pygad Module¶
This section of the PyGAD’s library documentation discusses thepygad module.
Using thepygad module, instances of the genetic algorithm can becreated, run, saved, and loaded. Single-objective and multi-objectiveoptimization problems can be solved.
pygad.GA Class¶
The first module available in PyGAD is namedpygad and contains aclass namedGA for building the genetic algorithm. The constructor,methods, function, and attributes within the class are discussed in thissection.
__init__()¶
For creating an instance of thepygad.GA class, the constructoraccepts several parameters that allow the user to customize the geneticalgorithm to different types of applications.
Thepygad.GA class constructor supports the following parameters:
num_generations: Number of generations.num_parents_mating: Number of solutions to be selected as parents.fitness_func: Accepts a function/method and returns the fitnessvalue(s) of the solution. If a function is passed, then it must accept3 parameters (1. the instance of thepygad.GAclass, 2. a singlesolution, and 3. its index in the population). If method, then itaccepts a fourth parameter representing the method’s class instance.Check thePreparing the fitness_funcParametersection for information about creating such a function. InPyGAD3.2.0,multi-objective optimization is supported. To consider the problem asmulti-objective, just return alist,tuple, ornumpy.ndarrayfrom the fitness function.fitness_batch_size=None: A new optional parameter calledfitness_batch_sizeis supported to calculate the fitness functionin batches. If it is assigned the value1orNone(default),then the normal flow is used where the fitness function is called foreach individual solution. If thefitness_batch_sizeparameter isassigned a value satisfying this condition1<fitness_batch_size<=sol_per_pop, then the solutions aregrouped into batches of sizefitness_batch_sizeand the fitnessfunction is called once for each batch. Check theBatch FitnessCalculationsection for more details and examples. Added in fromPyGAD2.19.0.initial_population: A user-defined initial population. It isuseful when the user wants to start the generations with a custominitial population. It defaults toNonewhich means no initialpopulation is specified by the user. In this case,PyGAD creates an initialpopulation using thesol_per_popandnum_genesparameters. Anexception is raised if theinitial_populationisNonewhileany of the 2 parameters (sol_per_popornum_genes) is alsoNone. Introduced inPyGAD2.0.0and higher.sol_per_pop: Number of solutions (i.e. chromosomes) within thepopulation. This parameter has no action ifinitial_populationparameter exists.num_genes: Number of genes in the solution/chromosome. Thisparameter is not needed if the user feeds the initial population totheinitial_populationparameter.gene_type=float: Controls the gene type. It can be assigned to asingle data type that is applied to all genes or can specify the datatype of each individual gene. It defaults tofloatwhich means allgenes are offloatdata type. Starting fromPyGAD2.9.0,thegene_typeparameter can be assigned to a numeric value of anyof these types:int,float, andnumpy.int/uint/float(8-64). Starting fromPyGAD2.14.0,it can be assigned to alist,tuple, or anumpy.ndarraywhich hold a data type for each gene (e.g.gene_type=[int,float,numpy.int8]). This helps to control thedata type of each individual gene. InPyGAD2.15.0,a precision for thefloatdata types can be specified (e.g.gene_type=[float,2].init_range_low=-4: The lower value of the random range from whichthe gene values in the initial population are selected.init_range_lowdefaults to-4. Available inPyGAD1.0.20and higher. This parameter has no action if theinitial_populationparameter exists.init_range_high=4: The upper value of the random range from whichthe gene values in the initial population are selected.init_range_highdefaults to+4. Available inPyGAD1.0.20and higher. This parameter has no action if theinitial_populationparameter exists.parent_selection_type="sss": The parent selection type. Supportedtypes aresss(for steady-state selection),rws(for roulettewheel selection),sus(for stochastic universal selection),rank(for rank selection),random(for random selection), andtournament(for tournament selection). A custom parent selectionfunction can be passed starting fromPyGAD2.16.0.Check theUser-Defined Crossover, Mutation, and Parent SelectionOperatorssection for more details about building a user-defined parentselection function.keep_parents=-1: Number of parents to keep in the currentpopulation.-1(default) means to keep all parents in the nextpopulation.0means keep no parents in the next population. Avaluegreaterthan0means keeps the specified number of parentsin the next population. Note that the value assigned tokeep_parentscannot be<-1or greater than the number ofsolutions within the populationsol_per_pop. Starting fromPyGAD2.18.0,this parameter have an effect only when thekeep_elitismparameteris0. Starting fromPyGAD2.20.0,the parents’ fitness from the last generation will not be re-used ifkeep_parents=0.keep_elitism=1: Added inPyGAD2.18.0.It can take the value0or a positive integer that satisfies(0<=keep_elitism<=sol_per_pop). It defaults to1whichmeans only the best solution in the current generation is kept in thenext generation. If assigned0, this means it has no effect. Ifassigned a positive integerK, then the bestKsolutions arekept in the next generation. It cannot be assigned a value greaterthan the value assigned to thesol_per_popparameter. If thisparameter has a value different than0, then thekeep_parentsparameter will have no effect.K_tournament=3: In case that the parent selection type istournament, theK_tournamentspecifies the number of parentsparticipating in the tournament selection. It defaults to3.crossover_type="single_point": Type of the crossover operation.Supported types aresingle_point(for single-point crossover),two_points(for two points crossover),uniform(for uniformcrossover), andscattered(for scattered crossover). Scatteredcrossover is supported from PyGAD2.9.0and higher. It defaults tosingle_point. A custom crossoverfunction can be passed starting fromPyGAD2.16.0.Check theUser-Defined Crossover, Mutation, and Parent SelectionOperatorssection for more details about creating a user-defined crossoverfunction. Starting fromPyGAD2.2.2and higher, ifcrossover_type=None, then the crossover step isbypassed which means no crossover is applied and thus no offspringwill be created in the next generations. The next generation will usethe solutions in the current population.crossover_probability=None: The probability of selecting a parentfor applying the crossover operation. Its value must be between 0.0and 1.0 inclusive. For each parent, a random value between 0.0 and 1.0is generated. If this random value is less than or equal to the valueassigned to thecrossover_probabilityparameter, then the parentis selected. Added inPyGAD2.5.0and higher.mutation_type="random": Type of the mutation operation. Supportedtypes arerandom(for random mutation),swap(for swapmutation),inversion(for inversion mutation),scramble(forscramble mutation), andadaptive(for adaptive mutation). Itdefaults torandom. A custom mutation function can be passedstarting fromPyGAD2.16.0.Check theUser-Defined Crossover, Mutation, and Parent SelectionOperatorssection for more details about creating a user-defined mutationfunction. Starting fromPyGAD2.2.2and higher, ifmutation_type=None, then the mutation step isbypassed which means no mutation is applied and thus no changes areapplied to the offspring created using the crossover operation. Theoffspring will be used unchanged in the next generation.Adaptivemutation is supported starting fromPyGAD2.10.0.For more information about adaptive mutation, go the theAdaptiveMutationsection. For example about using adaptive mutation, check theUseAdaptive Mutation inPyGADsection.mutation_probability=None: The probability of selecting a gene forapplying the mutation operation. Its value must be between 0.0 and 1.0inclusive. For each gene in a solution, a random value between 0.0 and1.0 is generated. If this random value is less than or equal to thevalue assigned to themutation_probabilityparameter, then thegene is selected. If this parameter exists, then there is no need forthe 2 parametersmutation_percent_genesandmutation_num_genes. Added inPyGAD2.5.0and higher.mutation_by_replacement=False: An optional bool parameter. Itworks only when the selected type of mutation is random(mutation_type="random"). In this case,mutation_by_replacement=Truemeans replace the gene by therandomly generated value. If False, then it has no effect and randommutation works by adding the random value to the gene. Supported inPyGAD2.2.2and higher. Check the changes inPyGAD2.2.2under the Release History section for an example.mutation_percent_genes="default": Percentage of genes to mutate.It defaults to the string"default"which is later translated intothe integer10which means 10% of the genes will be mutated. Itmust be>0and<=100. Out of this percentage, the number ofgenes to mutate is deduced which is assigned to themutation_num_genesparameter. Themutation_percent_genesparameter has no action ifmutation_probabilityormutation_num_genesexist. Starting fromPyGAD2.2.2and higher, this parameter has no action ifmutation_typeisNone.mutation_num_genes=None: Number of genes to mutate which defaultstoNonemeaning that no number is specified. Themutation_num_genesparameter has no action if the parametermutation_probabilityexists. Starting fromPyGAD2.2.2and higher, this parameter has no action ifmutation_typeisNone.random_mutation_min_val=-1.0: Forrandommutation, therandom_mutation_min_valparameter specifies the start value of therange from which a random value is selected to be added to the gene.It defaults to-1. Starting fromPyGAD2.2.2and higher, this parameter has no action ifmutation_typeisNone.random_mutation_max_val=1.0: Forrandommutation, therandom_mutation_max_valparameter specifies the end value of therange from which a random value is selected to be added to the gene.It defaults to+1. Starting fromPyGAD2.2.2and higher, this parameter has no action ifmutation_typeisNone.gene_space=None: It is used to specify the possible values foreach gene in case the user wants to restrict the gene values. It isuseful if the gene space is restricted to a certain range or todiscrete values. It accepts alist,range, ornumpy.ndarray. When all genes have the same global space, specifytheir values as alist/tuple/range/numpy.ndarray. Forexample,gene_space=[0.3,5.2,-4,8]restricts the gene valuesto the 4 specified values. If each gene has its own space, then thegene_spaceparameter can be nested like[[0.4,-5],[0.5,-3.2,8.2,-9],...]where the first sublistdetermines the values for the first gene, the second sublist for thesecond gene, and so on. If the nested list/tuple has aNonevalue,then the gene’s initial value is selected randomly from the rangespecified by the 2 parametersinit_range_lowandinit_range_highand its mutation value is selected randomly fromthe range specified by the 2 parametersrandom_mutation_min_valandrandom_mutation_max_val.gene_spaceis added inPyGAD2.5.0.Check theRelease History of PyGAD2.5.0section of the documentation for more details. InPyGAD2.9.0,NumPy arrays can be assigned to thegene_spaceparameter. InPyGAD2.11.0,thegene_spaceparameter itself or any of its elements can beassigned to a dictionary to specify the lower and upper limits of thegenes. For example,{'low':2,'high':4}means the minimum andmaximum values are 2 and 4, respectively. InPyGAD2.15.0,a new key called"step"is supported to specify the step of movingfrom the start to the end of the range specified by the 2 existingkeys"low"and"high".gene_constraint=None: A list of callables (i.e. functions) actingas constraints for the gene values. Before selecting a value for agene, the callable is called to ensure the candidate value is valid.Added inPyGAD3.5.0.Check theGeneConstraintsection for more information.sample_size=100: In some cases where a gene value is to beselected, this variable defines the size of the sample from which avalue is selected randomly. Useful if eitherallow_duplicate_genesorgene_constraintis used. If PyGAD failed to find a unique valueor a value that meets a gene constraint, it is recommended toincreases this parameter’s value. Added inPyGAD3.5.0.Check thesample_sizeParametersection for more information.on_start=None: Accepts a function/method to be called only oncebefore the genetic algorithm starts its evolution. If function, thenit must accept a single parameter representing the instance of thegenetic algorithm. If method, then it must accept 2 parameters wherethe second one refers to the method’s object. Added inPyGAD2.6.0.on_fitness=None: Accepts a function/method to be called aftercalculating the fitness values of all solutions in the population. Iffunction, then it must accept 2 parameters: 1) a list of allsolutions’ fitness values 2) the instance of the genetic algorithm. Ifmethod, then it must accept 3 parameters where the third one refers tothe method’s object. Added inPyGAD2.6.0.on_parents=None: Accepts a function/method to be called afterselecting the parents that mates. If function, then it must accept 2parameters: 1) the selected parents 2) the instance of the geneticalgorithm If method, then it must accept 3 parameters where the thirdone refers to the method’s object. Added inPyGAD2.6.0.on_crossover=None: Accepts a function to be called each time thecrossover operation is applied. This function must accept 2parameters: the first one represents the instance of the geneticalgorithm and the second one represents the offspring generated usingcrossover. Added inPyGAD2.6.0.on_mutation=None: Accepts a function to be called each time themutation operation is applied. This function must accept 2 parameters:the first one represents the instance of the genetic algorithm and thesecond one represents the offspring after applying the mutation. AddedinPyGAD2.6.0.on_generation=None: Accepts a function to be called after eachgeneration. This function must accept a single parameter representingthe instance of the genetic algorithm. If the function returned thestringstop, then therun()method stops without completingthe other generations. Added inPyGAD2.6.0.on_stop=None: Accepts a function to be called only once exactlybefore the genetic algorithm stops or when it completes all thegenerations. This function must accept 2 parameters: the first onerepresents the instance of the genetic algorithm and the second one isa list of fitness values of the last population’s solutions. Added inPyGAD2.6.0.save_best_solutions=False: WhenTrue, then the best solutionafter each generation is saved into an attribute namedbest_solutions. IfFalse(default), then no solutions aresaved and thebest_solutionsattribute will be empty. Supported inPyGAD2.9.0.save_solutions=False: IfTrue, then all solutions in eachgeneration are appended into an attribute calledsolutionswhichis NumPy array. Supported inPyGAD2.15.0.suppress_warnings=False: A bool parameter to control whether thewarning messages are printed or not. It defaults toFalse.allow_duplicate_genes=True: Added inPyGAD2.13.0.IfTrue, then a solution/chromosome may have duplicate genevalues. IfFalse, then each gene will have a unique value in itssolution.stop_criteria=None: Some criteria to stop the evolution. Added inPyGAD2.15.0.Each criterion is passed asstrwhich has a stop word. The current2 supported words arereachandsaturate.reachstops therun()method if the fitness value is equal to or greater than agiven fitness value. An example forreachis"reach_40"whichstops the evolution if the fitness is >= 40.saturatemeans stopthe evolution if the fitness saturates for a given number ofconsecutive generations. An example forsaturateis"saturate_7"which means stop therun()method if the fitnessdoes not change for 7 consecutive generations.parallel_processing=None: Added inPyGAD2.17.0.IfNone(Default), this means no parallel processing is applied.It can accept a list/tuple of 2 elements [1) Can be either'process'or'thread'to indicate whether processes or threadsare used, respectively., 2) The number of processes or threads touse.]. For example,parallel_processing=['process',10]appliesparallel processing with 10 processes. If a positive integer isassigned, then it is used as the number of threads. For example,parallel_processing=5uses 5 threads which is equivalent toparallel_processing=["thread",5]. For more information, check theParallel Processing inPyGADsection.random_seed=None: Added inPyGAD2.18.0.It defines the random seed to be used by the random functiongenerators (we use random functions in the NumPy and random modules).This helps to reproduce the same results by setting the same randomseed (e.g.random_seed=2). If given the valueNone, then ithas no effect.logger=None: Accepts an instance of thelogging.Loggerclassto log the outputs. Any message is no longer printed usingprint()but logged. Iflogger=None, then a logger is created that usesStreamHandlerto logs the messages to the console. Added inPyGAD3.0.0.Check theLoggingOutputsfor more information.
The user doesn’t have to specify all of such parameters while creatingan instance of the GA class. A very important parameter you must careabout isfitness_func which defines the fitness function.
It is OK to set the value of any of the 2 parametersinit_range_lowandinit_range_high to be equal, higher, or lower than the otherparameter (i.e.init_range_low is not needed to be lower thaninit_range_high). The same holds for therandom_mutation_min_valandrandom_mutation_max_val parameters.
If the 2 parametersmutation_type andcrossover_type areNone, this disables any type of evolution the genetic algorithm canmake. As a result, the genetic algorithm cannot find a better solutionthat the best solution in the initial population.
The parameters are validated within the constructor. If at least aparameter is not correct, an exception is thrown.
Plotting Methods inpygad.GA Class¶
plot_fitness(): Shows how the fitness evolves by generation.plot_genes(): Shows how the gene value changes for eachgeneration.plot_new_solution_rate(): Shows the number of new solutionsexplored in each solution.
Class Attributes¶
supported_int_types: A list of the supported types for the integernumbers.supported_float_types: A list of the supported types for thefloating-point numbers.supported_int_float_types: A list of the supported types for allnumbers. It just concatenates the previous 2 lists.
Other Instance Attributes & Methods¶
All the parameters and functions passed to thepygad.GA classconstructor are used as class attributes and methods in the instances ofthepygad.GA class. In addition to such attributes, there are otherattributes and methods added to the instances of thepygad.GA class:
The next 2 subsections list such attributes and methods.
Other Attributes¶
generations_completed: Holds the number of the last completedgeneration.population: A NumPy array holding the initial population.valid_parameters: Set toTruewhen all the parameters passedin theGAclass constructor are valid.run_completed: Set toTrueonly after therun()methodcompletes gracefully.pop_size: The population size.best_solutions_fitness: A list holding the fitness values of thebest solutions for all generations.best_solution_generation: The generation number at which the bestfitness value is reached. It is only assigned the generation numberafter therun()method completes. Otherwise, its value is -1.best_solutions: A NumPy array holding the best solution per eachgeneration. It only exists when thesave_best_solutionsparameterin thepygad.GAclass constructor is set toTrue.last_generation_fitness: The fitness values of the solutions inthe last generation.Added in PyGAD2.12.0.previous_generation_fitness: At the end of each generation, thefitness of the most recent population is saved in thelast_generation_fitnessattribute. The fitness of the populationexactly preceding this most recent population is saved in thelast_generation_fitnessattribute. Thisprevious_generation_fitnessattribute is used to fetch thepre-calculated fitness instead of calling the fitness function foralready explored solutions.Added in PyGAD2.16.2.last_generation_parents: The parents selected from the lastgeneration.Added in PyGAD2.12.0.last_generation_offspring_crossover: The offspring generated afterapplying the crossover in the last generation.Added in PyGAD2.12.0.last_generation_offspring_mutation: The offspring generated afterapplying the mutation in the last generation.Added in PyGAD2.12.0.gene_type_single: A flag that is set toTrueif thegene_typeparameter is assigned to a single data type that isapplied to all genes. Ifgene_typeis assigned alist,tuple, ornumpy.ndarray, then the value ofgene_type_singlewill beFalse.Added in PyGAD2.14.0.last_generation_parents_indices: This attribute holds the indicesof the selected parents in the last generation. Supported inPyGAD2.15.0.last_generation_elitism: This attribute holds the elitism of thelast generation. It is effective only if thekeep_elitismparameter has a non-zero value. Supported inPyGAD2.18.0.last_generation_elitism_indices: This attribute holds the indicesof the elitism of the last generation. It is effective only if thekeep_elitismparameter has a non-zero value. Supported inPyGAD2.19.0.logger: This attribute holds the logger from theloggingmodule. Supported inPyGAD3.0.0.gene_space_unpacked: This is the unpacked version of thegene_spaceparameter. For example,range(1,5)is unpacked to[1,2,3,4]. For an infinite range like{'low':2,'high':4}, then it is unpacked to a limited number ofvalues (e.g. 100). Supported inPyGAD3.1.0.pareto_fronts: A new instance attribute namedpareto_frontsadded to thepygad.GAinstances that holds the pareto fronts whensolving a multi-objective problem. Supported inPyGAD3.2.0.
Note that the attributes with names starting withlast_generation_are updated after each generation.
Other Methods¶
cal_pop_fitness(): A method that calculates the fitness values forall solutions within the population by calling the function passed tothefitness_funcparameter for each solution.crossover(): Refers to the method that applies the crossoveroperator based on the selected type of crossover in thecrossover_typeproperty.mutation(): Refers to the method that applies the mutationoperator based on the selected type of mutation in themutation_typeproperty.select_parents(): Refers to a method that selects the parentsbased on the parent selection type specified in theparent_selection_typeattribute.adaptive_mutation_population_fitness(): Returns the averagefitness value used in the adaptive mutation to filter the solutions.summary(): Prints a Keras-like summary of the PyGAD lifecycle.This helps to have an overview of the architecture. Supported inPyGAD2.19.0.Check thePrint LifecycleSummarysection for more details and examples.4 methods with names starting with
run_. Their purpose is to keepthe main loop inside therun()method clean. The details insidethe loop are moved to 4 individual methods. Generally, any method witha name starting withrun_is meant to be called by PyGAD frominside therun()method. Supported inPyGAD3.3.1.run_select_parents(call_on_parents=True): Select the parentsand call the callableon_parents()if defined. Ifcall_on_parentsisTrue, then the callableon_parents()is called. It must beFalsewhen therun_select_parents()method is called to update the parents at the end of therun()method.run_crossover(): Apply crossover and call the callableon_crossover()if defined.run_mutation(): Apply mutation and call the callableon_mutation()if defined.run_update_population(): Update thepopulationattributeafter completing the processes of crossover and mutation.
There are many methods that are not designed for user usage. Some ofthem are listed above but this is not a comprehensive list. Thereleasehistorysection usually covers them. Moreover, you can check thePyGAD GitHubrepository tofind more.
The next sections discuss the methods available in thepygad.GAclass.
initialize_population()¶
It creates an initial population randomly as a NumPy array. The array issaved in the instance attribute namedpopulation.
Accepts the following parameters:
low: The lower value of the random range from which the genevalues in the initial population are selected. It defaults to -4.Available in PyGAD 1.0.20 and higher.high: The upper value of the random range from which the genevalues in the initial population are selected. It defaults to -4.Available in PyGAD 1.0.20.
This method assigns the values of the following 3 instance attributes:
pop_size: Size of the population.population: Initially, it holds the initial population and laterupdated after each generation.initial_population: Keeping the initial population.
cal_pop_fitness()¶
Thecal_pop_fitness() method calculates and returns the fitnessvalues of the solutions in the current population.
This function is optimized to save time by making fewer calls thefitness function. It follows this process:
If the
save_solutionsparameter is set toTrue, then itchecks if the solution is already explored and saved in thesolutionsinstance attribute. If so, then it just retrieves itsfitness from thesolutions_fitnessinstance attribute withoutcalling the fitness function.If
save_solutionsis set toFalseor if it isTruebutthe solution was not explored yet, then thecal_pop_fitness()method checks if thekeep_elitismparameter is set to a positiveinteger. If so, then it checks if the solution is saved into thelast_generation_elitisminstance attribute. If so, then itretrieves its fitness from theprevious_generation_fitnessinstance attribute.If neither of the above 3 conditions apply (1.
save_solutionsisset toFalseor 2. if it isTruebut the solution was notexplored yet or 3.keep_elitismis set to zero), then thecal_pop_fitness()method checks if thekeep_parentsparameteris set to-1or a positive integer. If so, then it checks if thesolution is saved into thelast_generation_parentsinstanceattribute. If so, then it retrieves its fitness from theprevious_generation_fitnessinstance attribute.If neither of the above 4 conditions apply, then we have to call thefitness function to calculate the fitness for the solution. This isby calling the function assigned to the
fitness_funcparameter.
This function takes into consideration:
The
parallel_processingparameter to check whether parallelprocessing is in effect.The
fitness_batch_sizeparameter to check if the fitness shouldbe calculated in batches of solutions.
It returns a vector of the solutions’ fitness values.
run()¶
Runs the genetic algorithm. This is the main method in which the geneticalgorithm is evolved through some generations. It accepts no parametersas it uses the instance to access all of its requirements.
For each generation, the fitness values of all solutions within thepopulation are calculated according to thecal_pop_fitness() methodwhich internally just calls the function assigned to thefitness_func parameter in thepygad.GA class constructor foreach solution.
According to the fitness values of all solutions, the parents areselected using theselect_parents() method. This method behaviour isdetermined according to the parent selection type in theparent_selection_type parameter in thepygad.GA classconstructor
Based on the selected parents, offspring are generated by applying thecrossover and mutation operations using thecrossover() andmutation() methods. The behaviour of such 2 methods is definedaccording to thecrossover_type andmutation_type parameters inthepygad.GA class constructor.
After the generation completes, the following takes place:
The
populationattribute is updated by the new population.The
generations_completedattribute is assigned by the number ofthe last completed generation.If there is a callback function assigned to the
on_generationattribute, then it will be called.
After therun() method completes, the following takes place:
The
best_solution_generationis assigned the generation number atwhich the best fitness value is reached.The
run_completedattribute is set toTrue.
Parent Selection Methods¶
TheParentSelection class in thepygad.utils.parent_selectionmodule has several methods for selecting the parents that will mate toproduce the offspring. All of such methods accept the same parameterswhich are:
fitness: The fitness values of the solutions in the currentpopulation.num_parents: The number of parents to be selected.
All of such methods return an array of the selected parents.
The next subsections list the supported methods for parent selection.
steady_state_selection()¶
Selects the parents using the steady-state selection technique.
rank_selection()¶
Selects the parents using the rank selection technique.
random_selection()¶
Selects the parents randomly.
tournament_selection()¶
Selects the parents using the tournament selection technique.
roulette_wheel_selection()¶
Selects the parents using the roulette wheel selection technique.
stochastic_universal_selection()¶
Selects the parents using the stochastic universal selection technique.
nsga2_selection()¶
Selects the parents for the NSGA-II algorithm to solve multi-objectiveoptimization problems. It selects the parents by ranking them based onnon-dominated sorting and crowding distance.
tournament_selection_nsga2()¶
Selects the parents for the NSGA-II algorithm to solve multi-objectiveoptimization problems. It selects the parents using the tournamentselection technique applied based on non-dominated sorting and crowdingdistance.
Crossover Methods¶
TheCrossover class in thepygad.utils.crossover module supportsseveral methods for applying crossover between the selected parents. Allof these methods accept the same parameters which are:
parents: The parents to mate for producing the offspring.offspring_size: The size of the offspring to produce.
All of such methods return an array of the produced offspring.
The next subsections list the supported methods for crossover.
single_point_crossover()¶
Applies the single-point crossover. It selects a point randomly at whichcrossover takes place between the pairs of parents.
two_points_crossover()¶
Applies the 2 points crossover. It selects the 2 points randomly atwhich crossover takes place between the pairs of parents.
uniform_crossover()¶
Applies the uniform crossover. For each gene, a parent out of the 2mating parents is selected randomly and the gene is copied from it.
scattered_crossover()¶
Applies the scattered crossover. It randomly selects the gene from oneof the 2 parents.
Mutation Methods¶
TheMutation class in thepygad.utils.mutation module supportsseveral methods for applying mutation. All of these methods accept thesame parameter which is:
offspring: The offspring to mutate.
All of such methods return an array of the mutated offspring.
The next subsections list the supported methods for mutation.
random_mutation()¶
Applies the random mutation which changes the values of some genesrandomly. The number of genes is specified according to either themutation_num_genes or themutation_percent_genes attributes.
For each gene, a random value is selected according to the rangespecified by the 2 attributesrandom_mutation_min_val andrandom_mutation_max_val. The random value is added to the selectedgene.
swap_mutation()¶
Applies the swap mutation which interchanges the values of 2 randomlyselected genes.
inversion_mutation()¶
Applies the inversion mutation which selects a subset of genes andinverts them.
scramble_mutation()¶
Applies the scramble mutation which selects a subset of genes andshuffles their order randomly.
adaptive_mutation()¶
Applies the adaptive mutation which selects the number/percentage ofgenes to mutate based on the solution’s fitness. If the fitness is high(i.e. solution quality is high), then small number/percentage of genesis mutated compared to a solution with a low fitness.
best_solution()¶
Returns information about the best solution found by the geneticalgorithm.
It accepts the following parameters:
pop_fitness=None: An optional parameter that accepts a list of thefitness values of the solutions in the population. IfNone, thenthecal_pop_fitness()method is called to calculate the fitnessvalues of the population.
It returns the following:
best_solution: Best solution in the current population.best_solution_fitness: Fitness value of the best solution.best_match_idx: Index of the best solution in the currentpopulation.
plot_fitness()¶
Previously namedplot_result(), this method creates, shows, andreturns a figure that summarizes how the fitness value evolves bygeneration.
It works only after completing at least 1 generation. If no generationis completed (at least 1), an exception is raised.
plot_new_solution_rate()¶
Theplot_new_solution_rate() method creates, shows, and returns afigure that shows the number of new solutions explored in eachgeneration. This method works only whensave_solutions=True in theconstructor of thepygad.GA class.
It works only after completing at least 1 generation. If no generationis completed (at least 1), an exception is raised.
plot_genes()¶
Theplot_genes() method creates, shows, and returns a figure thatdescribes each gene. It has different options to create the figureswhich helps to:
Explore the gene value for each generation by creating a normal plot.
Create a histogram for each gene.
Create a boxplot.
This is controlled by thegraph_type parameter.
It works only after completing at least 1 generation. If no generationis completed (at least 1), an exception is raised.
save()¶
Saves the genetic algorithm instance
Accepts the following parameter:
filename: Name of the file to save the instance. No extension isneeded.
Functions inpygad¶
Besides the methods available in thepygad.GA class, this sectiondiscusses the functions available inpygad. Up to this time, thereis only a single function namedload().
pygad.load()¶
Reads a saved instance of the genetic algorithm. This is not a methodbut a function that is indented under thepygad module. So, it couldbe called by the pygad module as follows:pygad.load(filename).
Accepts the following parameter:
filename: Name of the file holding the saved instance of thegenetic algorithm. No extension is needed.
Returns the genetic algorithm instance.
Steps to Usepygad¶
To use thepygad module, here is a summary of the required steps:
Preparing the
fitness_funcparameter.Preparing Other Parameters.
Import
pygad.Create an Instance of the
pygad.GAClass.Run the Genetic Algorithm.
Plotting Results.
Information about the Best Solution.
Saving & Loading the Results.
Let’s discuss how to do each of these steps.
Preparing thefitness_func Parameter¶
Even though some steps in the genetic algorithm pipeline can work thesame regardless of the problem being solved, one critical step is thecalculation of the fitness value. There is no unique way of calculatingthe fitness value and it changes from one problem to another.
PyGAD has a parameter calledfitness_func that allows the user tospecify a custom function/method to use when calculating the fitness.This function/method must be a maximization function/method so that asolution with a high fitness value returned is selected compared to asolution with a low value.
The fitness function is where the user can decide whether theoptimization problem is single-objective or multi-objective.
If the fitness function returns a numeric value, then the problem issingle-objective. The numeric data types supported by PyGAD are listedin the
supported_int_float_typesvariable of thepygad.GAclass.If the fitness function returns a
list,tuple, ornumpy.ndarray, then the problem is multi-objective. Even if thereis only one element, the problem is still considered multi-objective.Each element represents the fitness value of its correspondingobjective.
Using a user-defined fitness function allows the user to freely usePyGAD to solve any problem by passing the appropriate fitnessfunction/method. It is very important to understand the problem wellbefore creating it.
Let’s discuss an example:
Given the following function:y = f(w1:w6) = w1x1 + w2x2 + w3x3 + w4x4 + w5x5 + 6wx6where (x1,x2,x3,x4,x5,x6)=(4, -2, 3.5, 5, -11, -4.7) and y=44What are the best values for the 6 weights (w1 to w6)? We are goingto use the genetic algorithm to optimize this function.
So, the task is about using the genetic algorithm to find the bestvalues for the 6 weightW1 toW6. Thinking of the problem, it isclear that the best solution is that returning an output that is closeto the desired outputy=44. So, the fitness function/method shouldreturn a value that gets higher when the solution’s output is closer toy=44. Here is a function that does that:
function_inputs=[4,-2,3.5,5,-11,-4.7]# Function inputs.desired_output=44# Function output.deffitness_func(ga_instance,solution,solution_idx):output=numpy.sum(solution*function_inputs)fitness=1.0/numpy.abs(output-desired_output)returnfitness
Because the fitness function returns a numeric value, then the problemis single-objective.
Such a user-defined function must accept 3 parameters:
The instance of the
pygad.GAclass. This helps the user to fetchany property that helps when calculating the fitness.The solution(s) to calculate the fitness value(s). Note that thefitness function can accept multiple solutions only if the
fitness_batch_sizeis given a value greater than 1.The indices of the solutions in the population. The number of indicesalso depends on the
fitness_batch_sizeparameter.
If a method is passed to thefitness_func parameter, then it acceptsa fourth parameter representing the method’s instance.
The__code__ object is used to check if this function accepts therequired number of parameters. If more or fewer parameters are passed,an exception is thrown.
By creating this function, you did a very important step towards usingPyGAD.
Preparing Other Parameters¶
Here is an example for preparing the other parameters:
num_generations=50num_parents_mating=4fitness_function=fitness_funcsol_per_pop=8num_genes=len(function_inputs)init_range_low=-2init_range_high=5parent_selection_type="sss"keep_parents=1crossover_type="single_point"mutation_type="random"mutation_percent_genes=10
Theon_generation Parameter¶
An optional parameter namedon_generation is supported which allowsthe user to call a function (with a single parameter) after eachgeneration. Here is a simple function that just prints the currentgeneration number and the fitness value of the best solution in thecurrent generation. Thegenerations_completed attribute of the GAclass returns the number of the last completed generation.
defon_gen(ga_instance):print("Generation : ",ga_instance.generations_completed)print("Fitness of the best solution :",ga_instance.best_solution()[1])
After being defined, the function is assigned to theon_generationparameter of the GA class constructor. By doing that, theon_gen()function will be called after each generation.
ga_instance=pygad.GA(...,on_generation=on_gen,...)
After the parameters are prepared, we can import PyGAD and build aninstance of thepygad.GA class.
Importpygad¶
The next step is to import PyGAD as follows:
importpygad
Thepygad.GA class holds the implementation of all methods forrunning the genetic algorithm.
Create an Instance of thepygad.GA Class¶
Thepygad.GA class is instantiated where the previously preparedparameters are fed to its constructor. The constructor is responsiblefor creating the initial population.
ga_instance=pygad.GA(num_generations=num_generations,num_parents_mating=num_parents_mating,fitness_func=fitness_function,sol_per_pop=sol_per_pop,num_genes=num_genes,init_range_low=init_range_low,init_range_high=init_range_high,parent_selection_type=parent_selection_type,keep_parents=keep_parents,crossover_type=crossover_type,mutation_type=mutation_type,mutation_percent_genes=mutation_percent_genes)
Run the Genetic Algorithm¶
After an instance of thepygad.GA class is created, the next step isto call therun() method as follows:
ga_instance.run()
Inside this method, the genetic algorithm evolves over some generationsby doing the following tasks:
Calculating the fitness values of the solutions within the currentpopulation.
Select the best solutions as parents in the mating pool.
Apply the crossover & mutation operation
Repeat the process for the specified number of generations.
Plotting Results¶
There is a method namedplot_fitness() which creates a figuresummarizing how the fitness values of the solutions change with thegenerations.
ga_instance.plot_fitness()

Information about the Best Solution¶
The following information about the best solution in the last populationis returned using thebest_solution() method.
Solution
Fitness value of the solution
Index of the solution within the population
solution,solution_fitness,solution_idx=ga_instance.best_solution()print(f"Parameters of the best solution :{solution}")print(f"Fitness value of the best solution ={solution_fitness}")print(f"Index of the best solution :{solution_idx}")
Using thebest_solution_generation attribute of the instance fromthepygad.GA class, the generation number at which thebestfitness is reached could be fetched.
ifga_instance.best_solution_generation!=-1:print(f"Best fitness value reached after{ga_instance.best_solution_generation} generations.")
Saving & Loading the Results¶
After therun() method completes, it is possible to save the currentinstance of the genetic algorithm to avoid losing the progress made. Thesave() method is available for that purpose. Just pass the file nameto it without an extension. According to the next code, a file namedgenetic.pkl will be created and saved in the current directory.
filename='genetic'ga_instance.save(filename=filename)
You can also load the saved model using theload() function andcontinue using it. For example, you might run the genetic algorithm forsome generations, save its current state using thesave() method,load the model using theload() function, and then call therun() method again.
loaded_ga_instance=pygad.load(filename=filename)
After the instance is loaded, you can use it to run any method or accessany property.
print(loaded_ga_instance.best_solution())
Life Cycle of PyGAD¶
The next figure lists the different stages in the lifecycle of aninstance of thepygad.GA class. Note that PyGAD stops when eitherall generations are completed or when the function passed to theon_generation parameter returns the stringstop.
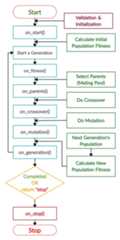
The next code implements all the callback functions to trace theexecution of the genetic algorithm. Each callback function prints itsname.
importpygadimportnumpyfunction_inputs=[4,-2,3.5,5,-11,-4.7]desired_output=44deffitness_func(ga_instance,solution,solution_idx):output=numpy.sum(solution*function_inputs)fitness=1.0/(numpy.abs(output-desired_output)+0.000001)returnfitnessfitness_function=fitness_funcdefon_start(ga_instance):print("on_start()")defon_fitness(ga_instance,population_fitness):print("on_fitness()")defon_parents(ga_instance,selected_parents):print("on_parents()")defon_crossover(ga_instance,offspring_crossover):print("on_crossover()")defon_mutation(ga_instance,offspring_mutation):print("on_mutation()")defon_generation(ga_instance):print("on_generation()")defon_stop(ga_instance,last_population_fitness):print("on_stop()")ga_instance=pygad.GA(num_generations=3,num_parents_mating=5,fitness_func=fitness_function,sol_per_pop=10,num_genes=len(function_inputs),on_start=on_start,on_fitness=on_fitness,on_parents=on_parents,on_crossover=on_crossover,on_mutation=on_mutation,on_generation=on_generation,on_stop=on_stop)ga_instance.run()
Based on the used 3 generations as assigned to thenum_generationsargument, here is the output.
on_start()on_fitness()on_parents()on_crossover()on_mutation()on_generation()on_fitness()on_parents()on_crossover()on_mutation()on_generation()on_fitness()on_parents()on_crossover()on_mutation()on_generation()on_stop()
Examples¶
This section gives the complete code of some examples that usepygad. Each subsection builds a different example.
Linear Model Optimization - Single Objective¶
This example is discussed in theSteps to UsePyGADsection which optimizes a linear model. Its complete code is listedbelow.
importpygadimportnumpy"""Given the following function: y = f(w1:w6) = w1x1 + w2x2 + w3x3 + w4x4 + w5x5 + 6wx6 where (x1,x2,x3,x4,x5,x6)=(4,-2,3.5,5,-11,-4.7) and y=44What are the best values for the 6 weights (w1 to w6)? We are going to use the genetic algorithm to optimize this function."""function_inputs=[4,-2,3.5,5,-11,-4.7]# Function inputs.desired_output=44# Function output.deffitness_func(ga_instance,solution,solution_idx):output=numpy.sum(solution*function_inputs)fitness=1.0/(numpy.abs(output-desired_output)+0.000001)returnfitnessnum_generations=100# Number of generations.num_parents_mating=10# Number of solutions to be selected as parents in the mating pool.sol_per_pop=20# Number of solutions in the population.num_genes=len(function_inputs)last_fitness=0defon_generation(ga_instance):globallast_fitnessprint(f"Generation ={ga_instance.generations_completed}")print(f"Fitness ={ga_instance.best_solution(pop_fitness=ga_instance.last_generation_fitness)[1]}")print(f"Change ={ga_instance.best_solution(pop_fitness=ga_instance.last_generation_fitness)[1]-last_fitness}")last_fitness=ga_instance.best_solution(pop_fitness=ga_instance.last_generation_fitness)[1]ga_instance=pygad.GA(num_generations=num_generations,num_parents_mating=num_parents_mating,sol_per_pop=sol_per_pop,num_genes=num_genes,fitness_func=fitness_func,on_generation=on_generation)# Running the GA to optimize the parameters of the function.ga_instance.run()ga_instance.plot_fitness()# Returning the details of the best solution.solution,solution_fitness,solution_idx=ga_instance.best_solution(ga_instance.last_generation_fitness)print(f"Parameters of the best solution :{solution}")print(f"Fitness value of the best solution ={solution_fitness}")print(f"Index of the best solution :{solution_idx}")prediction=numpy.sum(numpy.array(function_inputs)*solution)print(f"Predicted output based on the best solution :{prediction}")ifga_instance.best_solution_generation!=-1:print(f"Best fitness value reached after{ga_instance.best_solution_generation} generations.")# Saving the GA instance.filename='genetic'# The filename to which the instance is saved. The name is without extension.ga_instance.save(filename=filename)# Loading the saved GA instance.loaded_ga_instance=pygad.load(filename=filename)loaded_ga_instance.plot_fitness()
Linear Model Optimization - Multi-Objective¶
This is a multi-objective optimization example that optimizes these 2functions:
y1=f(w1:w6)=w1x1+w2x2+w3x3+w4x4+w5x5+6wx6y2=f(w1:w6)=w1x7+w2x8+w3x9+w4x10+w5x11+6wx12
Where:
(x1,x2,x3,x4,x5,x6)=(4,-2,3.5,5,-11,-4.7)andy=50(x7,x8,x9,x10,x11,x12)=(-2,0.7,-9,1.4,3,5)andy=30
The 2 functions use the same parameters (weights)w1 tow6.
The goal is to use PyGAD to find the optimal values for such weightsthat satisfy the 2 functionsy1 andy2.
To use PyGAD to solve multi-objective problems, the only adjustment isto return alist,tuple, ornumpy.ndarray from the fitnessfunction. Each element represents the fitness of an objective in order.That is the first element is the fitness of the first objective, thesecond element is the fitness for the second objective, and so on.
importpygadimportnumpy"""Given these 2 functions: y1 = f(w1:w6) = w1x1 + w2x2 + w3x3 + w4x4 + w5x5 + 6wx6 y2 = f(w1:w6) = w1x7 + w2x8 + w3x9 + w4x10 + w5x11 + 6wx12 where (x1,x2,x3,x4,x5,x6)=(4,-2,3.5,5,-11,-4.7) and y=50 and (x7,x8,x9,x10,x11,x12)=(-2,0.7,-9,1.4,3,5) and y=30What are the best values for the 6 weights (w1 to w6)? We are going to use the genetic algorithm to optimize these 2 functions.This is a multi-objective optimization problem.PyGAD considers the problem as multi-objective if the fitness function returns: 1) List. 2) Or tuple. 3) Or numpy.ndarray."""function_inputs1=[4,-2,3.5,5,-11,-4.7]# Function 1 inputs.function_inputs2=[-2,0.7,-9,1.4,3,5]# Function 2 inputs.desired_output1=50# Function 1 output.desired_output2=30# Function 2 output.deffitness_func(ga_instance,solution,solution_idx):output1=numpy.sum(solution*function_inputs1)output2=numpy.sum(solution*function_inputs2)fitness1=1.0/(numpy.abs(output1-desired_output1)+0.000001)fitness2=1.0/(numpy.abs(output2-desired_output2)+0.000001)return[fitness1,fitness2]num_generations=100num_parents_mating=10sol_per_pop=20num_genes=len(function_inputs1)ga_instance=pygad.GA(num_generations=num_generations,num_parents_mating=num_parents_mating,sol_per_pop=sol_per_pop,num_genes=num_genes,fitness_func=fitness_func,parent_selection_type='nsga2')ga_instance.run()ga_instance.plot_fitness(label=['Obj 1','Obj 2'])solution,solution_fitness,solution_idx=ga_instance.best_solution(ga_instance.last_generation_fitness)print(f"Parameters of the best solution :{solution}")print(f"Fitness value of the best solution ={solution_fitness}")prediction=numpy.sum(numpy.array(function_inputs1)*solution)print(f"Predicted output 1 based on the best solution :{prediction}")prediction=numpy.sum(numpy.array(function_inputs2)*solution)print(f"Predicted output 2 based on the best solution :{prediction}")
This is the result of the print statements. The predicted outputs areclose to the desired outputs.
Parametersofthebestsolution:[0.79676439-2.98823386-4.126776625.70539445-2.02797016-1.07243922]Fitnessvalueofthebestsolution=[1.68090829349.8591915]Predictedoutput1basedonthebestsolution:50.59491545442283Predictedoutput2basedonthebestsolution:29.99714270722312
This is the figure created by theplot_fitness() method. The fitnessof the first objective has the green color. The blue color is used forthe second objective fitness.
Reproducing Images¶
This project reproduces a single image using PyGAD by evolving pixelvalues. This project works with both color and gray images. Check thisproject atGitHub:https://github.com/ahmedfgad/GARI.
For more information about this project, read this tutorial titledReproducing Images using a Genetic Algorithm withPythonavailable at these links:
Heartbeat:https://heartbeat.fritz.ai/reproducing-images-using-a-genetic-algorithm-with-python-91fc701ff84
LinkedIn:https://www.linkedin.com/pulse/reproducing-images-using-genetic-algorithm-python-ahmed-gad
Project Steps¶
The steps to follow in order to reproduce an image are as follows:
Read an image
Prepare the fitness function
Create an instance of the pygad.GA class with the appropriateparameters
Run PyGAD
Plot results
Calculate some statistics
The next sections discusses the code of each of these steps.
Read an Image¶
There is an image namedfruit.jpg in theGARIproject which is read accordingto the next code.
importimageioimportnumpytarget_im=imageio.imread('fruit.jpg')target_im=numpy.asarray(target_im/255,dtype=float)
Here is the read image.

Based on the chromosome representation used in the example, the pixelvalues can be either in the 0-255, 0-1, or any other ranges.
Note that the range of pixel values affect other parameters like therange from which the random values are selected during mutation and alsothe range of the values used in the initial population. So, beconsistent.
Prepare the Fitness Function¶
The next code creates a function that will be used as a fitness functionfor calculating the fitness value for each solution in the population.This function must be a maximization function that accepts 3 parametersrepresenting the instance of thepygad.GA class, a solution, and itsindex. It returns a value representing the fitness value.
importgaritarget_chromosome=gari.img2chromosome(target_im)deffitness_fun(ga_instance,solution,solution_idx):fitness=numpy.sum(numpy.abs(target_chromosome-solution))# Negating the fitness value to make it increasing rather than decreasing.fitness=numpy.sum(target_chromosome)-fitnessreturnfitness
The fitness value is calculated using the sum of absolute differencebetween genes values in the original and reproduced chromosomes. Thegari.img2chromosome() function is called before the fitness functionto represent the image as a vector because the genetic algorithm canwork with 1D chromosomes.
The implementation of thegari module is available at theGARIGitHubproject andits code is listed below.
importnumpyimportfunctoolsimportoperatordefimg2chromosome(img_arr):returnnumpy.reshape(a=img_arr,newshape=(functools.reduce(operator.mul,img_arr.shape)))defchromosome2img(vector,shape):iflen(vector)!=functools.reduce(operator.mul,shape):raiseValueError(f"A vector of length{len(vector)} into an array of shape{shape}.")returnnumpy.reshape(a=vector,newshape=shape)
Create an Instance of thepygad.GA Class¶
It is very important to use random mutation and set themutation_by_replacement toTrue. Based on the range of pixelvalues, the values assigned to theinit_range_low,init_range_high,random_mutation_min_val, andrandom_mutation_max_val parameters should be changed.
If the image pixel values range from 0 to 255, then setinit_range_low andrandom_mutation_min_val to 0 as they are butchangeinit_range_high andrandom_mutation_max_val to 255.
Feel free to change the other parameters or add other parameters. Pleasecheck thePyGAD’s documentation forthe full list of parameters.
importpygadga_instance=pygad.GA(num_generations=20000,num_parents_mating=10,fitness_func=fitness_fun,sol_per_pop=20,num_genes=target_im.size,init_range_low=0.0,init_range_high=1.0,mutation_percent_genes=0.01,mutation_type="random",mutation_by_replacement=True,random_mutation_min_val=0.0,random_mutation_max_val=1.0)
Run PyGAD¶
Simply, call therun() method to run PyGAD.
ga_instance.run()
Plot Results¶
After therun() method completes, the fitness values of allgenerations can be viewed in a plot using theplot_fitness() method.
ga_instance.plot_fitness()
Here is the plot after 20,000 generations.

Calculate Some Statistics¶
Here is some information about the best solution.
# Returning the details of the best solution.solution,solution_fitness,solution_idx=ga_instance.best_solution()print(f"Fitness value of the best solution ={solution_fitness}")print(f"Index of the best solution :{solution_idx}")ifga_instance.best_solution_generation!=-1:print(f"Best fitness value reached after{ga_instance.best_solution_generation} generations.")result=gari.chromosome2img(solution,target_im.shape)matplotlib.pyplot.imshow(result)matplotlib.pyplot.title("PyGAD & GARI for Reproducing Images")matplotlib.pyplot.show()
Evolution by Generation¶
The solution reached after the 20,000 generations is shown below.

After more generations, the result can be enhanced like what shownbelow.

The results can also be enhanced by changing the parameters passed tothe constructor of thepygad.GA class.
Here is how the image is evolved from generation 0 to generation20,000s.
Generation 0

Generation 1,000

Generation 2,500

Generation 4,500

Generation 7,000

Generation 8,000

Generation 20,000

Clustering¶
For a 2-cluster problem, the code is availablehere.For a 3-cluster problem, the code ishere.The 2 examples are using artificial samples.
Soon a tutorial will be published atPaperspace to explain howclustering works using the genetic algorithm with examples in PyGAD.
CoinTex Game Playing using PyGAD¶
The code is available theCoinTex GitHubproject.CoinTex is an Android game written in Python using the Kivy framework.Find CoinTex atGooglePlay:https://play.google.com/store/apps/details?id=coin.tex.cointexreactfast
Check thisPaperspacetutorialfor how the genetic algorithm plays CoinTex:https://blog.paperspace.com/building-agent-for-cointex-using-genetic-algorithm.Check also thisYouTube video showingthe genetic algorithm while playing CoinTex.