USP14-regulated allostery of the human proteasome by time-resolved cryo-EM
- PMID:35477760
- PMCID: PMC9117149
- DOI: 10.1038/s41586-022-04671-8
USP14-regulated allostery of the human proteasome by time-resolved cryo-EM
Abstract
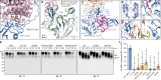
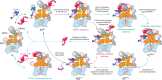
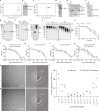
Proteasomal degradation of ubiquitylated proteins is tightly regulated at multiple levels1-3. A primary regulatory checkpoint is the removal of ubiquitin chains from substrates by the deubiquitylating enzyme ubiquitin-specific protease 14 (USP14), which reversibly binds the proteasome and confers the ability to edit and reject substrates. How USP14 is activated and regulates proteasome function remain unknown4-7. Here we present high-resolution cryo-electron microscopy structures of human USP14 in complex with the 26S proteasome in 13 distinct conformational states captured during degradation of polyubiquitylated proteins. Time-resolved cryo-electron microscopy analysis of the conformational continuum revealed two parallel pathways of proteasome state transitions induced by USP14, and captured transient conversion of substrate-engaged intermediates into substrate-inhibited intermediates. On the substrate-engaged pathway, ubiquitin-dependent activation of USP14 allosterically reprograms the conformational landscape of the AAA-ATPase motor and stimulates opening of the core particle gate8-10, enabling observation of a near-complete cycle of asymmetric ATP hydrolysis around the ATPase ring during processive substrate unfolding. Dynamic USP14-ATPase interactions decouple the ATPase activity from RPN11-catalysed deubiquitylation11-13 and kinetically introduce three regulatory checkpoints on the proteasome, at the steps of ubiquitin recognition, substrate translocation initiation and ubiquitin chain recycling. These findings provide insights into the complete functional cycle of the USP14-regulated proteasome and establish mechanistic foundations for the discovery of USP14-targeted therapies.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










Comment in
- DUB-le vision: snapshots of the proteasome during substrate processing.Schnell HM, Hanna J.Schnell HM, et al.Trends Biochem Sci. 2022 Nov;47(11):903-905. doi: 10.1016/j.tibs.2022.07.007. Epub 2022 Aug 10.Trends Biochem Sci. 2022.PMID:35963751Free PMC article.
Similar articles
- In vitro analysis of proteasome-associated USP14 activity for substrate degradation and deubiquitylation.Muniyappan S, Lee BH.Muniyappan S, et al.Methods Enzymol. 2019;619:249-268. doi: 10.1016/bs.mie.2018.12.028. Epub 2019 Feb 1.Methods Enzymol. 2019.PMID:30910023
- The deubiquitinating enzyme Usp14 allosterically inhibits multiple proteasomal activities and ubiquitin-independent proteolysis.Kim HT, Goldberg AL.Kim HT, et al.J Biol Chem. 2017 Jun 9;292(23):9830-9839. doi: 10.1074/jbc.M116.763128. Epub 2017 Apr 17.J Biol Chem. 2017.PMID:28416611Free PMC article.
- Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.Dong Y, Zhang S, Wu Z, Li X, Wang WL, Zhu Y, Stoilova-McPhie S, Lu Y, Finley D, Mao Y.Dong Y, et al.Nature. 2019 Jan;565(7737):49-55. doi: 10.1038/s41586-018-0736-4. Epub 2018 Nov 12.Nature. 2019.PMID:30479383Free PMC article.
- Structure, Dynamics and Function of the 26S Proteasome.Mao Y.Mao Y.Subcell Biochem. 2021;96:1-151. doi: 10.1007/978-3-030-58971-4_1.Subcell Biochem. 2021.PMID:33252727Review.
- Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes.Lee MJ, Lee BH, Hanna J, King RW, Finley D.Lee MJ, et al.Mol Cell Proteomics. 2011 May;10(5):R110.003871. doi: 10.1074/mcp.R110.003871. Epub 2010 Sep 7.Mol Cell Proteomics. 2011.PMID:20823120Free PMC article.Review.
Cited by
- The Proteasome-Family-Members-Based Prognostic Model Improves the Risk Classification for Adult Acute Myeloid Leukemia.Sheng G, Tao J, Jin P, Li Y, Jin W, Wang K.Sheng G, et al.Biomedicines. 2024 Sep 22;12(9):2147. doi: 10.3390/biomedicines12092147.Biomedicines. 2024.PMID:39335660Free PMC article.
- An Unsupervised Classification Algorithm for Heterogeneous Cryo-EM Projection Images Based on Autoencoders.Wang X, Lu Y, Lin X, Li J, Zhang Z.Wang X, et al.Int J Mol Sci. 2023 May 6;24(9):8380. doi: 10.3390/ijms24098380.Int J Mol Sci. 2023.PMID:37176089Free PMC article.
- Loss of the proteasomal deubiquitinase USP14 induces growth defects and a senescence phenotype in colorectal cancer cells.Gubat J, Sjöstrand L, Selvaraju K, Telli K, D'Arcy P.Gubat J, et al.Sci Rep. 2024 Jun 6;14(1):13037. doi: 10.1038/s41598-024-63791-5.Sci Rep. 2024.PMID:38844605Free PMC article.
- AlphaFold, allosteric, and orthosteric drug discovery: Ways forward.Nussinov R, Zhang M, Liu Y, Jang H.Nussinov R, et al.Drug Discov Today. 2023 Jun;28(6):103551. doi: 10.1016/j.drudis.2023.103551. Epub 2023 Mar 11.Drug Discov Today. 2023.PMID:36907321Free PMC article.Review.
- Proteasome substrate receptors and their therapeutic potential.Osei-Amponsa V, Walters KJ.Osei-Amponsa V, et al.Trends Biochem Sci. 2022 Nov;47(11):950-964. doi: 10.1016/j.tibs.2022.06.006. Epub 2022 Jul 9.Trends Biochem Sci. 2022.PMID:35817651Free PMC article.Review.
References
- Mao, Y. In Macromolecular Protein Complexes III: Structure and Function (eds Harris J. R. & Marles-Wright, J.) Ch. 1 (Springer, 2021).
Publication types
MeSH terms
Substances
Related information
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
