EAGER: efficient ancient genome reconstruction
- PMID:27036623
- PMCID: PMC4815194
- DOI: 10.1186/s13059-016-0918-z
EAGER: efficient ancient genome reconstruction
Abstract
Background: The automated reconstruction of genome sequences in ancient genome analysis is a multifaceted process.
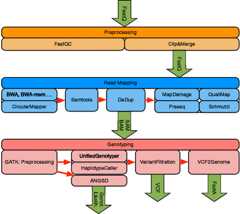
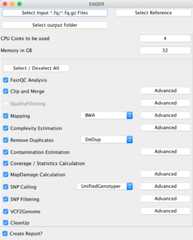
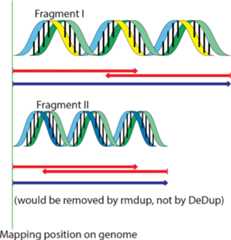
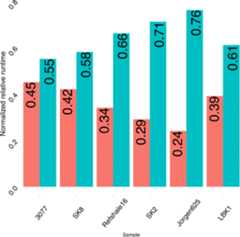
Results: Here we introduce EAGER, a time-efficient pipeline, which greatly simplifies the analysis of large-scale genomic data sets. EAGER provides features to preprocess, map, authenticate, and assess the quality of ancient DNA samples. Additionally, EAGER comprises tools to genotype samples to discover, filter, and analyze variants.
Conclusions: EAGER encompasses both state-of-the-art tools for each step as well as new complementary tools tailored for ancient DNA data within a single integrated solution in an easily accessible format.
Keywords: Authentication; Bioinformatics; Genome reconstruction; aDNA; aDNA analysis.
Figures
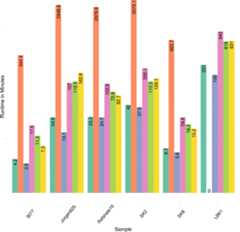

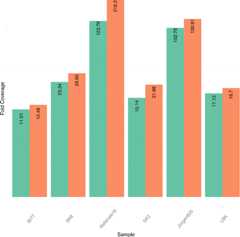
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
