Functional interactions of alcohol-sensitive sites in the N-methyl-D-aspartate receptor M3 and M4 domains
- PMID:18208816
- PMCID: PMC2276391
- DOI: 10.1074/jbc.M705933200
Functional interactions of alcohol-sensitive sites in the N-methyl-D-aspartate receptor M3 and M4 domains
Abstract
The N-methyl-D-aspartate receptor is an important mediator of the behavioral effects of ethanol in the central nervous system. Previous studies have demonstrated sites in the third and fourth membrane-associated (M) domains of the N-methyl-D-aspartate receptor NR2A subunit that influence alcohol sensitivity and ion channel gating. We investigated whether two of these sites, Phe-637 in M3 and Met-823 in M4, interactively regulate the ethanol sensitivity of the receptor by testing dual substitution mutants at these positions. A majority of the mutations decreased steady-state glutamate EC(50) values and maximal steady-state to peak current ratios (I(ss)/I(p)), whereas only two mutations altered peak glutamate EC(50) values. Steady-state glutamate EC(50) values were correlated with maximal glutamate I(ss)/I(p) values, suggesting that changes in glutamate potency were attributable to changes in desensitization. In addition, there was a significant interaction between the substituents at positions 637 and 823 with respect to glutamate potency and desensitization. IC(50) values for ethanol among the mutants varied over the approximate range 100-325 mm. The sites in M3 and M4 significantly interacted in regulating ethanol sensitivity, although this was apparently dependent upon the presence of methionine in position 823. Molecular dynamics simulations of the NR2A subunit revealed possible binding sites for ethanol near both positions in the M domains. Consistent with this finding, the sum of the molecular volumes of the substituents at the two positions was not correlated with ethanol IC(50) values. Thus, there is a functional interaction between Phe-637 and Met-823 with respect to glutamate potency, desensitization, and ethanol sensitivity, but the two positions do not appear to form a unitary site of alcohol action.
Figures






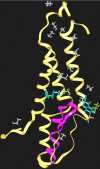
References
- Franks, N. P., and Lieb, W. R. (1994) Nature 367 607–614 - PubMed
- Peoples, R. W., Li, C., and Weight, F. F. (1996) Annu. Rev. Pharmacol. Toxicol. 36 185–201 - PubMed
- Lovinger, D. M., White, G., and Weight, F. F. (1989) Science 243 1721–1724 - PubMed
- Hoffman, P. L., Rabe, C. S., Moses, F., and Tabakoff, B. (1989) J. Neurochem. 52 1937–1940 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
