Tracing the legacy of the early Hainan Islanders--a perspective from mitochondrial DNA
- PMID:21324107
- PMCID: PMC3048540
- DOI: 10.1186/1471-2148-11-46
Tracing the legacy of the early Hainan Islanders--a perspective from mitochondrial DNA
Abstract
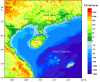
Background: Hainan Island is located around the conjunction of East Asia and Southeast Asia, and during the Last Glacial Maximum (LGM) was connected with the mainland. This provided an opportunity for the colonization of Hainan Island by modern human in the Upper Pleistocene. Whether the ancient dispersal left any footprints in the contemporary gene pool of Hainan islanders is debatable.
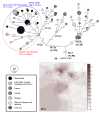
Results: We collected samples from 285 Li individuals and analyzed mitochondrial DNA (mtDNA) variations of hypervariable sequence I and II (HVS-I and II), as well as partial coding regions. By incorporating previously reported data, the phylogeny of Hainan islanders was reconstructed. We found that Hainan islanders showed a close relationship with the populations in mainland southern China, especially from Guangxi. Haplotype sharing analyses suggested that the recent gene flow from the mainland might play important roles in shaping the maternal pool of Hainan islanders. More importantly, haplogroups M12, M7e, and M7c1* might represent the genetic relics of the ancient population that populated this region; thus, 14 representative complete mtDNA genomes were further sequenced.
Conclusions: The detailed phylogeographic analyses of haplogroups M12, M7e, and M7c1* indicated that the early peopling of Hainan Island by modern human could be traced back to the early Holocene and/or even the late Upper Pleistocene, around 7-27 kya. These results correspond to both Y-chromosome and archaeological studies.
Figures




References
- Huang ZG, Zhang WQ, Chai FX, Xu QH. On the lowest sea level during the culmination of the lastest glacial period in South China. Acta Geogr Sin. 1995;50:385–393.
- Yao YT, Harff J, Meyer M, Zhan WH. Reconstruction of paleocoastlines for the northwestern South China Sea since the Last Glacial Maximum. Sci China Ser D-Earth Sci. 2009;52:1127–1136. doi: 10.1007/s11430-009-0098-8. - DOI
- Hao SD, Wang DX. Hainan archaeology: retrospect and prospect. Kaogu. 2003. pp. 291–299.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
