StreamHandler()# Set the handler level to debug.stream_handler.setLevel(logging.DEBUG)# Create a formatterformatter=logging.Formatter('%(message)s')# Add the formatter to handler.stream_handler.setFormatter(formatter)# Add the stream handler to the loggerlogger.addHandler(stream_handler)
Now, we can log messages to the console with the format specified in theFormatter.
logger.debug('Debug message.')logger.info('Info message.')logger.warning('Warn message.')logger.error('Error message.')logger.critical('Critical message.')
The outputs are identical to those returned using theprint()statement.
Debugmessage.Infomessage.Warnmessage.Errormessage.Criticalmessage.
By changing the format of the output messages, we can have moreinformation about each message.
formatter=logging.Formatter('%(asctime)s%(levelname)s:%(message)s',datefmt='%Y-%m-%d %H:%M:%S')
This is a sample output.
2023-04-0318:46:27DEBUG:Debugmessage.2023-04-0318:46:27INFO:Infomessage.2023-04-0318:46:27WARNING:Warnmessage.2023-04-0318:46:27ERROR:Errormessage.2023-04-0318:46:27CRITICAL:Criticalmessage.
Note that you may need to clear the handlers after finishing theexecution. This is to make sure no cached handlers are used in the nextrun. If the cached handlers are not cleared, then the single outputmessage may be repeated.
logger.handlers.clear()
Logging to a File¶
This is another example to log the messages to a file namedlogfile.txt. The formatter prints the following about each message:
The date and time at which the message is logged.
The log level.
The message.
The path of the file.
The lone number of the log message.
importlogginglevel=logging.DEBUGname='logfile.txt'logger=logging.getLogger(name)logger.setLevel(level)file_handler=logging.FileHandler(name,'a+','utf-8')file_handler.setLevel(logging.DEBUG)file_format=logging.Formatter('%(asctime)s%(levelname)s:%(message)s -%(pathname)s:%(lineno)d',datefmt='%Y-%m-%d %H:%M:%S')file_handler.setFormatter(file_format)logger.addHandler(file_handler)
This is how the outputs look like.
2023-04-0318:54:03DEBUG:Debugmessage.-c:\users\agad069\desktop\logger\example2.py:462023-04-0318:54:03INFO:Infomessage.-c:\users\agad069\desktop\logger\example2.py:472023-04-0318:54:03WARNING:Warnmessage.-c:\users\agad069\desktop\logger\example2.py:482023-04-0318:54:03ERROR:Errormessage.-c:\users\agad069\desktop\logger\example2.py:492023-04-0318:54:03CRITICAL:Criticalmessage.-c:\users\agad069\desktop\logger\example2.py:50
Consider clearing the handlers if necessary.
logger.handlers.clear()
Log to Both the Console and a File¶
This is an example to create a single Logger associated with 2 handlers:
A file handler.
A stream handler.
importlogginglevel=logging.DEBUGname='logfile.txt'logger=logging.getLogger(name)logger.setLevel(level)file_handler=logging.FileHandler(name,'a+','utf-8')file_handler.setLevel(logging.DEBUG)file_format=logging.Formatter('%(asctime)s%(levelname)s:%(message)s -%(pathname)s:%(lineno)d',datefmt='%Y-%m-%d %H:%M:%S')file_handler.setFormatter(file_format)logger.addHandler(file_handler)console_handler=logging.StreamHandler()console_handler.setLevel(logging.INFO)console_format=logging.Formatter('%(message)s')console_handler.setFormatter(console_format)logger.addHandler(console_handler)
When a log message is executed, then it is both printed to the consoleand saved in thelogfile.txt.
Consider clearing the handlers if necessary.
logger.handlers.clear()
PyGAD Example¶
To use the logger in PyGAD, just create your custom logger and pass itto thelogger parameter.
importloggingimportpygadimportnumpylevel=logging.DEBUGname='logfile.txt'logger=logging.getLogger(name)logger.setLevel(level)file_handler=logging.FileHandler(name,'a+','utf-8')file_handler.setLevel(logging.DEBUG)file_format=logging.Formatter('%(asctime)s%(levelname)s:%(message)s',datefmt='%Y-%m-%d %H:%M:%S')file_handler.setFormatter(file_format)logger.addHandler(file_handler)console_handler=logging.StreamHandler()console_handler.setLevel(logging.INFO)console_format=logging.Formatter('%(message)s')console_handler.setFormatter(console_format)logger.addHandler(console_handler)equation_inputs=[4,-2,8]desired_output=2671.1234deffitness_func(ga_instance,solution,solution_idx):output=numpy.sum(solution*equation_inputs)fitness=1.0/(numpy.abs(output-desired_output)+0.000001)returnfitnessdefon_generation(ga_instance):ga_instance.logger.info(f"Generation ={ga_instance.generations_completed}")ga_instance.logger.info(f"Fitness ={ga_instance.best_solution(pop_fitness=ga_instance.last_generation_fitness)[1]}")ga_instance=pygad.GA(num_generations=10,sol_per_pop=40,num_parents_mating=2,keep_parents=2,num_genes=len(equation_inputs),fitness_func=fitness_func,on_generation=on_generation,logger=logger)ga_instance.run()logger.handlers.clear()
By executing this code, the logged messages are printed to the consoleand also saved in the text file.
2023-04-0319:04:27INFO:Generation=12023-04-0319:04:27INFO:Fitness=0.000380869603680762762023-04-0319:04:27INFO:Generation=22023-04-0319:04:27INFO:Fitness=0.000382148714080108532023-04-0319:04:27INFO:Generation=32023-04-0319:04:27INFO:Fitness=0.00038327959079746782023-04-0319:04:27INFO:Generation=42023-04-0319:04:27INFO:Fitness=0.000383986120550171962023-04-0319:04:27INFO:Generation=52023-04-0319:04:27INFO:Fitness=0.000384423488908675162023-04-0319:04:27INFO:Generation=62023-04-0319:04:27INFO:Fitness=0.00038544060391377632023-04-0319:04:27INFO:Generation=72023-04-0319:04:27INFO:Fitness=0.000386460831740632842023-04-0319:04:27INFO:Generation=82023-04-0319:04:27INFO:Fitness=0.00038751691930249362023-04-0319:04:27INFO:Generation=92023-04-0319:04:27INFO:Fitness=0.00038888167273110212023-04-0319:04:27INFO:Generation=102023-04-0319:04:27INFO:Fitness=0.000389832593101348
Solve Non-Deterministic Problems¶
PyGAD can be used to solve both deterministic and non-deterministicproblems. Deterministic are those that return the same fitness for thesame solution. For non-deterministic problems, a different fitness valuewould be returned for the same solution.
By default, PyGAD settings are set to solve deterministic problems.PyGAD can save the explored solutions and their fitness to reuse in thefuture. These instances attributes can save the solutions:
solutions: Exists ifsave_solutions=True.best_solutions: Exists ifsave_best_solutions=True.last_generation_elitism: Exists ifkeep_elitism> 0.last_generation_parents: Exists ifkeep_parents> 0 orkeep_parents=-1.
To configure PyGAD for non-deterministic problems, we have to disablesaving the previous solutions. This is by setting these parameters:
keep_elitism=0keep_parents=0keep_solutions=Falsekeep_best_solutions=False
importpygad...ga_instance=pygad.GA(...,keep_elitism=0,keep_parents=0,save_solutions=False,save_best_solutions=False,...)
This way PyGAD will not save any explored solution and thus the fitnessfunction have to be called for each individual solution.
Reuse the Fitness instead of Calling the Fitness Function¶
It may happen that a previously explored solution in generation X isexplored again in another generation Y (where Y > X). For some problems,calling the fitness function takes much time.
For deterministic problems, it is better to not call the fitnessfunction for an already explored solutions. Instead, reuse the fitnessof the old solution. PyGAD supports some options to help you save timecalling the fitness function for a previously explored solution.
The parameters explored in this section can be set in the constructor ofthepygad.GA class.
Thecal_pop_fitness() method of thepygad.GA class checks theseparameters to see if there is a possibility of reusing the fitnessinstead of calling the fitness function.
1.save_solutions¶
It defaults toFalse. If set toTrue, then the population ofeach generation is saved into thesolutions attribute of thepygad.GA instance. In other words, every single solution is saved inthesolutions attribute.
2.save_best_solutions¶
It defaults toFalse. IfTrue, then it only saves the bestsolution in every generation.
3.keep_elitism¶
It accepts an integer and defaults to 1. If set to a positive integer,then it keeps the elitism of one generation available in the nextgeneration.
4.keep_parents¶
It accepts an integer and defaults to -1. It set to-1 or a positiveinteger, then it keeps the parents of one generation available in thenext generation.
Why the Fitness Function is not Called for Solution at Index 0?¶
PyGAD has a parameter calledkeep_elitism which defaults to 1. Thisparameter defines the number of best solutions in generationX tokeep in the next generationX+1. The best solutions are just copiedfrom generationX to generationX+1 without making any change.
ga_instance=pygad.GA(...,keep_elitism=1,...)
The best solutions are copied at the beginning of the population. Ifkeep_elitism=1, this means the best solution in generation X is keptin the next generation X+1 at index 0 of the population. Ifkeep_elitism=2, this means the 2 best solutions in generation X arekept in the next generation X+1 at indices 0 and 1 of the population ofgeneration 1.
Because the fitness of these best solutions are already calculated ingeneration X, then their fitness values will not be recalculated atgeneration X+1 (i.e. the fitness function will not be called for thesesolutions again). Instead, their fitness values are just reused. This iswhy you see that no solution with index 0 is passed to the fitnessfunction.
To force calling the fitness function for each solution in everygeneration, consider settingkeep_elitism andkeep_parents to 0.Moreover, keep the 2 parameterssave_solutions andsave_best_solutions to their default valueFalse.
ga_instance=pygad.GA(...,keep_elitism=0,keep_parents=0,save_solutions=False,save_best_solutions=False,...)
Batch Fitness Calculation¶
InPyGAD2.19.0,a new optional parameter calledfitness_batch_size is supported. Anew optional parameter calledfitness_batch_size is supported tocalculate the fitness function in batches. Thanks toLinanQiu for opening theGitHub issue#136.
Its values can be:
1orNone: If thefitness_batch_sizeparameter is assignedthe value1orNone(default), then the normal flow is usedwhere the fitness function is called for each individual solution.That is if there are 15 solutions, then the fitness function is called15 times.1<fitness_batch_size<=sol_per_pop: If thefitness_batch_sizeparameter is assigned a value satisfying thiscondition1<fitness_batch_size<=sol_per_pop, then thesolutions are grouped into batches of sizefitness_batch_sizeandthe fitness function is called once for each batch. In this case, thefitness function must return a list/tuple/numpy.ndarray with a lengthequal to the number of solutions passed.
Example withoutfitness_batch_size Parameter¶
This is an example where thefitness_batch_size parameter is giventhe valueNone (which is the default value). This is equivalent tousing the value1. In this case, the fitness function will be calledfor each solution. This means the fitness functionfitness_func willreceive only a single solution. This is an example of the passedarguments to the fitness function:
solution:[2.52860734,-0.94178795,2.97545704,0.84131987,-3.78447118,2.41008358]solution_idx:3
The fitness function also must return a single numeric value as thefitness for the passed solution.
As we have a population of20 solutions, then the fitness functionis called 20 times per generation. For 5 generations, then the fitnessfunction is called20*5=100 times. In PyGAD, the fitness functionis called after the last generation too and this adds additional 20times. So, the total number of calls to the fitness function is20*5+20=120.
Note that thekeep_elitism andkeep_parents parameters are setto0 to make sure no fitness values are reused and to force callingthe fitness function for each individual solution.
importpygadimportnumpyfunction_inputs=[4,-2,3.5,5,-11,-4.7]desired_output=44number_of_calls=0deffitness_func(ga_instance,solution,solution_idx):globalnumber_of_callsnumber_of_calls=number_of_calls+1output=numpy.sum(solution*function_inputs)fitness=1.0/(numpy.abs(output-desired_output)+0.000001)returnfitnessga_instance=pygad.GA(num_generations=5,num_parents_mating=10,sol_per_pop=20,fitness_func=fitness_func,fitness_batch_size=None,# fitness_batch_size=1,num_genes=len(function_inputs),keep_elitism=0,keep_parents=0)ga_instance.run()print(number_of_calls)
120Example withfitness_batch_size Parameter¶
This is an example where thefitness_batch_size parameter is usedand assigned the value4. This means the solutions will be groupedinto batches of4 solutions. The fitness function will be calledonce for each patch (i.e. called once for each 4 solutions).
This is an example of the arguments passed to it:
solutions:[[3.1129432-0.691235891.937924142.23772968-1.54616001-0.53930799][3.385081210.198908121.937924142.23095014-3.089555973.10194128][2.37079504-0.888198032.975457041.41742256-3.955940552.45028256][2.52860734-0.941787952.975457040.84131987-3.784471182.41008358]]solutions_indices:[16,17,18,19]
As we have 20 solutions, then there are20/4=5 patches. As aresult, the fitness function is called only 5 times per generationinstead of 20. For each call to the fitness function, it receives abatch of 4 solutions.
As we have 5 generations, then the function will be called5*5=25times. Given the call to the fitness function after the last generation,then the total number of calls is5*5+5=30.
importpygadimportnumpyfunction_inputs=[4,-2,3.5,5,-11,-4.7]desired_output=44number_of_calls=0deffitness_func_batch(ga_instance,solutions,solutions_indices):globalnumber_of_callsnumber_of_calls=number_of_calls+1batch_fitness=[]forsolutioninsolutions:output=numpy.sum(solution*function_inputs)fitness=1.0/(numpy.abs(output-desired_output)+0.000001)batch_fitness.append(fitness)returnbatch_fitnessga_instance=pygad.GA(num_generations=5,num_parents_mating=10,sol_per_pop=20,fitness_func=fitness_func_batch,fitness_batch_size=4,num_genes=len(function_inputs),keep_elitism=0,keep_parents=0)ga_instance.run()print(number_of_calls)
30When batch fitness calculation is used, then we saved120-30=90calls to the fitness function.
Use Functions and Methods to Build Fitness and Callbacks¶
In PyGAD 2.19.0, it is possible to pass user-defined functions ormethods to the following parameters:
fitness_funcon_starton_fitnesson_parentson_crossoveron_mutationon_generationon_stop
This section gives 2 examples to assign these parameters user-defined:
Functions.
Methods.
Assign Functions¶
This is a dummy example where the fitness function returns a randomvalue. Note that the instance of thepygad.GA class is passed as thelast parameter of all functions.
importpygadimportnumpydeffitness_func(ga_instanse,solution,solution_idx):returnnumpy.random.rand()defon_start(ga_instanse):print("on_start")defon_fitness(ga_instanse,last_gen_fitness):print("on_fitness")defon_parents(ga_instanse,last_gen_parents):print("on_parents")defon_crossover(ga_instanse,last_gen_offspring):print("on_crossover")defon_mutation(ga_instanse,last_gen_offspring):print("on_mutation")defon_generation(ga_instanse):print("on_generation\n")defon_stop(ga_instanse,last_gen_fitness):print("on_stop")ga_instance=pygad.GA(num_generations=5,num_parents_mating=4,sol_per_pop=10,num_genes=2,on_start=on_start,on_fitness=on_fitness,on_parents=on_parents,on_crossover=on_crossover,on_mutation=on_mutation,on_generation=on_generation,on_stop=on_stop,fitness_func=fitness_func)ga_instance.run()
Assign Methods¶
The next example has all the method defined inside the classTest.All of the methods accept an additional parameter representing themethod’s object of the classTest.
All methods acceptself as the first parameter and the instance ofthepygad.GA class as the last parameter.
importpygadimportnumpyclassTest:deffitness_func(self,ga_instanse,solution,solution_idx):returnnumpy.random.rand()defon_start(self,ga_instanse):print("on_start")defon_fitness(self,ga_instanse,last_gen_fitness):print("on_fitness")defon_parents(self,ga_instanse,last_gen_parents):print("on_parents")defon_crossover(self,ga_instanse,last_gen_offspring):print("on_crossover")defon_mutation(self,ga_instanse,last_gen_offspring):print("on_mutation")defon_generation(self,ga_instanse):print("on_generation\n")defon_stop(self,ga_instanse,last_gen_fitness):print("on_stop")ga_instance=pygad.GA(num_generations=5,num_parents_mating=4,sol_per_pop=10,num_genes=2,on_start=Test().on_start,on_fitness=Test().on_fitness,on_parents=Test().on_parents,on_crossover=Test().on_crossover,on_mutation=Test().on_mutation,on_generation=Test().on_generation,on_stop=Test().on_stop,fitness_func=Test().fitness_func)ga_instance.run()