ITGA6 is directly regulated by hypoxia-inducible factors and enriches for cancer stem cell activity and invasion in metastatic breast cancer models
- PMID:27001172
- PMCID: PMC4802728
- DOI: 10.1186/s12943-016-0510-x
ITGA6 is directly regulated by hypoxia-inducible factors and enriches for cancer stem cell activity and invasion in metastatic breast cancer models
Abstract
Background: Hypoxia-inducible factors (HIFs) are well-established mediators of tumor growth, the epithelial to mesenchymal transition (EMT) and metastasis. In several types of solid tumors, including breast cancers, the HIFs play a critical role in maintaining cancer stem cell (CSC) activity. Thus, we hypothesized that HIFs may also regulate transcription of markers of breast CSC activity. One approach to enrich for breast cells with stem-like phenotypes is FACS sorting, in which sub-populations of live cells are gated based on the expression of cell surface antigens, including various integrin subunits. Integrin alpha 6 (ITGA6; CD49f) is routinely used in combination with other integrin subunits to enrich for breast stem cells by FACS. Integrins not only mediate interactions with the extracellular matrix (ECM), but also drive intracellular signaling events that communicate from the tumor microenvironment to inside of the tumor cell to alter phenotypes including migration and invasion.
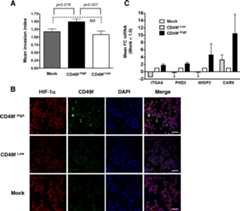
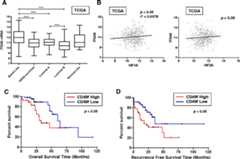
Methods: We used two models of metastatic breast cancer (MBC), polyoma middle T (MMTV-PyMT) and MDA-MB-231 cells, to compare the expression of ITGA6 in wild type and knockout (KO) or knockdown cells. Chromatin immunoprecipitation (ChIP) and luciferase reporter assays verified that ITGA6 is a direct HIF transcriptional target. We also used FACS sorting to enrich for CD49f (+) cells to compare tumorsphere formation, tumor initiating cell activity, invasion and HIF activity relative to CD49f(neg or low) cells. Knockdown of ITGA6 significantly reduced invasion, whereas re-expression of ITGA6 in the context of HIF knockdown partially rescued invasion. A search of public databases also revealed that ITGA6 expression is an independent prognostic factor of survival in breast cancer patients.
Results: We report that ITGA6 is a HIF-dependent target gene and that high ITGA6 expression enhances invasion and tumor-initiating cell activities in models of MBC. Moreover, cells that express high levels of ITGA6 are enriched for HIF-1α expression and the expression of HIF-dependent target genes.
Conclusions: Our data suggest that HIF-dependent regulation of ITGA6 is one mechanism by which sorting for CD49f (+) cells enhances CSC and metastatic phenotypes in breast cancers. Our results are particularly relevant to basal-like breast cancers which express higher levels of the HIFα subunits, core HIF-dependent target genes and ITGA6 relative to other molecular subtypes.
Keywords: Breast cancer; CD49f; Cancer stem cells (CSC); Hypoxia; Hypoxia-inducible factor (HIF); Invasion; Metastasis.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
