Between Lake Baikal and the Baltic Sea: genomic history of the gateway to Europe
- PMID:29297395
- PMCID: PMC5751809
- DOI: 10.1186/s12863-017-0578-3
Between Lake Baikal and the Baltic Sea: genomic history of the gateway to Europe
Abstract
Background: The history of human populations occupying the plains and mountain ridges separating Europe from Asia has been eventful, as these natural obstacles were crossed westward by multiple waves of Turkic and Uralic-speaking migrants as well as eastward by Europeans. Unfortunately, the material records of history of this region are not dense enough to reconstruct details of population history. These considerations stimulate growing interest to obtain a genetic picture of the demographic history of migrations and admixture in Northern Eurasia.
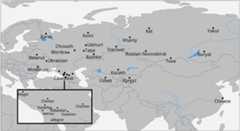
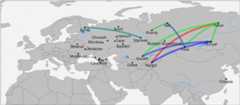
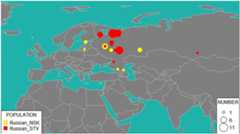
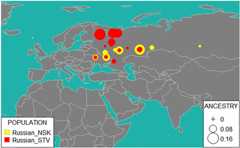
Results: We genotyped and analyzed 1076 individuals from 30 populations with geographical coverage spanning from Baltic Sea to Baikal Lake. Our dense sampling allowed us to describe in detail the population structure, provide insight into genomic history of numerous European and Asian populations, and significantly increase quantity of genetic data available for modern populations in region of North Eurasia. Our study doubles the amount of genome-wide profiles available for this region. We detected unusually high amount of shared identical-by-descent (IBD) genomic segments between several Siberian populations, such as Khanty and Ket, providing evidence of genetic relatedness across vast geographic distances and between speakers of different language families. Additionally, we observed excessive IBD sharing between Khanty and Bashkir, a group of Turkic speakers from Southern Urals region. While adding some weight to the "Finno-Ugric" origin of Bashkir, our studies highlighted that the Bashkir genepool lacks the main "core", being a multi-layered amalgamation of Turkic, Ugric, Finnish and Indo-European contributions, which points at intricacy of genetic interface between Turkic and Uralic populations. Comparison of the genetic structure of Siberian ethnicities and the geography of the region they inhabit point at existence of the "Great Siberian Vortex" directing genetic exchanges in populations across the Siberian part of Asia. Slavic speakers of Eastern Europe are, in general, very similar in their genetic composition. Ukrainians, Belarusians and Russians have almost identical proportions of Caucasus and Northern European components and have virtually no Asian influence. We capitalized on wide geographic span of our sampling to address intriguing question about the place of origin of Russian Starovers, an enigmatic Eastern Orthodox Old Believers religious group relocated to Siberia in seventeenth century. A comparative reAdmix analysis, complemented by IBD sharing, placed their roots in the region of the Northern European Plain, occupied by North Russians and Finno-Ugric Komi and Karelian people. Russians from Novosibirsk and Russian Starover exhibit ancestral proportions close to that of European Eastern Slavs, however, they also include between five to 10 % of Central Siberian ancestry, not present at this level in their European counterparts.
Conclusions: Our project has patched the hole in the genetic map of Eurasia: we demonstrated complexity of genetic structure of Northern Eurasians, existence of East-West and North-South genetic gradients, and assessed different inputs of ancient populations into modern populations.
Keywords: Admixture; Biogeography; Eastern Europe; IBD; Population genetics; Siberia.
Conflict of interest statement
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committee of the Research Institute of Medical Genetics, Tomsk National Medical Research Center of the Russian Academy of Science, the Ethics Committee of the Institute of Biochemistry and Genetics of Ufa Scientific Centre Russian Academy of Science, and the Ethics Committee of the Center “Bioengineering” (BNG) Russian Academy of Science. Written informed consent was obtained from all participants in the study. For each participant, information about geographic and ethnic origins of their parents and grandparents was recorded.
Consent for publication
Not applicable
Competing interests
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
- Cavalli-Sforza LL, Menozzi P, Piazza A, Princeton NJ. The history and geography of human genes, vol. 541: Princeton University Press; SRC - GoogleScholar. 1994.
- Cavalli-Sforza LL. The human genome diversity project: past, present and future. Nat Rev Genet. 2005;6(4):333–340. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
