Nuclear expression of a group II intron is consistent with spliceosomal intron ancestry
- PMID:20351053
- PMCID: PMC2854396
- DOI: 10.1101/gad.1905010
Nuclear expression of a group II intron is consistent with spliceosomal intron ancestry
Abstract
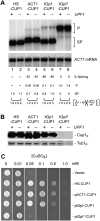
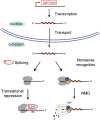
Group II introns are self-splicing RNAs found in eubacteria, archaea, and eukaryotic organelles. They are mechanistically similar to the metazoan nuclear spliceosomal introns; therefore, group II introns have been invoked as the progenitors of the eukaryotic pre-mRNA introns. However, the ability of group II introns to function outside of the bacteria-derived organelles is debatable, since they are not found in the nuclear genomes of eukaryotes. Here, we show that the Lactococcus lactis Ll.LtrB group II intron splices accurately and efficiently from different pre-mRNAs in a eukaryote, Saccharomyces cerevisiae. However, a pre-mRNA harboring a group II intron is spliced predominantly in the cytoplasm and is subject to nonsense-mediated mRNA decay (NMD), and the mature mRNA from which the group II intron is spliced is poorly translated. In contrast, a pre-mRNA bearing the Tetrahymena group I intron or the yeast spliceosomal ACT1 intron at the same location is not subject to NMD, and the mature mRNA is translated efficiently. Thus, a group II intron can splice from a nuclear transcript, but RNA instability and translation defects would have favored intron loss or evolution into protein-dependent spliceosomal introns, consistent with the bacterial group II intron ancestry hypothesis.
Figures

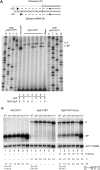
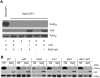

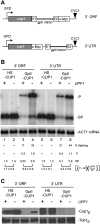
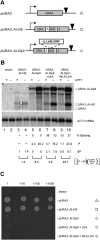
Similar articles
- RNA-RNA interactions and pre-mRNA mislocalization as drivers of group II intron loss from nuclear genomes.Qu G, Dong X, Piazza CL, Chalamcharla VR, Lutz S, Curcio MJ, Belfort M.Qu G, et al.Proc Natl Acad Sci U S A. 2014 May 6;111(18):6612-7. doi: 10.1073/pnas.1404276111. Epub 2014 Apr 10.Proc Natl Acad Sci U S A. 2014.PMID:24722636Free PMC article.
- Bacterial group II introns generate genetic diversity by circularization and trans-splicing from a population of intron-invaded mRNAs.LaRoche-Johnston F, Monat C, Coulombe S, Cousineau B.LaRoche-Johnston F, et al.PLoS Genet. 2018 Nov 21;14(11):e1007792. doi: 10.1371/journal.pgen.1007792. eCollection 2018 Nov.PLoS Genet. 2018.PMID:30462638Free PMC article.
- Recent horizontal transfer, functional adaptation and dissemination of a bacterial group II intron.LaRoche-Johnston F, Monat C, Cousineau B.LaRoche-Johnston F, et al.BMC Evol Biol. 2016 Oct 20;16(1):223. doi: 10.1186/s12862-016-0789-7.BMC Evol Biol. 2016.PMID:27765015Free PMC article.
- Organellar maturases: A window into the evolution of the spliceosome.Schmitz-Linneweber C, Lampe MK, Sultan LD, Ostersetzer-Biran O.Schmitz-Linneweber C, et al.Biochim Biophys Acta. 2015 Sep;1847(9):798-808. doi: 10.1016/j.bbabio.2015.01.009. Epub 2015 Jan 24.Biochim Biophys Acta. 2015.PMID:25626174Review.
- Intron specificity in pre-mRNA splicing.Mishra SK, Thakran P.Mishra SK, et al.Curr Genet. 2018 Aug;64(4):777-784. doi: 10.1007/s00294-017-0802-8. Epub 2018 Jan 3.Curr Genet. 2018.PMID:29299619Review.
Cited by
- Localization of a bacterial group II intron-encoded protein in human cells.Reinoso-Colacio M, García-Rodríguez FM, García-Cañadas M, Amador-Cubero S, García Pérez JL, Toro N.Reinoso-Colacio M, et al.Sci Rep. 2015 Aug 5;5:12716. doi: 10.1038/srep12716.Sci Rep. 2015.PMID:26244523Free PMC article.
- Categorizing 161 plant (streptophyte) mitochondrial group II introns into 29 families of related paralogues finds only limited links between intron mobility and intron-borne maturases.Zumkeller S, Knoop V.Zumkeller S, et al.BMC Ecol Evol. 2023 Mar 13;23(1):5. doi: 10.1186/s12862-023-02108-y.BMC Ecol Evol. 2023.PMID:36915058Free PMC article.
- The spliceosomal catalytic core arose in the RNA world… or did it?Doolittle WF.Doolittle WF.Genome Biol. 2013 Dec 13;14(12):141. doi: 10.1186/gb4145.Genome Biol. 2013.PMID:24330866Free PMC article.
- Evolution of introns in the archaeal world.Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP.Tocchini-Valentini GD, et al.Proc Natl Acad Sci U S A. 2011 Mar 22;108(12):4782-7. doi: 10.1073/pnas.1100862108. Epub 2011 Mar 7.Proc Natl Acad Sci U S A. 2011.PMID:21383132Free PMC article.
- The trouble with (group II) introns.Doolittle WF.Doolittle WF.Proc Natl Acad Sci U S A. 2014 May 6;111(18):6536-7. doi: 10.1073/pnas.1405174111. Epub 2014 Apr 22.Proc Natl Acad Sci U S A. 2014.PMID:24757059Free PMC article.No abstract available.
References
- Belfort M, Derbyshire V, Cousineau B, Lambowitz A 2002. Mobile introns: Pathways and proteins. In Mobile DNA II (ed. Craig N et al.), pp. 761–783 ASM Press, Washington, DC
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 - PubMed
- Brehm SL, Cech TR 1983. Fate of an intervening sequence ribonucleic acid: Excision and cyclization of the Tetrahymena ribosomal ribonucleic acid intervening sequence in vivo. Biochemistry 22: 2390–2397 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases