Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants
- PMID:27226176
- PMCID: PMC4880914
- DOI: 10.1038/srep26685
Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants
Abstract
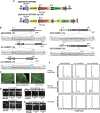
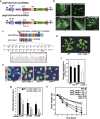
Genome editing using the CRISPR/Cas9 system can be used to modify plant genomes, however, improvements in specificity and applicability are still needed in order for the editing technique to be useful in various plant species. Here, using genome editing mediated by a truncated gRNA (tru-gRNA)/Cas9 combination, we generated new alleles for OST2, a proton pump in Arabidopsis, with no off-target effects. By following expression of Cas9 and the tru-gRNAs, newly generated mutations in CRIPSR/Cas9 transgenic plants were detected with high average mutation rates of up to 32.8% and no off-target effects using constitutive promoter. Reducing nuclear localization signals in Cas9 decreased the mutation rate. In contrast, tru-gRNA Cas9 cassettes driven by meristematic- and reproductive-tissue-specific promoters increased the heritable mutation rate in Arabidopsis, showing that high expression in the germ line can produce bi-allelic mutations. Finally, the new mutant alleles obtained for OST2 exhibited altered stomatal closing in response to environmental conditions. These results suggest further applications in molecular breeding to improve plant function using optimized plant CRISPR/Cas9 systems.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
