Massive migration from the steppe was a source for Indo-European languages in Europe
- PMID:25731166
- PMCID: PMC5048219
- DOI: 10.1038/nature14317
Massive migration from the steppe was a source for Indo-European languages in Europe
Abstract
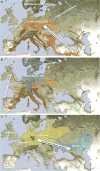
We generated genome-wide data from 69 Europeans who lived between 8,000-3,000 years ago by enriching ancient DNA libraries for a target set of almost 400,000 polymorphisms. Enrichment of these positions decreases the sequencing required for genome-wide ancient DNA analysis by a median of around 250-fold, allowing us to study an order of magnitude more individuals than previous studies and to obtain new insights about the past. We show that the populations of Western and Far Eastern Europe followed opposite trajectories between 8,000-5,000 years ago. At the beginning of the Neolithic period in Europe, ∼8,000-7,000 years ago, closely related groups of early farmers appeared in Germany, Hungary and Spain, different from indigenous hunter-gatherers, whereas Russia was inhabited by a distinctive population of hunter-gatherers with high affinity to a ∼24,000-year-old Siberian. By ∼6,000-5,000 years ago, farmers throughout much of Europe had more hunter-gatherer ancestry than their predecessors, but in Russia, the Yamnaya steppe herders of this time were descended not only from the preceding eastern European hunter-gatherers, but also from a population of Near Eastern ancestry. Western and Eastern Europe came into contact ∼4,500 years ago, as the Late Neolithic Corded Ware people from Germany traced ∼75% of their ancestry to the Yamnaya, documenting a massive migration into the heartland of Europe from its eastern periphery. This steppe ancestry persisted in all sampled central Europeans until at least ∼3,000 years ago, and is ubiquitous in present-day Europeans. These results provide support for a steppe origin of at least some of the Indo-European languages of Europe.
Figures






Comment in
- Human evolution: ancient DNA steps into the language debate.Novembre J.Novembre J.Nature. 2015 Jun 11;522(7555):164-5. doi: 10.1038/522164a.Nature. 2015.PMID:26062506No abstract available.
References
- Keller A, et al. New insights into the Tyrolean Iceman's origin and phenotype as inferred by whole-genome sequencing. Nat Commun. 2012;3:698. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
