22474-20028.40 COMPOSITIONS AND METHODS FOR MULTIPLEXED ACTIVATION AND REPRESSION OF T CELL GENE EXPRESSION Cross-Reference to Related Applications [0001] This application claims priority from U.S. provisional application No.63/530,049 filed July 31, 2023, U.S. provisional application No.63/581,952 filed September 11, 2023, and U.S. provisional application No.63/570,751 filed March 27, 2024, the contents of which are incorporated by reference in their entireties. Incorporation by Reference of Sequence Listing [0002] The present application is being filed along with a Sequence Listing in electronic format. The Sequence Listing is provided as a file entitled 22474-2002840_SeqList.xml, created July 252024, which is 954,065 bytes in size. The information in the electronic format of the Sequence Listing is herein incorporated by reference in its entirety. Field [0003] The present disclosure relates in some aspects to epigenetic-modifying DNA- targeting systems, such as CRISPR-Cas/guide RNA (gRNA) systems, that bind to or target multiple genes or regulatory elements thereof, for instance in a T cell. In some aspects, the provided epigenetic-modifying DNA-targeting systems promote increased transcription of one or more first genes (also referred to herein as activation genes) and promote repressed transcription of one or more second genes (also referred to herein as repression genes). In some aspects, the provided epigenetic-modifying DNA-targeting systems modulate a T cell function, such as a T cell phenotype or activity. The present disclosure also relates in some aspects to polynucleotides, vectors, cells, methods, and uses related to the provided epigenetic-modifying DNA-targeting systems, for example in modulating T cells, such as in connection with methods of adoptive T cell therapy. Background [0004] The administration of T cells targeting a specific antigen, also known as Adoptive Cell Therapy (ACT), is a promising approach for treating diseases such as cancer. However, sf-6059407 22474-20028.40 current ACT treatments face challenges, including suboptimal T cell function, expansion, and persistence. Therefore, there is a need for new and improved methods to overcome these challenges. The present disclosure addresses these and other needs. Summary [0005] Provided herein in some embodiments is an epigenetic-modifying DNA-targeting system comprising a plurality of DNA-targeting modules comprising at least one activator DNA-targeting module and at least one repressor DNA-targeting module, wherein: the at least one activator DNA-targeting module is for increasing transcription of one or more activation genes in a T cell, and each activator DNA-targeting module comprises a fusion protein comprising (i) a first DNA-binding domain for targeting to a target site of one of the one or more activation genes and (ii) at least one transcriptional activator domain; and the at least one repressor DNA-targeting module is for repressing transcription of one or more repression genes in the T cell, and each repressor DNA-targeting module comprises a fusion protein comprising (i) a second DNA-binding domain for targeting to a target site of one of the one or more repression genes and (ii) at least one transcriptional repressor domain; wherein the first DNA- binding domain or domains are all different from the second DNA-binding domain or domains. [0006] In some of any embodiments, the one or more activation genes are selected from the group consisting of BATF, CD28, EOMES, IL-2, IL-2RB, IRF4, LAT, LCP2, TBX21, and VAV1. In some of any embodiments, the one or more repression genes are selected from the group consisting of CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and RASA2. [0007] Also provided herein in some embodiments is an epigenetic-modifying DNA- targeting system comprising a plurality of DNA-targeting modules comprising at least one activator DNA-targeting module and at least one repressor DNA-targeting module, wherein: the at least one activator DNA-targeting module is for increasing transcription of one or more activation genes, and each activator DNA-targeting module comprises a fusion protein comprising (i) a first DNA-binding domain for targeting to a target site of one of the one or more activation genes and (ii) at least one transcriptional activator domain, wherein the one or more activation genes are selected from the group consisting of BATF, CD28, EOMES, IL-2, IL-2RB, IRF4, LAT, LCP2, TBX21, and VAV1; and the at least one repressor DNA-targeting module is for repressing transcription of one or more repression genes, and each repressor DNA- sf-6059407 22474-20028.40 targeting module comprises a fusion protein comprising (i) a second DNA-binding domain for targeting to a target site of one of the one or more repression genes and (ii) at least one transcriptional repressor domain, wherein the one or more repression genes are selected from the group consisting of CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and RASA2; wherein the first DNA-binding domain or domains are all different from the second DNA-binding domain or domains. [0008] In some of any embodiments, the epigenetic-modifying DNA-targeting system does not introduce a genetic disruption or a DNA break. [0009] In some of any embodiments, at least one, optionally each first DNA-binding domain is a Clustered Regularly Interspaced Short Palindromic Repeats associated (Cas) protein; a zinc finger protein (ZFP); a transcription activator-like effector (TALE); a meganuclease; a homing endonuclease; or an I-SceI enzyme. In some of any embodiments, at least one, optionally each first DNA-binding domain is catalytically inactive. [0010] In some of any embodiments, at least one, optionally each second DNA-binding domain is a Cas protein; a ZFP; a TALE; a meganuclease; a homing endonuclease; or an I-SceI enzyme. In some of any embodiments, at least one, optionally each second DNA-binding domain is catalytically inactive. [0011] In some of any embodiments, at least one, optionally each first DNA-binding domain is a first Cas protein, and at least one, optionally each of the at least one activator DNA-targeting module further comprises a gRNA for targeting the first Cas protein to a target site of one of the one or more activation genes. In some of any embodiments, each first DNA-binding domain is a first Cas protein, and each of the at least one activator DNA-targeting module further comprises a gRNA for targeting the first Cas protein to a target site of one of the one or more activation genes. [0012] In some of any embodiments, at least one, optionally each second DNA-binding domain is a ZFP. [0013] In some of any embodiments, at least one, optionally each second DNA-binding domain is a second Cas protein, and at least one, optionally each of the at least one repressor DNA-targeting module further comprises a gRNA for targeting the second Cas protein to a target site of one of the one or more repression genes. In some of any embodiments, each second DNA-binding domain is a second Cas protein, and each of the at least one repressor DNA- sf-6059407 22474-20028.40 targeting module further comprises a gRNA for targeting the second Cas protein to a target site of one of the one or more repression genes. [0014] In some of any embodiments, at least one, optionally each first DNA-binding domain is a ZFP. [0015] In some of any embodiments, at least one, optionally each first DNA-binding domain is a ZFP; and at least one, optionally each second DNA-binding domain is a Cas protein, and at least one, optionally each of the at least one repressor DNA-targeting module further comprises a gRNA for targeting the second Cas protein to a target site of one of the one or more repression genes. [0016] In some of any embodiments: at least one, optionally each first DNA-binding domain is a first Cas protein, and at least one, optionally each of the at least one activator DNA-targeting module further comprises a gRNA for targeting the first Cas protein to a target site of one of the one or more activation genes; and at least one, optionally each second DNA-binding domain is a second Cas protein, and at least one, optionally each of the at least one repressor DNA-targeting module further comprises a gRNA for targeting the second Cas protein to a target site of one of the one or more repression genes. [0017] In some of any embodiments: each first DNA-binding domain is a first Cas protein, and each of the at least one activator DNA-targeting module further comprises a gRNA for targeting the first Cas protein to a target site of one of the one or more activation genes; and each second DNA-binding domain is a second Cas protein, and each of the at least one repressor DNA-targeting module further comprises a gRNA for targeting the second Cas protein to a target site of one of the one or more repression genes. [0018] In some of any embodiments, the first Cas protein or proteins are from a different Staphylococcus species than the second Cas protein or proteins. [0019] Also provided herein in some embodiments is an epigenetic-modifying DNA- targeting system comprising a plurality of DNA-targeting modules comprising at least one activator DNA-targeting module and at least one repressor DNA-targeting module, wherein: the at least one activator DNA-targeting module is for increasing transcription of one or more activation genes, and each activator DNA-targeting module comprises (1) a fusion protein comprising (i) a first Cas protein and (ii) at least one transcriptional activator domain, and (2) a gRNA for targeting the first Cas protein to a target site of one of the one or more activation genes; wherein the one or more activation genes are selected from the group consisting of sf-6059407 22474-20028.40 BATF, CD28, EOMES, IL-2, IL-2RB, IRF4, LAT, LCP2, TBX21, and VAV1; and the at least one repressor DNA-targeting module is for repressing transcription of one or more repression genes, and each repressor DNA-targeting module comprises (1) a fusion protein comprising (i) a second Cas protein and (ii) at least one transcriptional repressor domain, and (2) a gRNA for targeting the second Cas protein to a target site of one of the one or more repression genes; wherein the one or more repression genes are selected from the group consisting of CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and RASA2; wherein the first Cas protein or proteins are from a different Staphylococcus species than the second Cas protein or proteins. [0020] Also provided herein in some embodiments is an epigenetic-modifying DNA- targeting system comprising a plurality of DNA-targeting modules comprising at least one activator DNA-targeting module and at least one repressor DNA-targeting module, wherein: the at least one activator DNA-targeting module is for increasing transcription of IL-2, and each activator DNA-targeting module comprises (1) a fusion protein comprising (i) an dSaCas protein and (ii) at least one transcriptional activator domain, and (2) a gRNA for targeting the dSaCas protein to a target site of IL-2; and the at least one repressor DNA-targeting module is for repressing transcription of one or more repression genes, and each repressor DNA-targeting module comprises (1) a fusion protein comprising (i) an dSpCas9 protein and (ii) at least one transcriptional repressor domain, and (2) a gRNA for targeting the dSpCas9 protein to a target site of one of the one or more repression genes; wherein the one or more repression genes are selected from the group consisting of CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and RASA2. [0021] In some of any embodiments, the first Cas protein or proteins do not bind to the protospacer-adjacent motifs (PAMs) that the second Cas protein or proteins bind to. [0022] Also provided herein in some embodiments is an epigenetic-modifying DNA- targeting system comprising a plurality of DNA-targeting modules comprising at least one activator DNA-targeting module and at least one repressor DNA-targeting module, wherein: the at least one activator DNA-targeting module is for increasing transcription of one or more activation genes, and each activator DNA-targeting module comprises (1) a fusion protein comprising (i) a first Cas protein and (ii) at least one transcriptional activator domain, and (2) a gRNA for targeting the first Cas protein to a target site of one of the one or more activation genes; wherein the one or more activation genes are selected from the group consisting of sf-6059407 22474-20028.40 BATF, CD28, EOMES, IL-2, IL-2RB, IRF4, LAT, LCP2, TBX21, and VAV1; and the at least one repressor DNA-targeting module is for repressing transcription of one or more repressor genes, and each repressor DNA-targeting module comprises (1) a fusion protein comprising (i) a second Cas protein and (ii) at least one transcriptional repressor domain, and (2) a gRNA for targeting the second Cas protein to a target site of one of the one or more repression genes; wherein the one or more repression genes are selected from the group consisting of CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and RASA2; wherein the first Cas protein or proteins do not bind to the protospacer- adjacent motifs (PAMs) that the second Cas protein or proteins bind to. [0023] In some of any embodiments, the first Cas protein or proteins are from a different Staphylococcus species than the second Cas protein or proteins. [0024] In some of any embodiments, at least one, optionally each of the first Cas protein or proteins bind to a first PAM 5’-NGG-3’ (SEQ ID NO: 224), wherein N is any nucleotide. In some of any embodiments, at least one, optionally each of the second Cas protein or proteins bind to a second PAM 5’- NNGRRT-3’ (SEQ ID NO: 225), wherein N is any nucleotide, and R is G or A. [0025] In some of any embodiments, the first Cas protein or proteins bind to a first PAM 5’- NGG-3’ (SEQ ID NO: 224), wherein N is any nucleotide. In some of any embodiments, the second Cas protein or proteins bind to a second PAM 5’- NNGRRT-3’ (SEQ ID NO: 225), wherein N is any nucleotide, and R is G or A. [0026] In some of any embodiments, at least one, optionally each of the first Cas protein or proteins bind to a first PAM 5’- NNGRRT-3’ (SEQ ID NO: 225), wherein N is any nucleotide, and R is G or A. In some of any embodiments, at least one, optionally each of the second Cas protein or proteins bind to a second PAM 5’-NGG-3’ (SEQ ID NO: 224), wherein N is any nucleotide. [0027] In some of any embodiments, the first Cas protein or proteins bind to a first PAM 5’- NNGRRT-3’ (SEQ ID NO: 225), wherein N is any nucleotide, and R is G or A. In some of any embodiments, the second Cas protein or proteins bind to a second PAM 5’-NGG-3’ (SEQ ID NO: 224), wherein N is any nucleotide. [0028] In some of any embodiments, the first Cas protein or proteins do not bind to the gRNA or gRNAs that target the one or more repression genes, and the second Cas protein or proteins do not bind to the gRNA or gRNAs that target the one or more activation genes. sf-6059407 22474-20028.40 [0029] In some of any embodiments, at least one, optionally each first Cas protein is a deactivated (dCas) protein. In some of any embodiments, at least one, optionally each first Cas protein lacks nuclease activity. In some of any embodiments, at least one, optionally each first Cas protein is a dCas9 protein. [0030] In some of any embodiments, at least one, optionally each first Cas protein is a Staphylococcus aureus dCas9 (dSaCas9) protein. In some of any embodiments, at least one, optionally each first Cas protein is a dSaCas9 protein comprising at least one amino acid mutation selected from D10A and N580A with reference to numbering of positions of SEQ ID NO:124. In some of any embodiments, at least one, optionally each first Cas protein comprises the sequence set forth in SEQ ID NO:125 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, at least one, optionally each first Cas protein is set forth in SEQ ID NO:125. [0031] In some of any embodiments, at least one, optionally each first Cas protein is a Streptococcus pyogenes dCas9 (dSpCas9) protein. In some of any embodiments, at least one, optionally each first Cas protein is a dSpCas9 protein comprising at least one amino acid mutation selected from D10A and H840A with reference to numbering of positions of SEQ ID NO:126. In some of any embodiments, at least one, optionally each first Cas protein comprises the sequence set forth in SEQ ID NO:127 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, at least one, optionally each first Cas protein is set forth in SEQ ID NO:127. [0032] In some of any embodiments, at least one, optionally each first Cas protein is a dCas12 protein. [0033] In some of any embodiments, the at least one activator DNA-targeting module is a single activator DNA-targeting module. In some of any embodiments, the at least one activator DNA-targeting module is a plurality of activator DNA-targeting modules. [0034] In some of any embodiments, at least one of the first DNA-binding domains is a ZFP that targets a target site of one of the one or more activation genes. In some of any embodiments, at least one of the second DNA-binding domains is a ZFP that targets a target site of one of the one or more repression genes. [0035] In some of any embodiments: the first DNA-binding domains of the at least one activator DNA-targeting module are different from one another. sf-6059407 22474-20028.40 [0036] In some of any embodiments, at least one, optionally each activator DNA-targeting module of the at least one activator DNA-targeting module is different. [0037] In some of any embodiments, the at least one activator DNA-targeting module is 2, 3, 4, 5, or 6 activator DNA-targeting modules. In some of any embodiments, the at least one activator DNA-targeting module is 2 activator DNA-targeting modules. [0038] In some of any embodiments, the one or more activation genes comprise IL-2. In some of any embodiments, the one or more activation genes is IL-2. [0039] In some of any embodiments, the at least one activator DNA-targeting module targets two or more activation genes. In some of any embodiments, the one or more activation genes comprise BATF and IL-2; BATF and VAV1; CD28 and BATF; CD28 and EOMES; CD28 and IL-2; CD28 and LCP2; CD28 and TBX21; CD28 and VAV1; EOMES and BATF; EOMES and LCP2; EOMES and TBX21; EOMES and VAV1; EOMES and IL-2; LCP2 and BATF; LCP2 and IL-2; LCP2 and TBX21; LCP2 and VAV1; TBX21 and BATF; TBX21 and IL-2; TBX21 and TBX21; TBX21 and VAV1; or VAV1 and IL-2. In some of any embodiments, the one or more activation genes comprise IL-2RB and VAV1. In some of any embodiments, the one or more activation genes comprise IL-2 and VAV1. [0040] In some of any embodiments, the target site for at least one, optionally each of the one or more activation genes is in a gene or a regulatory DNA element. In some of any embodiments, the target site for at least one, optionally each of the one or more activation genes is in a gene, an enhancer, or a promoter. In some of any embodiments, the target site for at least one, optionally each of the one or more activation genes is in an enhancer or a promoter. [0041] In some of any embodiments, the target site for at least one, optionally each of the one or more activation genes is within 1000 base pairs of a transcriptional start site of one of the one or more activation genes. In some of any embodiments, the target site for at least one, optionally each of the one or more activation genes is within 500 base pairs of a transcriptional start site of one of the one or more activation genes. [0042] In some of any embodiments, the target site for at least one of the one or more activation genes is selected from: (a) a target site for VAV1 having the sequence set forth in any one of SEQ ID NOS:7-9, 156, and 170, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (b) a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:78 and 388-403, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (c) a target site sf-6059407 22474-20028.40 for BATF having the sequence set forth in any one of SEQ ID NOS:172-174, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (d) a target site for CD28 having the sequence set forth in any one of SEQ ID NOS:144-146 and 189-191, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (e) a target site for EOMES having the sequence set forth in any one of SEQ ID NOS:147-149, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (f) a target site for IRF4 having the sequence set forth in any one of SEQ ID NOS:175-177, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (g) a target site for LAT having the sequence set forth in any one of SEQ ID NOS:184-186, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (h) a target site for LCP2 having the sequence set forth in any one of SEQ ID NOS:150-152 and 187-188, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; and (i) a target site for TBX21 having the sequence set forth in any one of SEQ ID NOS:153-155, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. [0043] In some of any embodiments, the target site for at least one of the one or more activation genes is selected from: (a) a target site for VAV1 having the sequence set forth in any one of SEQ ID NOS:7-9, 156, and 170, or a complementary sequence of any of the foregoing; (b) a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:78 and 388-403 or a complementary sequence of any of the foregoing; (c) a target site for BATF having the sequence set forth in any one of SEQ ID NOS:172-174, or a complementary sequence of any of the foregoing; (d) a target site for CD28 having the sequence set forth in any one of SEQ ID NOS:144-146 and 189-191, or a complementary sequence of any of the foregoing; (e) a target site for EOMES having the sequence set forth in any one of SEQ ID NOS:147-149, or a complementary sequence of any of the foregoing; (f) a target site for IRF4 having the sequence set forth in any one of SEQ ID NOS:175-177, or a complementary sequence of any of the foregoing; (g) a target site for LAT having the sequence set forth in any one of SEQ ID NOS:184-186, or a complementary sequence of any of the foregoing; (h) a target site for LCP2 having the sequence set forth in any one of SEQ ID NOS:150-152 and 187-188, or a complementary sequence of any of the foregoing; or (i) a target site for TBX21 having the sf-6059407 22474-20028.40 sequence set forth in any one of SEQ ID NOS:153-155, or a complementary sequence of any of the foregoing. [0044] In some of any embodiments, the target site for each of the one or more activation genes is selected from: (a) a target site for VAV1 having the sequence set forth in any one of SEQ ID NOS:7-9, 156, and 170, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (b) a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:78 and 388-403, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (c) a target site for BATF having the sequence set forth in any one of SEQ ID NOS:172-174, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (d) a target site for CD28 having the sequence set forth in any one of SEQ ID NOS:144-146 and 189-191, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (e) a target site for EOMES having the sequence set forth in any one of SEQ ID NOS:147-149, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (f) a target site for IRF4 having the sequence set forth in any one of SEQ ID NOS:175-177, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (g) a target site for LAT having the sequence set forth in any one of SEQ ID NOS:184-186, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (h) a target site for LCP2 having the sequence set forth in any one of SEQ ID NOS:150-152 and 187-188, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; or (i) a target site for TBX21 having the sequence set forth in any one of SEQ ID NOS:153-155, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. [0045] In some of any embodiments, the target site for each of the one or more activation genes is selected from: (a) a target site for VAV1 having the sequence set forth in any one of SEQ ID NOS:7-9, 156, and 170, or a complementary sequence of any of the foregoing; (b) a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:78 and 388-403 or a complementary sequence of any of the foregoing; (c) a target site for BATF having the sequence set forth in any one of SEQ ID NOS:172-174, or a complementary sequence of any of the foregoing; (d) a target site for CD28 having the sequence set forth in any one of SEQ ID NOS:144-146 and 189-191, or a complementary sequence of any of the foregoing; (e) a target sf-6059407 22474-20028.40 site for EOMES having the sequence set forth in any one of SEQ ID NOS:147-149, or a complementary sequence of any of the foregoing; (f) a target site for IRF4 having the sequence set forth in any one of SEQ ID NOS:175-177, or a complementary sequence of any of the foregoing; (g) a target site for LAT having the sequence set forth in any one of SEQ ID NOS:184-186, or a complementary sequence of any of the foregoing; (h) a target site for LCP2 having the sequence set forth in any one of SEQ ID NOS:150-152 and 187-188, or a complementary sequence of any of the foregoing; or (i) a target site for TBX21 having the sequence set forth in any one of SEQ ID NOS:153-155, or a complementary sequence of any of the foregoing. [0046] In some of any embodiments, the target site for at least one of the one or more activation genes is a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:78 and 388-403, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. In some of any embodiments, the target site for at least one of the one or more activation genes is a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:78 and 388-403 or a complementary sequence of any of the foregoing. In some of any embodiments, the target site for each of the one or more activation genes is a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:78 and 388-403, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. In some of any embodiments, the target site for each of the one or more activation genes is a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:78 and 388-403 or a complementary sequence of any of the foregoing. [0047] In some of any embodiments, the target site for at least one of the one or more activation genes is a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:404-412, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. In some of any embodiments, the target site for at least one of the one or more activation genes is a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:404-412 or a complementary sequence of any of the foregoing. In some of any embodiments, the target site for each of the one or more activation genes is a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:404-412, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. In some of any embodiments, the target site for each of the one or more activation genes is a target site for sf-6059407 22474-20028.40 IL-2 having the sequence set forth in any one of SEQ ID NO:404-412 or a complementary sequence of any of the foregoing. [0048] In some of any embodiments, the target site for at least one of the one or more activation genes is any one of the sequences set forth in any one of SEQ ID NO:451-453, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. In some of any embodiments, the target site for at least one of the one or more activation genes is any one of the sequences set forth in any one of SEQ ID NO:451-453 or a complementary sequence of any of the foregoing. In some of any embodiments, the target site for each of the one or more activation genes is any one of the sequences set forth in any one of SEQ ID NO:451-453, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. In some of any embodiments, the target site for each of the one or more activation genes is any one of the sequences set forth in any one of SEQ ID NO:451-453 or a complementary sequence of any of the foregoing. [0049] In some of any embodiments, the first DNA-binding domain is a ZFP that targets the target site of one or more activation genes set forth in SEQ ID NO: 451. In some of any embodiments, the ZFP comprises a zinc finger recognition region comprising six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, selected from F1-F6 as follows: F1: QNAHRKT (SEQ ID NO: 472), F2: RKYYLAK (SEQ ID NO: 473), F3: RSAHLSR (SEQ ID NO: 474), F4: QSGDLTR (SEQ ID NO: 475), F5: RSDHLTQ (SEQ ID NO: 476), and F6: DSANLSR (SEQ ID NO: 477). In some of any embodiments, the ZFP comprises the sequence set forth in SEQ ID NO: 458, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, the ZFP comprises the sequence set forth in SEQ ID NO: 458. In some of any embodiments, the ZFP is encoded by the sequence set forth in SEQ ID NO: 465 or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, the ZFP is encoded by the sequence set forth in SEQ ID NO: 465. [0050] In some of any embodiments, the first DNA-binding domain is a ZFP that targets the target site of one or more activation genes set forth in SEQ ID NO: 452. In some of any embodiments, the ZFP comprises a zinc finger recognition region comprising six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, selected from F1-F6 as follows: F1: DSSHLEL (SEQ ID NO: 478), F2: DRSNLTR (SEQ ID NO: 479), F3: RSDNLSE (SEQ ID sf-6059407 22474-20028.40 NO: 480), F4: VRRALSS (SEQ ID NO: 481), F5: QSGALAR (SEQ ID NO: 482), and F6: RLDWLPM (SEQ ID NO: 483). In some of any embodiments, the ZFP comprises the sequence set forth in SEQ ID NO: 459, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, the ZFP comprises the sequence set forth in SEQ ID NO: 459. In some of any embodiments, the ZFP is encoded by the sequence set forth in SEQ ID NO: 466 or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, the ZFP is encoded by the sequence set forth in SEQ ID NO: 466. In some of any embodiments, the first DNA-binding domain is a ZFP that targets the target site of one or more activation genes set forth in SEQ ID NO: 453. In some of any embodiments, the ZFP comprises a zinc finger recognition region comprising six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, selected from F1-F6 as follows: F1: RSDNLSV (SEQ ID NO: 484), F2: RSAHLSR (SEQ ID NO: 485), F3: QNAHRKT (SEQ ID NO: 486), F4: LRHHLTR (SEQ ID NO: 487), F5: TSSNRKT (SEQ ID NO: 488), and F6: TSSNLSR (SEQ ID NO: 489). In some of any embodiments, the ZFP comprises the sequence set forth in SEQ ID NO: 460, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, the ZFP comprises the sequence set forth in SEQ ID NO: 460. In some of any embodiments, the ZFP is encoded by the sequence set forth in SEQ ID NO: 467 or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, the ZFP is encoded by the sequence set forth in SEQ ID NO: 467. [0051] In some of any embodiments, at least one, optionally each gRNA for targeting a target site of one of the one or more activation genes comprises a gRNA spacer sequence that is complementary to a target site of one of the one or more activation genes. In some of any embodiments, at least one, optionally each gRNA for targeting a target site of one of the one or more activation genes comprises a gRNA spacer sequence between 14 nt and 24 nt or between 16 nt and 22 nt in length. In some of any embodiments, at least one, optionally each gRNA for targeting a target site of one of the one or more activation genes comprises a gRNA spacer sequence that is 18 nt, 19 nt, 20 nt, 21 nt, or 22 nt in length. sf-6059407 22474-20028.40 [0052] In some of any embodiments, at least one, optionally each gRNA for targeting a target site of one of the one or more activation genes further comprises a gRNA scaffold sequence set forth in SEQ ID NO:69. In some of any embodiments, at least one, optionally each gRNA for targeting a target site of one or the one or more activation genes further comprises a gRNA scaffold sequence set forth in SEQ ID NO:387. [0053] In some of any embodiments, at least one of the gRNA for targeting a target site of one or the one or more activation genes is selected from: (a) a gRNA targeting a target site for VAV1 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:41-43, 169, and 171, or a contiguous portion thereof of at least 14 nt; (b) a gRNA targeting a target site for IL-2 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NO:79 and 413-428, or a contiguous portion thereof of at least 14 nt; (c) a gRNA targeting a target site for BATF and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:178-180, or a contiguous portion thereof of at least 14 nt; (d) a gRNA targeting a target site for CD28 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:157-159 and 197- 199, or a contiguous portion thereof of at least 14 nt; (e) a gRNA targeting a target site for EOMES and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:160-162, or a contiguous portion thereof of at least 14 nt; (f) a gRNA targeting a target site for IRF4 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:181-183, or a contiguous portion thereof of at least 14 nt; (g) a gRNA targeting a target site for LAT and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:192-194, or a contiguous portion thereof of at least 14 nt; (h) a gRNA targeting a target site for LCP2 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:163-165 and 195-196, or a contiguous portion thereof of at least 14 nt; or (i) a gRNA targeting a target site for TBX21 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:166-168, or a contiguous portion thereof of at least 14 nt. [0054] In some of any embodiments, at least one of the gRNA for targeting a target site of one or the one or more activation genes is selected from: (a) a gRNA targeting a target site for VAV1 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:41-43, 169, and 171; (b) a gRNA targeting a target site for IL-2 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NO:79 and 413-428; (c) a gRNA targeting a target site sf-6059407 22474-20028.40 for BATF and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:178- 180; (d) a gRNA targeting a target site for CD28 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:157-159 and 197-199; (e) a gRNA targeting a target site for EOMES and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:160-162; (f) a gRNA targeting a target site for IRF4 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:181-183; (g) a gRNA targeting a target site for LAT and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:192-194; (h) a gRNA targeting a target site for LCP2 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:163-165 and 195-196; or (i) a gRNA targeting a target site for TBX21 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:166-168. [0055] In some of any embodiments, each gRNA for targeting a target site of one or the one or more activation genes is selected from: (a) a gRNA targeting a target site for VAV1 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:41-43, 169, and 171, or a contiguous portion thereof of at least 14 nt; (b) a gRNA targeting a target site for IL-2 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NO:79 and 413-428, or a contiguous portion thereof of at least 14 nt; (c) a gRNA targeting a target site for BATF and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:178-180, or a contiguous portion thereof of at least 14 nt; (d) a gRNA targeting a target site for CD28 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:157-159 and 197-199, or a contiguous portion thereof of at least 14 nt; (e) a gRNA targeting a target site for EOMES and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:160-162, or a contiguous portion thereof of at least 14 nt; (f) a gRNA targeting a target site for IRF4 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:181-183, or a contiguous portion thereof of at least 14 nt; (g) a gRNA targeting a target site for LAT and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:192-194, or a contiguous portion thereof of at least 14 nt; (h) a gRNA targeting a target site for LCP2 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:163-165 and 195-196, or a contiguous portion thereof of at least 14 nt; or (i) a gRNA targeting a target site for TBX21 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:166-168, or a contiguous portion thereof of at least 14 nt. sf-6059407 22474-20028.40 [0056] In some of any embodiments, each gRNA for targeting a target site of one or the one or more activation genes is selected from: (a) a gRNA targeting a target site for VAV1 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:41-43, 169, and 171; (b) a gRNA targeting a target site for IL-2 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NO:79 and 413-428; (c) a gRNA targeting a target site for BATF and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:178-180; (d) a gRNA targeting a target site for CD28 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:157-159 and 197-199; (e) a gRNA targeting a target site for EOMES and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:160-162; (f) a gRNA targeting a target site for IRF4 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:181-183; (g) a gRNA targeting a target site for LAT and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:192-194; (h) a gRNA targeting a target site for LCP2 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:163- 165 and 195-196; or (i) a gRNA targeting a target site for TBX21 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:166-168. [0057] In some of any embodiments, at least one of the gRNA for targeting a target site of one or the one or more activation genes is a gRNA targeting a target site for IL-2 and comprises a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NO:79 and 413-428, or a contiguous portion thereof of at least 14 nt. In some of any embodiments, at least one of the gRNA for targeting a target site of one or the one or more activation genes is a gRNA targeting a target site for IL-2 and comprises a gRNA spacer sequence set forth in any one of SEQ ID NO:79 and 413-428. In some of any embodiments, each gRNA for targeting a target site of one or the one or more activation genes is a gRNA targeting a target site for IL-2 and comprises a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NO:79 and 413-428, or a contiguous portion thereof of at least 14 nt. In some of any embodiments, each gRNA for targeting a target site of one or the one or more activation genes is a gRNA targeting a target site for IL-2 and comprises a gRNA spacer sequence set forth in any one of SEQ ID NO:79 and 413-428. [0058] In some of any embodiments, at least one of the gRNA for targeting a target site of one or the one or more activation genes is a gRNA targeting a target site for IL-2 and comprises a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NO:429-437, or a contiguous portion thereof of at least 14 nt. In some of any embodiments, at least one of the sf-6059407 22474-20028.40 gRNA for targeting a target site of one or the one or more activation genes is a gRNA targeting a target site for IL-2 and comprises a gRNA spacer sequence set forth in any one of SEQ ID NO:429-437. In some of any embodiments, each gRNA for targeting a target site of one or the one or more activation genes is a gRNA targeting a target site for IL-2 and comprises a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NO:429-437, or a contiguous portion thereof of at least 14 nt. In some of any embodiments, each gRNA targeting a target site of one or the one or more activation genes is a gRNA targeting a target site for IL-2 and comprises a gRNA spacer sequence set forth in any one of SEQ ID NO:429-437. [0059] In some of any embodiments, at least one, optionally each transcriptional activator domain epigenetically modifies a target site of one of the one or more activation genes to promote increased transcription of one of the one or more activation genes. [0060] In some of any embodiments, at least one, optionally each transcriptional activator domain is a VP64 domain, a p65 activation domain, a p300 domain, an Rta domain, a CBP domain, a VPR domain, a VPH domain, an HSF1 domain, a TET protein domain, optionally wherein the TET protein is TET1, a SunTag domain, or a domain, portion, variant, or truncation of any of the foregoing. In some of any embodiments, at least one, optionally each transcriptional activator domain comprises at least one VP16 domain or a variant or portion thereof that exhibits transcriptional activation activity. In some of any embodiments, at least one, optionally each transcriptional activator domain comprises a VP16 tetramer (VP64) domain or a variant or portion thereof that exhibits transcriptional activation activity. In some of any embodiments, at least one, optionally each transcriptional activator domain is a VP64 domain. [0061] In some of any embodiments, at least one, optionally each transcriptional activator domain comprises a NCOA3 domain or a variant or portion thereof that exhibits transcriptional activation activity. In some of any embodiments, at least one, optionally each transcriptional activator domain comprises a FOXO3 domain or a variant or portion thereof that exhibits transcriptional activation activity. In some of any embodiments, at least one, optionally each transcriptional activator domain comprises a NCOA3-FOXO3-NCOA3 domain. In some of any embodiments, at least one, optionally each transcriptional activator domain comprises a NCOA3-FOXO3-NCOA3 domain. In some of any embodiments, at least one, optionally each transcriptional activator domain is a NCOA3-FOXO3-NCOA3 domain. In some of any embodiments, at least one, optionally each transcriptional activator domain comprises a VP16 tetramer (VP64) domain and a NCOA3-FOXO3-NCOA3 domain. In some of any embodiments, sf-6059407 22474-20028.40 at least one, optionally each transcriptional activator domain is a VP16 tetramer (VP64) domain and a NCOA3-FOXO3-NCOA3 domain. [0062] In some of any embodiments, at least one, optionally each transcriptional activator domain comprises the sequence set forth in SEQ ID NO:142, a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. [0063] In some of any embodiments, at least one, optionally each fusion protein of the at least one activator DNA-targeting module comprises the sequence set forth in SEQ ID NO:77 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. [0064] In some of any embodiments, at least one, optionally each fusion protein of the at least one activator DNA-targeting module comprises the sequence set forth in SEQ ID NO:386 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. [0065] In some of any embodiments, at least one, optionally each fusion protein of the at least one activator DNA-targeting module comprises the sequence set forth in SEQ ID NO:528 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, at least one, optionally each fusion protein of the at least one activator DNA-targeting module comprises the sequence set forth in SEQ ID NO:529 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, at least one, optionally each fusion protein of the at least one activator DNA-targeting module comprises the sequence set forth in SEQ ID NO:514 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, at least one, optionally each fusion protein of the at least one activator DNA-targeting module comprises the sequence set forth in SEQ ID NO:515 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, at least one, optionally each fusion protein of the at least one activator DNA-targeting module comprises the sequence set forth in SEQ ID NO:516 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. sf-6059407 22474-20028.40 [0066] In some of any embodiments, at least one, optionally each second Cas protein is a deactivated (dCas) protein. In some of any embodiments, at least one, optionally each second Cas protein lacks nuclease activity. In some of any embodiments, at least one, optionally each second Cas protein is a dCas9 protein. [0067] In some of any embodiments, at least one, optionally each second Cas protein is a dSaCas9 protein. In some of any embodiments, at least one, optionally each second Cas protein is a dSaCas9 protein comprising at least one amino acid mutation selected from D10A and N580A with reference to numbering of positions of SEQ ID NO:124. In some of any embodiments, at least one, optionally each second Cas protein comprises the sequence set forth in SEQ ID NO:125 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, at least one, optionally each second Cas protein is set forth in SEQ ID NO:125. [0068] In some of any embodiments, at least one, optionally each second Cas protein is a dSpCas9 protein. In some of any embodiments, at least one, optionally each second Cas protein is a dSpCas9 protein comprising at least one amino acid mutation selected from D10A and H840A with reference to numbering of positions of SEQ ID NO:126. In some of any embodiments, at least one, optionally each second Cas protein comprises the sequence set forth in SEQ ID NO:127 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, at least one, optionally each second Cas protein is set forth in SEQ ID NO:127. [0069] In some of any embodiments, at least one, optionally each second Cas protein is a dCas12 protein. [0070] In some of any embodiments, the at least one repressor DNA-targeting module is a single repressor DNA-targeting module. In some of any embodiments, the at least one repressor DNA-targeting module is a plurality of repressor DNA-targeting modules. [0071] In some of any embodiments, the second DNA-binding domains of the at least one repressor DNA-targeting module are different from one another. [0072] In some of any embodiments, at least one, optionally each repressor DNA-targeting module of the at least one repressor DNA-targeting module is different. [0073] In some of any embodiments, the at least one repressor DNA-targeting module is 2, 3, 4, 5, or 6 repressor DNA-targeting modules. In some of any embodiments, the at least one repressor DNA-targeting module is 2 repressor DNA-targeting modules. sf-6059407 22474-20028.40 [0074] In some of any embodiments, the one or more repression genes comprise MED12. In some of any embodiments, the one or more repression genes is MED12. [0075] In some of any embodiments, the at least one repressor DNA-targeting modules targets two or more repression genes. In some of any embodiments, the one or more repression genes comprise CBLB and CCNC; CBLB and CD5; CBLB and CISH; CBLB and DGKZ; CBLB and ELOB; CBLB and FAS; CBLB and Fli1; CBLB and GATA3; CBLB and KDM1A; CBLB and MED12; CBLB and MYB; CBLB and PRDM1; CBLB and RASA2; CD5 and CISH; CD5 and MYB; CISH and DGKZ; CISH and MYB; CISH and RASA2; GATA3 and CD5; GATA3 and CISH; GATA3 and MYB; MED12 and CBLB; MED12 and CD5; MED12 and CISH; MED12 and DGKZ; MED12 and ELOB; MED12 and GATA3; MED12 and MYB; MED12 and PRDM1; MED12 and RASA2; MYB and RASA2; PRDM1 and CISH; PRDM1 and GATA3; PRDM1 and MYB; PRDM1 and RASA2; CD5, CISH, and MYB; GATA3, CBLB, and MYB; GATA3, CD5, and MYB; or PRDM1, GATA3, and CISH. In some of any embodiments, the two or more repression genes comprise CBLB and MYB. In some of any embodiments, the two or more repression genes comprise CBLB and MED12. In some of any embodiments, the two or more repression genes comprise CBLB and CCNC. In some eof any embodiments, the two or more repression genes comprise MED12 and TGFBR2. [0076] In some of any embodiments, the at least one repressor DNA-targeting modules targets three or more repression genes. In some embodiments, the three or more repression genes comprise MED12, TGFBR2 and CISH. [0077] In some of any embodiments, the target site for at least one, optionally each of the one or more repression genes is in a gene or a regulatory DNA element. In some of any embodiments, the target site for at least one, optionally each of the one or more repression genes is in a gene, an enhancer, or a promoter. In some of any embodiments, the target site for at least one, optionally each of the one or more repression genes is in an enhancer or a promoter. [0078] In some of any embodiments, the target site for at least one, optionally each of the one or more repression genes is within 1000 base pairs of a transcriptional start site of one of the one or more repression genes. In some of any embodiments, the target site for at least one, optionally each of the one or more repression genes is within 500 base pairs of a transcriptional start site of one of the one or more repression genes. [0079] In some of any embodiments, the target site for at least one of the one or more repression genes is selected from: (a) a target site for CD5 having the sequence set forth in any sf-6059407 22474-20028.40 one of SEQ ID NOS:1-3, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (b) a target site for KDM1A having the sequence set forth in any one of SEQ ID NOS:4-6, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (c) a target site for CBLB having the sequence set forth in any one of SEQ ID NOS:10-12, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (d) a target site for DGKZ having the sequence set forth in any one of SEQ ID NOS:13-15, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (e) a target site for MYB having the sequence set forth in any one of SEQ ID NOS:16-18, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (f) a target site for RASA2 having the sequence set forth in any one of SEQ ID NOS:19-21, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (g) a target site for ELOB having the sequence set forth in any one of SEQ ID NOS:22-24, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (h) a target site for GATA3 having the sequence set forth in any one of SEQ ID NOS:25-27, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (i) a target site for CISH having the sequence set forth in any one of SEQ ID NOS:28-30, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (j) a target site for PRDM1 having the sequence set forth in any one of SEQ ID NOS:31-33, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (k) a target site for MED12 having the sequence set forth in any one of SEQ ID NOS:80-90, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (l) a target site for CCNC having the sequence set forth in any one of SEQ ID NOS:102-112, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (m) a target site for FAS having the sequence set forth in any one of SEQ ID NOS:200-205 and 292-295, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (n) a target site for Fli1 having the sequence set forth in any one of SEQ ID NOS:206-211, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; and (o) a target site for TGFBR2 having the sequence set forth sf-6059407 22474-20028.40 in any one of SEQ ID NOS:300-302 and 306-308, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. [0080] In some of any embodiments, the target site for at least one of the one or more repression genes is selected from: (a) a target site for CD5 having the sequence set forth in any one of SEQ ID NOS:1-3 or a complementary sequence thereof; (b) a target site for KDM1A having the sequence set forth in any one of SEQ ID NOS:4-6 or a complementary sequence thereof; (c) a target site for CBLB having the sequence set forth in any one of SEQ ID NOS:10- 12 or a complementary sequence thereof; (d) a target site for DGKZ having the sequence set forth in any one of SEQ ID NOS:13-15 or a complementary sequence of any of the foregoing; (e) a target site for MYB having the sequence set forth in any one of SEQ ID NOS:16-18 or a complementary sequence of any of the foregoing; (f) a target site for RASA2 having the sequence set forth in any one of SEQ ID NOS:19-21 or a complementary sequence of any of the foregoing; (g) a target site for ELOB having the sequence set forth in any one of SEQ ID NOS:22-24 or a complementary sequence of any of the foregoing; (h) a target site for GATA3 having the sequence set forth in any one of SEQ ID NOS:25-27 or a complementary sequence of any of the foregoing; (i) a target site for CISH having the sequence set forth in any one of SEQ ID NOS:28-30 or a complementary sequence of any of the foregoing; (j) a target site for PRDM1 having the sequence set forth in any one of SEQ ID NOS:31-33 or a complementary sequence of any of the foregoing; (k) a target site for MED12 having the sequence set forth in any one of SEQ ID NOS:80-90 or a complementary sequence of any of the foregoing; (l) a target site for CCNC having the sequence set forth in any one of SEQ ID NOS:102-112 or a complementary sequence of any of the foregoing; (m) a target site for FAS having the sequence set forth in any one of SEQ ID NOS:200-205 and 292-295 or a complementary sequence of any of the foregoing; (n) a target site for Fli1 having the sequence set forth in any one of SEQ ID NOS:206-211 or a complementary sequence of any of the foregoing; and (o) a target site for TGFBR2 having the sequence set forth in any one of SEQ ID NOS:300-302 and 306-308 or a complementary sequence of any of the foregoing. [0081] In some of any embodiments, the target site for each of the one or more repression genes is selected from: (a) a target site for CD5 having the sequence set forth in any one of SEQ ID NOS:1-3, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (b) a target site for KDM1A having the sequence set forth in any one of SEQ ID NOS:4-6, a contiguous portion thereof of at least 14 nucleotides (nt), or a sf-6059407 22474-20028.40 complementary sequence of any of the foregoing; (c) a target site for CBLB having the sequence set forth in any one of SEQ ID NOS:10-12, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (d) a target site for DGKZ having the sequence set forth in any one of SEQ ID NOS:13-15, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (e) a target site for MYB having the sequence set forth in any one of SEQ ID NOS:16-18, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (f) a target site for RASA2 having the sequence set forth in any one of SEQ ID NOS:19-21, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (g) a target site for ELOB having the sequence set forth in any one of SEQ ID NOS:22-24, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (h) a target site for GATA3 having the sequence set forth in any one of SEQ ID NOS:25-27, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (i) a target site for CISH having the sequence set forth in any one of SEQ ID NOS:28-30, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (j) a target site for PRDM1 having the sequence set forth in any one of SEQ ID NOS:31-33, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (k) a target site for MED12 having the sequence set forth in any one of SEQ ID NOS:80-90, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (l) a target site for CCNC having the sequence set forth in any one of SEQ ID NOS:102-112, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (m) a target site for FAS having the sequence set forth in any one of SEQ ID NOS:200-205 and 292-295, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (n) a target site for Fli1 having the sequence set forth in any one of SEQ ID NOS:206-211, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; and (o) a target site for TGFBR2 having the sequence set forth in any one of SEQ ID NOS:300-302 and 306-308, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. [0082] In some of any embodiments, the target site for each of the one or more repression genes is selected from: (a) a target site for CD5 having the sequence set forth in any one of SEQ sf-6059407 22474-20028.40 ID NOS:1-3 or a complementary sequence thereof; (b) a target site for KDM1A having the sequence set forth in any one of SEQ ID NOS:4-6 or a complementary sequence thereof; (c) a target site for CBLB having the sequence set forth in any one of SEQ ID NOS:10-12 or a complementary sequence thereof; (d) a target site for DGKZ having the sequence set forth in any one of SEQ ID NOS:13-15 or a complementary sequence of any of the foregoing; (e) a target site for MYB having the sequence set forth in any one of SEQ ID NOS:16-18 or a complementary sequence of any of the foregoing; (f) a target site for RASA2 having the sequence set forth in any one of SEQ ID NOS:19-21 or a complementary sequence of any of the foregoing; (g) a target site for ELOB having the sequence set forth in any one of SEQ ID NOS:22-24 or a complementary sequence of any of the foregoing; (h) a target site for GATA3 having the sequence set forth in any one of SEQ ID NOS:25-27 or a complementary sequence of any of the foregoing; (i) a target site for CISH having the sequence set forth in any one of SEQ ID NOS:28-30 or a complementary sequence of any of the foregoing; (j) a target site for PRDM1 having the sequence set forth in any one of SEQ ID NOS:31-33 or a complementary sequence of any of the foregoing; (k) a target site for MED12 having the sequence set forth in any one of SEQ ID NOS:80-90 or a complementary sequence of any of the foregoing; (l) a target site for CCNC having the sequence set forth in any one of SEQ ID NOS:102-112 or a complementary sequence of any of the foregoing; (m) a target site for FAS having the sequence set forth in any one of SEQ ID NOS:200-205 and 292-295 or a complementary sequence of any of the foregoing; (n) a target site for Fli1 having the sequence set forth in any one of SEQ ID NOS:206-211 or a complementary sequence of any of the foregoing; and (o) a target site for TGFBR2 having the sequence set forth in any one of SEQ ID NOS:300-302 and 306-308 or a complementary sequence of any of the foregoing. [0083] In some of any embodiments, the target site for at least one of the one or more repression genes is a target site for MED12 having the sequence set forth in any one of SEQ ID NOS:80-90, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. In some of any embodiments, the target site for at least one of the one or more repression genes is a target site for MED12 having the sequence set forth in any one of SEQ ID NOS:80-90 or a complementary sequence of any of the foregoing. In some of any embodiments, the target site for each of the one or more repression genes is a target site for MED12 having the sequence set forth in any one of SEQ ID NOS:80-90, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. In sf-6059407 22474-20028.40 some of any embodiments, the target site for each of the one or more repression genes is a target site for MED12 having the sequence set forth in any one of SEQ ID NOS:80-90 or a complementary sequence of any of the foregoing. [0084] In some of any embodiments, the target site of one of the one or more repression genes is any one of the sequences set forth in any one of SEQ ID NO:454-457, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. In some of any embodiments, the target site of one of the one or more repression genes is any one of the sequences set forth in any one of SEQ ID NO:454-457 or a complementary sequence of any of the foregoing. [0085] In some of any embodiments, the second DNA-binding domain is a ZFP or that targets the target site of one or more repression genes set forth in SEQ ID NO: 454. In some of any embodiments, the ZFP comprises a zinc finger recognition region comprising six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, selected from F1-F6 as follows: F1: DRSHLTR (SEQ ID NO: 490), F2: DRSYRNT (SEQ ID NO: 491), F3: QRRSLPH (SEQ ID NO: 492), F4: RSADLSR (SEQ ID NO: 493), F5: RSDTLSE (SEQ ID NO: 494), and F6: NRRGRWS (SEQ ID NO: 495). In some of any embodiments, the ZFP comprises the sequence set forth in SEQ ID NO: 461, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, the ZFP comprises the sequence set forth in SEQ ID NO: 461. In some of any embodiments, the ZFP is encoded by the sequence set forth in SEQ ID NO: 468 or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, the ZFP is encoded by the sequence set forth in SEQ ID NO: 468. [0086] In some of any embodiments, the second DNA-binding domain is a ZFP that targets the target site of one or more repression genes set forth in SEQ ID NO: 455. In some of any embodiments, the ZFP comprises a zinc finger recognition region comprising six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, selected from F1-F6 as follows: F1: RSANLAR (SEQ ID NO: 496), F2: DRSALAR (SEQ ID NO: 497), F3: RSDALST (SEQ ID NO: 498), F4: QSATRTK (SEQ ID NO: 499), F5: RSDTLSE (SEQ ID NO: 500), and F6: FRYARQY (SEQ ID NO: 501). In some of any embodiments, the ZFP comprises the sequence set forth in SEQ ID NO: 462, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some sf-6059407 22474-20028.40 of any embodiments, the ZFP comprises the sequence set forth in SEQ ID NO: 462. In some of any embodiments, the ZFP is encoded by the sequence set forth in SEQ ID NO: 469 or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, the ZFP is encoded by the sequence set forth in SEQ ID NO: 469. [0087] In some of any embodiments, the second DNA-binding domain is a ZFP that targets the target site of one or more repression genes set forth in in SEQ ID NO: 456. In some of any embodiments, the ZFP comprises a zinc finger recognition region comprising six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, selected from F1-F6 as follows: F1: DQSNLRA (SEQ ID NO: 502), F2: QNAHRKT (SEQ ID NO: 503), F3: TSGSLSR (SEQ ID NO: 504), F4: DRSNLSS (SEQ ID NO: 505), F5: RSAHLSR (SEQ ID NO: 506), and F6: RSDHLTQ (SEQ ID NO: 507). In some of any embodiments, the ZFP comprises the sequence set forth in SEQ ID NO: 463, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, the ZFP comprises the sequence set forth in SEQ ID NO: 463. In some of any embodiments, the ZFP is encoded by the sequence set forth in SEQ ID NO: 470 or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, the ZFP is encoded by the sequence set forth in SEQ ID NO: 470. [0088] In some of any embodiments, the second DNA-binding domain is a ZFP that targets the target site of one or more repression genes set forth in SEQ ID NO: 457. In some of any embodiments, the ZFP comprises a zinc finger recognition region comprising six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, selected from F1-F6 as follows: F1: RSDHLSA (SEQ ID NO: 508), F2: QSSDLRR (SEQ ID NO: 509), F3: HHNNRTH (SEQ ID NO: 510), F4: RNASRTR (SEQ ID NO: 511), F5: RSDHLSA (SEQ ID NO: 512), and F6: RSANLTR (SEQ ID NO: 513). In some of any embodiments, the ZFP comprises the sequence set forth in SEQ ID NO: 464, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, the ZFP comprises the sequence set forth in SEQ ID NO: 464. In some of any embodiments, the ZFP is encoded by the sequence set forth in SEQ ID NO: 471 or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, sf-6059407 22474-20028.40 97%, 98%, or 99% sequence identity thereto. In some of any embodiments, the ZFP is encoded by the sequence set forth in SEQ ID NO: 471. [0089] In some of any embodiments, at least one, optionally each gRNA for targeting a target site of one of the one or more repression genes comprises a gRNA spacer sequence that is complementary to a target site of one of the one or more repression genes. In some of any embodiments, at least one, optionally each gRNA for targeting a target site of one of the one or more repression genes comprises a gRNA spacer sequence between 14 nt and 24 nt or between 16 nt and 22 nt in length. In some of any embodiments, at least one, optionally each gRNA for targeting a target site of one of the one or more repression genes comprises a gRNA spacer sequence that is 18 nt, 19 nt, 20 nt, 21 nt, or 22 nt in length. [0090] In some of any embodiments, at least one, optionally each gRNA for targeting a target site of one of the one or more repression genes further comprises a gRNA scaffold sequence set forth in SEQ ID NO:69. In some of any embodiments, at least one, optionally each gRNA for targeting a target site of one of the one or more repression genes further comprises a gRNA scaffold sequence set forth in SEQ ID NO:387. [0091] In some of any embodiments, at least one of the gRNA for targeting a target site of one of the one or more repression genes is selected from: (a) a gRNA targeting a target site for CD5 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:35-37, or a contiguous portion thereof of at least 14 nt; (b) a gRNA targeting a target site for KDM1A and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:38-40, or a contiguous portion thereof of at least 14 nt; (c) a gRNA targeting a target site for CBLB and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:44-46, or a contiguous portion thereof of at least 14 nt; (d) a gRNA targeting a target site for DGKZ and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:47-49, or a contiguous portion thereof of at least 14 nt; (e) a gRNA targeting a target site for MYB and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:50-52, or a contiguous portion thereof of at least 14 nt; (f) a gRNA targeting a target site for RASA2 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:53-55, or a contiguous portion thereof of at least 14 nt; (g) a gRNA targeting a target site for ELOB and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:56-58, or a contiguous portion thereof of at least 14 nt; (h) a gRNA sf-6059407 22474-20028.40 targeting a target site for GATA3 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:59-61, or a contiguous portion thereof of at least 14 nt; (i) a gRNA targeting a target site for CISH and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:62-64, or a contiguous portion thereof of at least 14 nt; (j) a gRNA targeting a target site for PRDM1 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:65-67, or a contiguous portion thereof of at least 14 nt; (k) a gRNA targeting a target site for MED12 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:91-101, or a contiguous portion thereof of at least 14 nt; (l) a gRNA targeting a target site for CCNC and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:113-123, or a contiguous portion thereof of at least 14 nt; (m) a gRNA targeting a target site for FAS and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:212-217 and 296-299, or a contiguous portion thereof of at least 14 nt; (n) a gRNA targeting a target site for Fli1 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:218-223, or a contiguous portion thereof of at least 14 nt; and (o) a gRNA targeting a target site for TGFBR2 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:303-305 and 309-311, or a contiguous portion thereof of at least 14 nt. [0092] In some of any embodiments, at least one of the gRNA for targeting a target site of one of the one or more repression genes is selected from: (a) a gRNA targeting a target site for CD5 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:35-37; (b) a gRNA targeting a target site for KDM1A and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:38-40; (c) a gRNA targeting a target site for CBLB and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:44-46; (d) a gRNA targeting a target site for DGKZ and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:47-49; (e) a gRNA targeting a target site for MYB and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:50-52; (f) a gRNA targeting a target site for RASA2 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:53-55; (g) a gRNA targeting a target site for ELOB and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:56-58; (h) a gRNA targeting a target site for GATA3 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:59-61; (i) a gRNA targeting a target site for CISH and comprising a gRNA spacer sequence set forth in any one of sf-6059407 22474-20028.40 SEQ ID NOS:62-64; (j) a gRNA targeting a target site for PRDM1 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:65-67; (k) a gRNA targeting a target site for MED12 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:91- 101; (l) a gRNA targeting a target site for CCNC and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:113-123; (m) a gRNA targeting a target site for FAS and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:212-217 and 296-299; (n) a gRNA targeting a target site for Fli1 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:218-223; and (o) a gRNA targeting a target site for TGFBR2 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:303-305 and 309-311. [0093] In some of any embodiments, each gRNA for targeting a target site of one of the one or more repression genes is selected from: (a) a gRNA targeting a target site for CD5 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:35-37, or a contiguous portion thereof of at least 14 nt; (b) a gRNA targeting a target site for KDM1A and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:38-40, or a contiguous portion thereof of at least 14 nt; (c) a gRNA targeting a target site for CBLB and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:44-46, or a contiguous portion thereof of at least 14 nt; (d) a gRNA targeting a target site for DGKZ and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:47-49, or a contiguous portion thereof of at least 14 nt; (e) a gRNA targeting a target site for MYB and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:50-52, or a contiguous portion thereof of at least 14 nt; (f) a gRNA targeting a target site for RASA2 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:53-55, or a contiguous portion thereof of at least 14 nt; (g) a gRNA targeting a target site for ELOB and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:56-58, or a contiguous portion thereof of at least 14 nt; (h) a gRNA targeting a target site for GATA3 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:59-61, or a contiguous portion thereof of at least 14 nt; (i) a gRNA targeting a target site for CISH and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:62-64, or a contiguous portion thereof of at least 14 nt; (j) a gRNA targeting a target site for PRDM1 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:65-67, or a sf-6059407 22474-20028.40 contiguous portion thereof of at least 14 nt; (k) a gRNA targeting a target site for MED12 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:91-101, or a contiguous portion thereof of at least 14 nt; (l) a gRNA targeting a target site for CCNC and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:113-123, or a contiguous portion thereof of at least 14 nt; (m) a gRNA targeting a target site for FAS and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:212-217 and 296-299, or a contiguous portion thereof of at least 14 nt; (n) a gRNA targeting a target site for Fli1 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:218-223, or a contiguous portion thereof of at least 14 nt; and (o) a gRNA targeting a target site for TGFBR2 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:303-305 and 309-311, or a contiguous portion thereof of at least 14 nt. [0094] In some of any embodiments, each gRNA for targeting a target site of one of the one or more repression genes is selected from: (a) a gRNA targeting a target site for CD5 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:35-37; (b) a gRNA targeting a target site for KDM1A and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:38-40; (c) a gRNA targeting a target site for CBLB and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:44-46; (d) a gRNA targeting a target site for DGKZ and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:47-49; (e) a gRNA targeting a target site for MYB and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:50-52; (f) a gRNA targeting a target site for RASA2 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:53-55; (g) a gRNA targeting a target site for ELOB and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:56-58; (h) a gRNA targeting a target site for GATA3 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:59-61; (i) a gRNA targeting a target site for CISH and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:62-64; (j) a gRNA targeting a target site for PRDM1 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:65-67; (k) a gRNA targeting a target site for MED12 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:91-101; (l) a gRNA targeting a target site for CCNC and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:113-123; (m) a gRNA targeting a target site for FAS and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:212-217 and 296-299; (n) a gRNA targeting a sf-6059407 22474-20028.40 target site for Fli1 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:218-223; and (o) a gRNA targeting a target site for TGFBR2 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:303-305 and 309-311. [0095] In some of any embodiments, at least one of the gRNA for targeting a target site of one of the one or more repression genes is a gRNA targeting a target site for MED12 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:91-101, or a contiguous portion thereof of at least 14 nt. In some of any embodiments, at least one of the gRNA for targeting a target site of one of the one or more repression genes is a gRNA targeting a target site for MED12 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:91-101. In some of any embodiments, each gRNA for targeting a target site of one of the one or more repression genes is a gRNA targeting a target site for MED12 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:91-101, or a contiguous portion thereof of at least 14 nt. In some of any embodiments, each gRNA for targeting a target site of one of the one or more repression genes is a gRNA targeting a target site for MED12 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:91-101. [0096] In some of any embodiments, at least one, optionally each transcriptional repressor domain epigenetically modifies a target site of one of the one or more repression genes to promote reduced transcription of one of the one or more repression genes. [0097] In some of any embodiments, at least one, optionally each transcriptional repressor domain is a KRAB domain, a DNMT3A domain, a DNMT3L domain, a DNMT3B domain, a DNMT3A-DNMT3L fusion protein domain, an ERF repressor domain, an Mxi1 repressor domain, a SID4X repressor domain, a Mad-SID repressor domain, an LSD1 repressor domain, an EZH2 repressor domain, a SunTag domain, a variant or portion of any of the foregoing, or a combination of any of the foregoing. In some of any embodiments, at least one, optionally each transcriptional repressor domain is a KRAB domain, a DNMT3A domain, a DNMT3L domain, or a combination of any of the foregoing. [0098] In some of any embodiments, at least one, optionally each transcriptional repressor domain comprises a KRAB domain or a variant or portion thereof that exhibits transcriptional repressor activity. In some of any embodiments, at least one, optionally each transcriptional repressor domain comprises the sequence set forth in any one of SEQ ID NOS:70, 235, and 355- sf-6059407 22474-20028.40 358, a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. [0099] In some of any embodiments, at least one, optionally each transcriptional repressor domain comprises a DNMT3A domain or a variant or portion thereof that exhibits transcriptional repressor activity. In some of any embodiments, at least one, optionally each transcriptional repressor domain comprises the sequence set forth in SEQ ID NO:131 or 238, a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. [0100] In some of any embodiments, at least one, optionally each transcriptional repressor domain comprises a DNMT3L domain or a variant or portion thereof that exhibits transcriptional repressor activity. In some of any embodiments, at least one, optionally each transcriptional repressor domain comprises the sequence set forth in any one of SEQ ID NOS:133 and 240-242, a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. [0101] In some of any embodiments, at least one, optionally each transcriptional repressor domain is a DNMT3A-DNMT3L fusion protein domain, a DNMT3B-DNMT3L fusion protein domain, or a variant thereof that exhibits transcriptional repressor activity. In some of any embodiments, at least one, optionally each transcriptional repressor domain comprises the sequence set forth in any one of SEQ ID NOS:135, 137, or 363, a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. [0102] In some of any embodiments, at least one, optionally each fusion protein of the at least one repressor DNA-targeting module comprises the sequence set forth in any one of SEQ ID NOS:138-141, 332-351, and 365-384, a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. [0103] In some of any embodiments, at least one, optionally each fusion protein of the at least one repressor DNA-targeting module comprises the sequence set forth in any one of SEQ ID NOS:517-520, a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. [0104] In some of any embodiments, delivery of the epigenetic-modifying DNA-targeting system to the T cell increases transcription of the one or more activation genes and decreases sf-6059407 22474-20028.40 transcription of the one or more repression genes, compared to a T cell that has not been delivered the epigenetic-modifying DNA-targeting system. [0105] In some of any embodiments, delivery of the epigenetic-modifying DNA-targeting system to the T cell promotes increased T cell effector function upon T cell stimulation, compared to a T cell that has not been delivered the epigenetic-modifying DNA-targeting system. [0106] In some of any embodiments, the delivery is transient delivery of the epigenetic- modifying DNA-targeting system to the T cell. In some of any embodiments, transient delivery of the epigenetic-modifying DNA-targeting system to the T cell increases transcription of the one or more activation genes and decreases transcription of the one or more repression genes, compared to a T cell that has not been delivered the epigenetic-modifying DNA-targeting system. In some of any embodiments, transient delivery of the epigenetic-modifying DNA- targeting system to the T cell promotes increased T cell effector function upon T cell stimulation, compared to a T cell that has not been delivered the epigenetic-modifying DNA- targeting system. [0107] In some of any embodiments, the T cell effector function is selected from the group consisting of IL-2 production, IFN-gamma production, TNF-alpha production, T cell proliferation, and a combination of any of the foregoing. In some of any embodiments, the T cell effector function is IL-2 production. In some of any embodiments, the T cell effector function is IFN-gamma production. In some of any embodiments, the T cell effector function is IL-2 production and IFN-gamma production. In some of any embodiments, the T cell effector function is polyfunctional production of IL-2, IFN-gamma, and TNF-alpha. [0108] In some of any embodiments, the T cell effector function further comprises T cell proliferation. In some of any embodiments, the T cell effector function further comprises killing of target cells. In some of any embodiments, the T cell effector function further comprises T cell persistence. [0109] In some of any embodiments, the increased T cell effector function occurs 48 hours or more after the delivery of the epigenetic-modifying DNA-targeting system to the T cell. In some of any embodiments, the increased T cell effector function occurs up to 6 days, up to 9 days, up to 12 days, up to 15 days, up to 21 days, up to 28 days, up to 35 days, up to 42 days, up to 49 days, up to 56 days, up to 63 days, or up to 71 days after the delivery of the epigenetic- modifying DNA-targeting system to the T cell. In some of any embodiments, the increased T sf-6059407 22474-20028.40 cell effector function occurs for up to 6 days, for up to 9 days, for up to 12 days, for up to 15 days, for up to 21 days, for up to 28 days, for up to 35 days, for up to 42 days, for up to 49 days, for up to 56 days, for up to 63 days, or for up to 71 days after the delivery of the epigenetic- modifying DNA-targeting system to the T cell. [0110] In some of any embodiments, the T cell stimulation is with an anti-CD3 and anti- CD28 activation reagent. [0111] In some of any embodiments, the T cell expresses a recombinant receptor. In some of any embodiments, the recombinant receptor is a chimeric antigen receptor (CAR) or a T cell receptor (eTCR). In some of any embodiments, the recombinant receptor is a CAR. In some of any embodiments, the recombinant receptor is directed against an antigen, and the T cell stimulation is an antigen-specific stimulation of the recombinant receptor. In some of any embodiments, the T cell stimulation is with antigen-expressing target cells. [0112] In some of any embodiments, the T cell stimulation is a restimulation after at least one prior T cell stimulation of the T cells. [0113] Also provided herein in some embodiments is a polynucleotide encoding any of the provided epigenetic-modifying DNA-targeting systems. [0114] Also provided herein in some embodiments is two or more polynucleotides together encoding any of the provided epigenetic-modifying DNA-targeting systems. [0115] Also provided herein in some embodiments is a polynucleotide comprising (a) a promoter sequence, (b) a first nucleic acid encoding: at least one activator DNA-targeting module comprising a fusion protein comprising (i) a first zinc finger protein (ZFP) for targeting to a target site of one of the one or more activation genes and (ii) at least one transcriptional activator domain; and (c) a second nucleic acid encoding at least one repressor DNA-targeting module comprising a fusion protein comprising (i) a second zinc finger protein (ZFP) for targeting to a target site of one of the one or more repression genes and (ii) at least one transcriptional repressor domain; and (d) a cleavable linker sequence that is located between the first nucleic acid sequence and the second nucleic acid sequence, wherein the promoter is operably linked to the first nucleic and and second nucleic acid to control their expression. In some embodiments, the cleavable linker sequence encodes a self-cleaving peptide. In some of any embodiments, the first ZFP targets the target site of one or more activation genes set forth in any one of SEQ ID NOs: 451-453; and the second ZFP targets the target site set forth in any one of SEQ ID NOs: 454-457. In some of any embodiments, the first ZFP comprises the sequence sf-6059407 22474-20028.40 set forth in any one of SEQ ID NOs: 458-460; and the second ZFP comprises the sequence set forth in any one of SEQ ID NOs: 461-464. In some of any embodiments, the at least one transcriptional activator domain comprises a sequence set forth in SEQ ID NO: 549; and the at least one transcriptional repressor domain comprises sequences set forth in SEQ ID NO: 70 and SEQ ID NO: 135. In some of any embodiments, the at least one activator DNA-targeting module comprises the sequence set forth in any one of SEQ ID NOs: 514-516; and the at least one repressor DNA-targeting module comprises the sequence set forth in any one of SEQ ID NOs: 517-520. In some of any embodiments, the at least one activator DNA-targeting module comprises SEQ ID NO: 516; and the at least one repressor DNA-targeting module comprises SEQ ID NO: 517. In some of any embodiments, the polynucleotide, from N-terminus to C- terminus, comprises: (a) the promoter sequence; (b) the first nucleic acid sequence encoding the at least one activator DNA-targeting module, (c) the cleavable linker sequence, and (d) the second nucleic acid encoding the at least one repressor DNA-targeting module. In some of any embodiments, the polynucleotide, from N-terminus to C-termionus, comprises: (a) the promoter sequence; (b) the second nucleic acid sequence encoding the at least one repressor DNA- targeting module, (c) the cleavable linker sequence, and (d) the first nucleic acid encoding the at least one activator DNA-targeting module. In some embodiments, the self-cleaving peptide is a 2A sequence, such as a P2A or T2A sequence. In some of any embodiments, the self-cleaving peptide comprises the sequence set forth in SEQ ID NO: 352. In some embodiments, the promoter sequence is a constitutive promoter, such as a CMV promoter, a EF-1α promoter, or a SV40 promoter. [0116] Also provided herein in some embodiments is a vector comprising any of the provided polynucleotides. Also provided herein in some embodiments is a vector comprising any of the provided two or more polynucleotides. [0117] In some of any embodiments, the vector is a viral vector. In some of any embodiments, the vector is an adeno-associated virus (AAV) vector. In some of any embodiments, the vector is an AAV1, AAV2, AAV3, AAV4, AAV5, AAV6, AAV7, AAV8, or AAV9 vector. [0118] In some of any embodiments, the vector is a non-viral vector. In some of any embodiments, the vector is a lipid nanoparticle, a liposome, an exosome, or a cell penetrating peptide. In some of any embodiments, the vector is a lipid nanoparticle. sf-6059407 22474-20028.40 [0119] In some of any embodiments, the vector exhibits immune cell tropism. In some of any embodiments, the vector exhibits T cell tropism. [0120] Also provided herein in some embodiments is two or more vectors together comprising any of the provided two or more polynucleotides. [0121] In some of any embodiments, at least one, optionally each of the two or more vectors are viral vectors. In some of any embodiments, at least one, optionally each of the two or more vectors are AAV vectors. In some of any embodiments, at least one, optionally each vector is an AAV1, AAV2, AAV3, AAV4, AAV5, AAV6, AAV7, AAV8, or AAV9 vector. [0122] In some of any embodiments, at least one, optionally each of the two or more vectors are non-viral vectors. In some of any embodiments, at least one, optionally each vector is a lipid nanoparticle, a liposome, an exosome, or a cell penetrating peptide. In some of any embodiments, at least one, optionally each of the two or more vectors are lipid nanoparticles. [0123] In some of any embodiments, the two or more vectors exhibit immune cell tropism. In some of any embodiments, the two or more vectors exhibit T cell tropism. [0124] Also provided herein in some embodiments is a modified T cell comprising any of the provided DNA-targeting systems, any of the provided polynucleotides, or any of the provided two or more polynucleotidess. [0125] Also provided herein in some embodiments is a modified T cell comprising an epigenetic or phenotypic modification resulting from being contacted by any of the provided DNA-targeting systems, any of the provided polynucleotides, any of the provided two or more polynucleotides, any of the provided vectors, or any of the provided two or more vectors. [0126] In some of any embodiments, the modified T cell is derived from a cell from a subject. In some of any embodiments, the modified T cell is derived from a primary T cell. In some of any embodiments, the modified T cell is derived from a T cell progenitor, a pluripotent stem cell, or an induced pluripotent stem cell. [0127] In some of any embodiments, the modified T cell is a tumor infiltrating lymphocyte (TIL) or is an engineered T cell that further comprises an eTCR or CAR. [0128] Also provided herein in some embodiments is a method of modulating transcription in a T cell, the method comprising introducing into a T cell any of the provided DNA-targeting systems, any of the provided polynucleotides, any of the provided two or more polynucleotides, any of the provided vectors, or any of the provided two or more vectors. sf-6059407 22474-20028.40 [0129] Also provided herein in some embodiments is a method of increasing T cell effector function, the method comprising introducing into a T cell any of the provided DNA-targeting systems, any of the provided polynucleotides, any of the provided two or more polynucleotides, any of the provided vectors, or any of the provided two or more vectors. [0130] In some of any embodiments, T cell effector function of the T cell is increased upon T cell stimulation compared to a T cell that has not been introduced to the DNA-targeting system, the polynucleotide, the two or more polynucleotides, the vector, or the two or more vectors. [0131] In some of any embodiments, the method further comprises stimulating the T cell. [0132] In some of any embodiments, the T cell effector function is selected from the group consisting of IL-2 production, IFN-gamma production, TNF-alpha production, T cell proliferation, and a combination of any of the foregoing. In some of any embodiments, the T cell effector function is IL-2 production. In some of any embodiments, the T cell effector function is IFN-gamma production. In some of any embodiments, the T cell effector function is IL-2 production and IFN-gamma production. In some of any embodiments, the T cell effector function is polyfunctional production of IL-2, IFN-gamma, and TNF-alpha. [0133] In some of any embodiments, the T cell effector function further comprises T cell proliferation. In some of any embodiments, the T cell effector function further comprises killing of target cells. In some of any embodiments, the T cell effector function further comprises T cell persistence. [0134] In some of any embodiments, the increased T cell effector function occurs 48 hours or more after the introducing to the T cell. In some of any embodiments, the increased T cell effector function occurs up to 6 days, up to 9 days, up to 12 days, up to 15 days, up to 21 days, up to 28 days, up to 35 days, up to 42 days, up to 49 days, up to 56 days, up to 63 days, or up to 71 days after the introducing. In some of any embodiments, the increased T cell effector function occurs for up to 6 days, for up to 9 days, for up to 12 days, for up to 15 days, for up to 21 days, for up to 28 days, for up to 35 days, for up to 42 days, for up to 49 days, for up to 56 days, for up to 63 days, or for up to 71 days after the delivery of the epigenetic-modifying DNA- targeting system to the T cell. [0135] In some of any embodiments, the T cell stimulation is with an anti-CD3 and anti- CD28 activation reagent. sf-6059407 22474-20028.40 [0136] In some of any embodiments, the T cell is a tumor infiltrating lymphocyte (TIL) therapy. [0137] In some of any embodiments, the T cell expresses a recombinant receptor. In some of any embodiments, the recombinant receptor is a CAR or a eTCR. In some of any embodiments, the recombinant receptor is directed against an antigen, and the T cell stimulation is an antigen- specific stimulation of the recombinant receptor. In some of any embodiments, the T cell stimulation is with antigen-expressing target cells. [0138] In some of any embodiments, the T cell stimulation is a restimulation after at least one prior T cell stimulation of the T cells. [0139] In some of any embodiments, the T cell is a T cell in a subject, and the method is carried out in vivo. [0140] In some of any embodiments, the T cell is a T cell from a subject or derived from a cell from the subject, and the method is carried out ex vivo. [0141] In some of any embodiments, the T cell is a primary T cell. In some of any embodiments, the T cell is derived from a T cell progenitor, a pluripotent stem cell, or an induced pluripotent stem cell. [0142] In some of any embodiments, the introducing is by transient delivery into the T cell. [0143] In some of any embodiments, the introducing comprises electroporation, transfection, or transduction. [0144] In some of any embodiments, the DNA-targeting system, the polynucleotide, the two or more polynucleotides, the vector, or the two or more vectors are transiently present in the T cell. [0145] In some of any embodiments, the introducing increases transcription of the one or more activation genes in the T cell. In some of any embodiments, the introducing represses transcription of the one or more repression genes in the T cell. [0146] Also provided herein in some embodiments is a modified T cell produced by any of the provided methods. [0147] Also provided herein in some embodiments is a method of treating a disease or condition in a subject, the method comprising administering to the subject any of the provided modified T cells. [0148] Also provided herein in some embodiments is a method of increasing T cell persistence in T cells of a subject, the method comprising administering to the subject any of the sf-6059407 22474-20028.40 provided DNA-targeting systems, any of the provided polynucleotides, any of the provided two or more polynucleotides, any of the provided vectors, or any of the provided two or more vectors. [0149] In some of any embodiments, the T cells are from an adoptive T cell therapy for treating a disease or condition in the subject. In some of any embodiments, the adoptive T cell therapy comprises T cells expressing a recombinant receptor directed against an antigen associated with the disease or condition. [0150] In some of any embodiments, the administration is carried out prior to, concurrently with, or after administration of the adoptive T cell therapy. In some of any embodiments, the administration is carried out after administration of the adoptive T cell therapy to the subject and at a time after the numbers or effector function of T cells of the adoptive T cell therapy are reduced, or are suspected of being reduced, in the subject. [0151] Also provided herein in some embodiments is a method of treating a disease or condition in a subject, the method comprising administering to a subject: an adoptive T cell therapy for treating the disease or condition; and any of the provided DNA-targeting systems, any of the provided polynucleotides, any of the provided two or more polynucleotides, any of the provided vectors, or any of the provided two or more vectors. [0152] In some of any embodiments, the adoptive T cell therapy is a tumor infiltrating lymphocyte (TIL) therapy. In some of any embodiments, the adoptive T cell therapy comprises T cells expressing a recombinant receptor directed against an antigen associated with the disease or condition. In some of any embodiments, the recombinant receptor is an eTCR or CAR. [0153] In some of any embodiments, the antigen is a tumor antigen. [0154] In some of any embodiments, the disease or condition is a cancer. In some of any embodiments, the cancer is a hematological cancer or is a solid tumor. [0155] In some of any embodiments, the disease or condition is an autoimmune condition and/or an inflammatory condition. [0156] In some of any embodiments, the administering of the DNA-targeting system, the polynucleotide, the two or more polynucleotides, the vector, or the two or more vectors results in transient delivery to the T cells of the DNA-targeting system, the polynucleotide, the two or more polynucleotides, the vector, or the two or more vectors. sf-6059407 22474-20028.40 [0157] In some of any embodiments, the administering of the DNA-targeting system, the polynucleotide, the two or more polynucleotides, the vector, or the two or more vectors increases transcription of the one or more activation genes in the T cells. [0158] In some of any embodiments, the administering of the DNA-targeting system, the polynucleotide, the two or more polynucleotides, the vector, or the two or more vectors represses transcription of the one or more repression genes in the T cells. Brief Description of the Drawings [0159] FIG.1A shows an exemplary workflow for a transient CRISPR screen. Cells are transfected with a gRNA library. Following enrichment of transfected cells, an epi-editor, such as a dCas fused to an effector domain (e.g., transcriptional repressor, transcriptional activator, etc.), is transiently transfected into the cells, such as by electroporation. Cells are then screened for the desired phenotype. FIG.1B shows an exemplary plot showing expression of IL-2 and IFNg in cells as assessed by flow cytometry, with boxes indicating populations to be sorted according to different phenotypes. [0160] FIG.2 shows a plot of an exemplary CRISPRi screen for gRNAs and genes that modulate IL-2 expression, IFNg expression, and/or proliferation. In particular, the plot shows gRNAs affecting IL-2 expression. Dots (designated by an “x”) represent individual gRNAs. gRNAs on the left are those that target genes whose inhibition results in decreased IL-2 expression, while gRNAs on the right are those that target genes whose inhibition result in upregulation of IL-2 expression. X-axis represents log2 fold change of gRNA abundance in IL- 2+ sorted cells versus unsorted cells. Y-axis represents significance (-log10 adjusted p-value). [0161] FIG.3 shows a plot from an exemplary CRISPRi screen for gRNAs and genes that modulate IL-2 expression, IFNg expression, and/or proliferation. In particular, the plot shows gRNAs affecting proliferation. Dots (designated by an “x”) represent individual gRNAs. gRNAs on the left are those that target genes whose inhibition results in decreased proliferation, while gRNAs on the right are those that target genes whose inhibition result in increased proliferation. X-axis represents log2 fold change of gRNA abundance in unsorted cells 6 days after electroporation versus unsorted cells before electroporation with a transiently expressed epi- editor for targeted transcriptional repression. Y-axis represents significance (-log10 adjusted p- value). sf-6059407 22474-20028.40 [0162] FIG.4 shows numbers of gRNA hits from indicated conditions at day 9, day 12, or both day 9 and day 12 of a CRISPRi screen for gRNAs and genes modulating T cell phenotypes. [0163] FIG.5 shows a plot from an exemplary CRISPRa screen for gRNAs and genes that modulate IL-2 expression, IFNg expression, and/or proliferation. In particular, the plot shows gRNAs affecting IL-2 expression. Dots (designated by an “x”) represent individual gRNAs. gRNAs on the left are those that target genes whose activation results in decreased IL-2 expression, while gRNAs on the right are those that target genes whose activation results in increased IL-2 expression. X-axis represents log2 fold change of gRNA abundance in IL-2+ sorted cells versus unsorted cells. Y-axis represents significance (-log10 adjusted p-value). [0164] FIG.6A shows percent knockdown (% KD) of MED12 expression, as assessed by RT-qPCR following transient delivery of dSpCas9-KRAB and indicated MED12-targeting gRNAs or non-targeting gRNAs (NT1, NT2), or dSpCas9-KRAB alone. Results are shown for 48 hours (left) and 6 days (right) after transfection, and for T cells from two different donors. [0165] FIG.6B shows MED12 expression, as assessed by RT-qPCR following transient delivery of dSpCas9-KRAB and indicated MED12-targeting gRNAs in T cells (left) and CAR T cells (right). Control conditions for left panel include cells delivered with dSpCas9-KRAB and non-targeting gRNAs (NT1, NT2), and dSpCas9-KRAB alone. Control conditions for right panel include cells not expressing a CAR (Mock), and cells not delivered with a DNA-targeting system. Results are shown for 48 hours (left) and 72 hours and 7 days after transfection. Results are normalized to dSpCas9-KRAB only condition (left panel) or Mock condition (right panel). [0166] FIG.6C shows percent knockdown (% KD) of CCNC expression, as assessed by RT-qPCR following transient delivery of dSpCas9-KRAB and indicated CCNC-targeting gRNAs or non-targeting gRNAs (NT1, NT2), or dSpCas9-KRAB alone. Results are shown for 48 hours (left) and 6 days (right) after transfection, and for T cells from two different donors. [0167] FIG.6D shows percent knockdown (% KD) of FAS expression, as assessed by intracellular cytokine staining (ICS) and flow cytometry following transient delivery of dSpCas9-KRAB and indicated FAS-targeting gRNAs, dSpCas9-KRAB alone, or mock delivery. Results are shown for 72 hours (bottom panel, left) and 7 days (bottom panel, right) after delivery, and for T cells from two different donors. Top panel shows an exemplary flow cytometry plot for assessing FAS expression in CD3+ T cells following delivery with dSpCas9- KRAB only, or dSpCas9-KRAB and a FAS-targeting gRNA. sf-6059407 22474-20028.40 [0168] FIG.7A shows flow cytometry plots for assessing IL-2 expression in CD3+ Her2 CAR T cells after stimulation and transient delivery of dSpCas9-2xVP64 and an IL-2-targeting gRNA (IL-2_1, targeting SEQ ID NO:78). Results are shown for CAR T cells derived from 2 different donors. CAR T cells were stimulated with SKOV3 or 143B cells. Different control conditions excluded the CAR, the gRNA, or stimulation, as indicated. [0169] FIG.7B shows quantified IL-2 expression (% IL-2+ cells as assessed by flow cytometry) in Her2 CAR T cells after stimulation and transient delivery of dSpCas9-2xVP64 and an IL-2-targeting gRNA (IL-2_1, targeting SEQ ID NO:78). Control conditions excluded the CAR (mock transduction), the gRNA, or stimulation, as indicated. [0170] FIG.7C shows IL-2 expression in CAR T cells following delivery of a DNA- targeting system for IL-2 activation. Left panel shows IL-2 expression in Her2 CAR T cells as assessed RT-qPCR at 72 hours and 7 days following transient delivery with a DNA-targeting system containing dSpCas9-2xVP64 and an IL-2 targeting gRNA (IL-2_1, targeting SEQ ID NO:78). Control conditions included cells not expressing the CAR (mock), or CAR T cells not delivered with a DNA-targeting system (CAR Only). Expression levels were normalized to the mock control cells. Right panel shows IL-2 expression in control cells (CAR only) and CAR T cells delivered with the DNA-targeting system for IL-2 activation following stimulation with Her2 antigen-expressing tumor cells, as assessed by ICS and flow cytometry. [0171] FIGS. 8A-8C show flow cytometry plots for assessing IL-2 (FIG.8A), IFNg (FIG. 8B), or TNFa (FIG.8C) expression in CD3+ Her2 CAR T cells after stimulation and transient delivery of dSpCas9-2xVP64 and a VAV1-targeting gRNA (VAV1_5, targeting SEQ ID NO:170). Results are shown for CAR T cells derived from 2 different donors. CAR T cells were stimulated with SKOV3 or 143B cells. Different control conditions excluded the CAR, the gRNA, or stimulation, as indicated. [0172] FIG.9 shows percentage polyfunctional cells (IL-2+/IFNg+/TNFa+ cells) as assessed by flow cytometry in Her2 CAR T cells after stimulation and transient delivery of dSpCas9-2xVP64 and an IL-2-targeting gRNA (IL-2_1, targeting SEQ ID NO:78) or a VAV1- targeting gRNA (VAV1_5, targeting SEQ ID NO:170). Control conditions excluded the CAR (mock transduction), the gRNA, or stimulation, as indicated. [0173] FIG.10 shows flow cytometry plots for assessing IL-2 and IFNg expression in CD4+ (top) or CD8+ (bottom) Her2 CAR T cells after stimulation and transient delivery of dSpCas9-2xVP64 and indicated CBLB-targeting gRNAs or a control non-targeting gRNA. Flow sf-6059407 22474-20028.40 cytometry plots allow for quantification of percentage of cells with specific phenotypes (e.g. IL- 2+), and for quantification of mean fluorescence intensity (MFI; corresponding to average expression levels), as shown in right panels. [0174] FIG.11A shows a heatmap indicating relative levels of intracellular cytokine expression (as assessed by ICS and flow cytometry) in Her2 CAR T cells after stimulation and transient delivery of dSpCas9-2xVP64 and indicated gRNAs. Cytokine expression was quantified according to % Her2 CAR T cells with the indicated phenotypes (e.g. CD4+/IL-2+, CD4+/IFNg+/TNFa+) as assessed by flow cytometry, and log 2 fold change was calculated with respect to the control condition with a non-targeting gRNA (non-targeting_sp_1). Darker shades correspond to higher cytokine expression, as indicated in the legend. Each condition was assigned a cumulative score according to all cytokine expression measurements and ranked from low to high cytokine expression. [0175] FIG.11B shows exemplary results from a serial stimulation killing assay. Growth of antigen-expressing target cells expressing a fluorescent marker was quantified over time based on overall fluorescence using an Incucyte automated tracking system. Antigen-expressing target cells were co-cultured with CAR T cells transiently delivered with a DNA-targeting system for activation of IL-2, CAR T cells transiently delivered with a DNA-targeting system for repression of MED12, CAR T cells with no DNA-targeting system (CAR alone), or cells not expressing a CAR (Mock). Serial stimulations (shown as stim 1, stim 2, and stim 3) were performed by replating CAR T cells with fresh target cells at a 1:4 ratio of CAR T cells:target cells. [0176] FIG.11C shows quantification of fold expansion (left) and IL-2 secretion (right) by CAR T cells and control cells from experiment shown in FIG.11B, following the second stimulation. [0177] FIG.12A depicts flow cytometry plots of intracellular cytokine staining (ICS) for IL-2 and IFN-g expression after transient transfection of T cells with control dSpCas9-2xVP64 effector only (without targeting gRNA) or with dSpCas9-2xVP64 in combination with a gRNA targeting either VAV1 (VAV1_5) or IL-2 (IL-2_1) individually. [0178] FIG.12B depicts an ICS flow cytometry plot for IL-2 and IFN-g after transient transfection of T cells with dSpCas9-2xVP64 effector and gRNAs targeting both VAV1 and IL-

22474-20028.40 [0179] FIG.12C depicts the percent of IL-2+ cells (left panel) or IL-2+, IFNg+ and TNFalpha+ cells (right panel) after transient transfection of T cells from either of two donors with dSpCas9-2xVP64 effector only (without targeting gRNA), dSpCas9-2xVP64 in combination with VAV1 or IL-2, or dSpCas9-2xVP64 in combination with gRNAs targeting both VAV1 and IL-2. [0180] FIGS. 13A-13B show heatmaps indicating relative levels of intracellular cytokine expression, cytokine secretion, proliferation, and target cell killing activity in Her2 CAR T cells after stimulation and transient delivery of dSpCas9-2xVP64 or dSpCas9-KRAB and the indicated gRNAs, for Her2 CAR T cells derived from a first donor (FIG.13A) and a second donor (FIG.13B). Multiple rounds of stimulation were performed, and cells electroporated with the different DNA-targeting systems were assessed based on multiple readouts of T cell effector function after the stimulations, including intracellular cytokine expression (by ICS), cytokine secretion, proliferation, and killing of target cells, as described in Example 5. The readouts of T cell effector function were quantified after a first, second, and/or third stimulation (shown as stim 1, stim 2, or stim 3 in figures). ICS was performed after a first stimulation only. Results are shown as Log 2 fold-change in comparison to control cells delivered with a non-targeting gRNA (NT). Darker shades correspond to increased measured T cell effector functions, as indicated in the legend. Each condition was assigned a cumulative score according to all measurements and ranked from low to high T cell effector function. [0181] FIG.14 shows a heatmap indicating relative levels of intracellular cytokine expression in Her2 CAR T cells after stimulation and transient delivery of dSpCas9-2xVP64 and a combination of 2 gRNAs targeting the indicated genes. gRNAs for targeting each gene are shown next to the heatmap. Where the genes targeted are the same, the same gRNA was delivered at twice the concentration. Cytokine expression was quantified according to % Her2 CAR T cells with the indicated phenotypes (e.g. CD8+/IL-2+/IFNg+/TNFa+) as assessed by ICS and flow cytometry, and log 2 fold change was calculated with respect to the control condition with a non-targeting gRNAs (NT+NT). Darker shades correspond to higher cytokine expression, as indicated in the legend. Each condition was assigned a cumulative score according to all cytokine expression measurements and ranked from low to high cytokine expression. [0182] FIGS. 15A-15B show heatmaps indicating relative levels of proliferation and cytokine expression in Her2 CAR T cells after stimulation and transient delivery of dSpCas9- 2xVP64 or dSpCas9-KRAB and gRNAs or combinations thereof targeting the indicated genes. sf-6059407 22474-20028.40 gRNAs for targeting each gene are indicated in the accompanying tables. Cytokine expression was quantified according to % Her2 CAR T cells with the indicated phenotypes (e.g. CD4+/IL- 2+/IFNg+/TNFa+) as assessed by ICS and flow cytometry. Proliferation was quantified according to fold change of Her2 CAR T cell numbers before and after 45timulateon, as described herein. For each cytokine or proliferation phenotype value, log 2 fold change was calculated with respect to a control condition with a non-targeting gRNA (non-targeting_sp_1 for FIG.15A; dCas only for FIG.15B). Darker shades correspond to increased proliferation and cytokine expression, as indicated in the legend. Each condition was assigned a cumulative score according to all cytokine and proliferation measurements and ranked from low to high cytokine expression and proliferation. [0183] FIG.16 shows fold expansion (i.e. proliferation) of Her2 CAR T cells after stimulation and transient delivery of dSpCas9-2xVP64 or dSpCas9-KRAB and indicated gRNAs. Results are shown for Her2 CAR T cells derived from two different donors, after a first stimulation with antigen-expressing target cells. Negative control conditions included CAR T cells delivered with a dCas-effector and no gRNA, CAR T cells not delivered with a DNA- targeting system (CAR only), and cells not expressing a CAR (mock). [0184] FIG.17 shows levels of secreted cytokines IL-2 or IFNg in Her2 CAR T cells after stimulation and transient delivery of dSpCas9-2xVP64 or dSpCas9-KRAB and indicated gRNAs. Results are shown for Her2 CAR T cells derived from two different donors, after a first stimulation with antigen-expresing target cells. Negative control conditions included CAR T cells delivered with a dCas-effector and no gRNA, CAR T cells not delivered with a DNA- targeting system (CAR only), and cells not expressing a CAR (mock). [0185] FIG.18 shows killing activity based on calculated killing index in Her2 CAR T cells after transient delivery of dSpCas9-2xVP64 or dSpCas9-KRAB and indicated gRNAs, after a third stimulation with antigen-expressing target cells. Negative control conditions included CAR T cells delivered with a dCas-effector and no gRNA, CAR T cells not delivered with a DNA- targeting system (CAR only), and cells not expressing a CAR (mock). [0186] FIGS. 19A-19B show heatmaps indicating relative levels of quantified T cell effector functions in Her2 CAR T cells after stimulation and transient delivery of dSpCas9- 2xVP64 or dSpCas9-KRAB and indicated gRNAs. Results are shown for cells derived from a first donor (FIG.19A) and second donor (FIG.19B). Assessed T cell effector functions included intracellular cytokine expression, secreted cytokine expression, proliferation, and sf-6059407 22474-20028.40 killing, which were measured as described in Example 5. The T cell effector functions were quantified after a first, second, and/or third stimulation with antigen-expressing target cells, as indicated in the figure. For each quantified T cell effector function, results are shown as log 2 fold change as calculated with respect to the negative control condition (CAR alone, i.e. CAR T cells not delivered with a DNA-targeting system). Darker shades correspond to increased measured T cell effector functions, as indicated in the legend. Each condition was assigned a cumulative score according to all measurements and ranked from low to high T cell effector function. [0187] FIGS. 20A-20C show results from CAR T cells electroporated with gRNAs targeting indicated genes and mRNA encoding either dSpCas9-2xVP64 for activation or dSpCas9-KRAB for repression. For all results, experiments were performed using CAR T cells from two different donors, and results are plotted for each of the two donors (triangles), with average value indicated by vertical line. FIG.20A shows CAR T fold expansion following electroporation of the DNA-targeting systems (production), and following a first, second, and third round of stimulation with Her2 antigen-expressing target cells. FIG.20B shows CAR T cell target cell killing index following the first and second round of stimulation. FIG.20C shows CAR T cell cytokine production as assessed by ICS and flow cytometry at Day 9 post- electroporation with the DNA-targeting systems, and after a first stimulation with Her2 antigen- expressing target cells. [0188] FIGS. 21A-21C show results from stimulated CAR T cells delivered with DNA- targeting systems for repression of TGF-beta receptor 2 (TGFBR2). DNA-targeting systems included indicated TGFBR2-targeting gRNAs, and either dSpCas9-KRAB (SEQ ID NO:332) or DNMT3A/L-XTEN80-dSpCas9-KRAB (SEQ ID NO:337). Control cells included dSpCas9 only, cells not expressing a CAR (Mock), or CAR T cells not stimulated with antigen-expressing cells (CAR Alone). FIG.21A shows expression of TGFBR2 at 48 hours post-electroporation with the DNA-targeting systems. FIG.21B shows secreted IFNg from the CAR T cells 24 hours after a second stimulation with Her2 antigen-expressing cells, in the presence of 10ng/mL TGFb. FIG.21C shows fold expansion of the stimulated CAR T cells after the second stimulation with Her2 antigen-expressing cells in the presence of 10ng/mL TGFb. [0189] FIGS. 21D-21G show results from stimulated CAR T cells delivered with DNA- targeting systems for repression of TGF-beta receptor 2 (TGFBR2). CAR T cells were electroporated for transient expression of DNA-targeting systems composed of dSpCas9-KRAB- sf-6059407 22474-20028.40 DNMT3A/L and indicated gRNAs targeting TGFBR2. Negative control cells were electroporated with dSpCas9-KRAB-DNMT3A/L and a non-targeting gRNA (non-targeting), not electroporated with a DNA-targeting system (CAR only), or did not express a CAR (mock). FIG.21D shows % TGFBR2 negative cells after indicated days post-electroporation. FIG.21E shows fold cell expansion of CAR T cells after stimulation with anti-CD3/anti-CD28 coated wells and exposure to indicated concentrations of TGF-beta, with expansion normalized to conditions with 0 ng/mL TGF-beta. FIG.21F shows production of IFNg by CAR T cells in response to exposure to 10 ng/mL TGF-beta. Dotted horizontal line indicates CAR only control condition. FIG.21G shows levels of secreted cytokines in stimulated CAR T cells after exposure to indicated concentrations of TGF-beta, with secreted cytokine levels normalized to conditions with 0 ng/mL. [0190] FIGS. 22A-22C show time course (FIG.22A) and results (FIG.22B) from an experiment for in vivo assessment of CAR T cells delivered with a DNA-targeting system for IL-2 activation (FIG.22B) or MED12 repression (FIG.22C). The in vivo experiments were carried out in immune-deficient mice transplanted implanted with antigen-expressing tumor cells (NCI-H1975) and injected with CAR T cells. FIG.22A shows timing and details of tumor implantation, CAR T cell injection, and tracking. FIG.22B and FIG.22C show results from experiments, including animal survival (left panels), tumor growth (middle panels), and levels of circulating CAR T cells (right panels) for CAR T cells delivered with the DNA-targeting system for IL-2 activation (dSpCas9-2xVP64 and gRNA IL-2_1, targeting SEQ ID NO:78) or the DNA-targeting system for MED12 repression (dSpCas9-KRAB and gRNA MED12_2, targeting SEQ ID NO:81), respectively. Control mice were injected with CAR T cells not delivered with a DNA-targeting system (CAR alone), were injected with T cells not expressing a CAR (Mock T Cells), or were not injected with T cells (Tumor Alone). [0191] FIG.23 shows MED12 expression as assessed by RT-qPCR at day 4 and day 21 post-electroporation with a MED12-targeting gRNA (MED12_2, targeting SEQ ID NO:81) and mRNA encoding dSpCas9 (control; no transcriptional repressor effector domain), dSpCas9- KRAB (SEQ ID NO:332), or DNMT3A/L-XTEN80-dSpCas9-KRAB (SEQ ID NO: 337). Expression levels (shown as fold-change in expression) are normalized to expression levels in T cells electroporated with the same fusion protein but with a non-targeting gRNA (dotted line at 1.0). sf-6059407 22474-20028.40 [0192] FIGS. 24A-24F show results from CAR T cells delivered with DNA-targeting systems comprising indicated MED12-targeting gRNAs and dSpCas9-KRAB (SEQ ID NO:332), or DNMT3A/L-XTEN80-dSpCas9-KRAB (SEQ ID NO: 337). Control cells included cells delivered with DNMT3A/L-XTEN80-dSpCas9-KRAB and no gRNA, DNMT3A/L- XTEN80-dSpCas9-KRAB and a non-targeting gRNA, and T cells not expressing a CAR (Mock). FIG.24A shows MED12 expression as assessed by qRT-PCR at day 10 post- electroporation with the DNA-targeting systems. FIG.24B shows MED12 expression at days 2, 7, 10, and 14 post-electroporation. FIG.24C shows CD25 expression as assessed by flow cytometry in CAR T cells at days 3, 7, 10, and 14 post-electroporation. FIG.24D shows secreted IFN-gamma expression, and FIG.24E shows secreted IL-2 expression, at 24 hours after a second stimulation of the CAR T cells with Her2-positive NCI-H1975 tumor cells. FIG. 24F shows proliferation (fold expansion normalized to fold expansion of CAR alone controls) after the second stimulation with the antigen-expressing cells. [0193] FIG.25 shows a timecourse of expression of an exemplary dSpCas9 protein following electroporation of mRNA encoding the protein. Results are shown for electroporation of the dSpCas9 protein with a non-targeting gRNA, a gRNA targeting a gene in the cell, or no electroporation (control). Expression was assessed based on an associated GFP tag, with results indicating percent GFP+ cells as assessed by flow cytometry at indicated time points. [0194] FIGS. 26A-26B show arrangements of fusion proteins for targeted transcriptional repression (FIG.26A), and results from experiments testing ability of the fusion proteins to mediate targeted and sustained gene repression (FIG.26B). FIG.26A shows 4 different arrangements of the dSpCas9 fusion proteins from N-terminal to C-terminal, with the fusion proteins including various domains selected from a DNMT3A, DNMT3B, DNMT3L, and a KRAB or EZH2 domain (shown as [KRAB]). FIG.26B shows MED12 expression at 4 days and 21 days in T cells electroporated with a gRNA targeting MED12 (MED12_2, targeting SEQ ID NO:81), and mRNA encoding the fusion proteins having each of the 4 different arrangements, and comprising a repression domain (shown as [KRAB]) selected from: a KRAB domain from KOX1 (KOX1(2-99)) (SEQ ID NO:355; KRAB domain used in dSpCas9 fusion proteins of preceding Examples), a KRAB domain from KOX1 (KOX1(1-72)) (SEQ ID NO:356), a KRAB domain from ZIM3 (SEQ ID NO:357), a KRAB domain from ZNF324 (SEQ ID NO:358), and an EZH2 domain (SEQ ID NO:359). The mRNA encoding the fusion proteins further included an N-terminal FLAG epitope and a C-terminal P2A-mCherry domain to assess expression of the sf-6059407 22474-20028.40 fusion protein. For each experimental condition, MED 12 expression was normalized to expression levels in T cells electroporated with the same fusion protein but with a non-targeting gRNA. [0195] FIG.27A shows IL-2 locus within human genome assembly GRCh38 (hg38) genomic coordinates chr4:122,451,261-122,593,946, with 7 annotated distinct regions. FIG. 27B shows a log 2 fold change (log2fc) to log2 fold change plot between two different donors. Large black dors, which have been boxed, represent verified hits. FIG.27C shows the distribution of target site hits across various regions within the IL-2 locus. [0196] FIG.28A shows the percent IL-2 expression following a first stimulation of Her2 CAR T cells that were delivered mRNA encoding a dSpCas9-2xVP64 effector fusion protein and various SpCas9 IL-2-targeting gRNA. FIG. 28B shows percent IL-2 expression following a second stimulation of the same Her2 CAR T cells shown in FIG.28A. FIG.28C shows percent IL-2 expression following a third stimulation of the same Her2 CAR T cells shown in FIG.28A. The solid line demarcates the percent IL-2 expression when using the control guide RNA IL-2 gRNA-1. The dashed line demarcates the percent IL-2 expression of Her2 CAR T cells (“CAR”). [0197] FIG.29A shows the change in percent IL-2 expression between the first stimulation and third stimulation for gRNA IL-2_H, gRNA IL-2 gRNA-1 and gRNA SpNT as well as other gRNAs. FIG.29B shows the fold IL-2+ CAR T cells over the non-targeting gRNA spNT between the first stimulation and third stimulation for gRNA IL-2_H, gRNA IL-2 gRNA-1 and other gRNAs. Data points representative of gRNA IL-2_H and gRNA IL-2 gRNA-1 are denoted. The open circle data points represent data from using the gRNA spNT. [0198] FIG.30 shows the mean fluorescence intensity (MFI) of Her2 CAR T cells that transiently expressed dCas9 effector fusion proteins for activation of IL-2 targeted by guide RNA IL-2_H compared to other guides. Data points of gRNA IL-2_H and gRNA IL-2 gRNA-1 are denoted. The open circle data points represent data from using the gRNA spNT. [0199] FIG.31A shows the fold increase in IL-2+ cells (which is the ratio of the absolute count of edited CAR+ IL-2+ cells to the absolute count of NT control, CAR+ IL-2+ cells at the third round of serial killing) against the IL-2 activation durability in an exemplary donor. FIG. 31B and 31C similarly show the fold increase in IL-2+ cells against the IL-2 activation durability in two additional exemplary donors. Select data points representing select combinations of gRNAs are highlighted for each exemplary donor. sf-6059407 22474-20028.40 [0200] FIG.32A shows percent IL-2 expression following a first stimulation of Her2 CAR T cells that were delivered mRNA encoding a dSpCas9-2xVP64 effector fusion protein and various SpCas9 IL-2-targeting gRNA or a dSaCas9-2xVP64 effector fusion protein and various SaCas9 IL-2-targeting gRNA. FIG.32B shows percent IL-2 expression following a second stimulation of the same Her2 CAR T cells shown in FIG.32A. [0201] FIG.33A shows IL-2 expression following stimulation of Her2 CAR T cells that were delivered mRNA encoding a dSpCas9-KRAB-DNMT3A/L effector fusion protein, an SpCas9 MED12-targeting gRNA, and mRNA encoding a dSpCas9-2xVP64 effector fusion protein. [0202] FIG.33B shows IL-2 expression following stimulation of Her2 CAR T cells that were delivered mRNA encoding a dSpCas9-KRAB-DNMT3A/L effector fusion protein, an SpCas9 IL-2-targeting gRNA, and mRNA encoding a dSpCas9-2xVP64 effector fusion protein. [0203] FIG.34A and FIG.34B depict schemes for combined activation of a first gene, e.g., IL-2, and repression of a second gene, e.g., MED12, in order to increase T cell effector function. The genes can be targeted using CRISPR/Cas-based DNA-targeting systems in which a dCas protein from one species, e.g., dSaCas9, is used for targeting the first gene for activation, and a dCas protein from a different species, e.g., dSpCas9, is used for targeting the second gene for repression. Increased T cell effector function can include increased CD25 expression, such as via MED12 repression. Increased T cell effector function can also include improved T cell proliferation, such as via IL-2 signaling in the JAK/STAT pathway. Together, simultaneous activation of IL-2 and repression of MED12 can be used in some aspects to amplify homeostatic feedback and sustain enhanced T cell function. [0204] FIG.34C shows IL-2 expression following stimulation of Her2 CAR T cells that were delivered mRNA encoding an dSpCas9-KRAB-DNMT3A/L effector fusion protein, an SpCas9 MED12-targeting gRNA, mRNA encoding an dSaCas9-2xVP64 effector fusion protein, and the SaCas9 IL-2-targeting gRNA IL-2_X. [0205] FIG.34D shows IL-2 expression in terms of IL-2 frequency (left) and IL-2 median fluorescent intensity (MFI; right) following stimulation of Her2 CAR T cells that were delivered mRNA encoding an dSpCas9-KRAB-DNMT3A/L effector fusion protein, the SpCas9 MED12- targeting gRNA MED12_7, mRNA encoding an dSaCas9-2xVP64 effector fusion protein, and the SaCas9 IL-2-targeting gRNA IL-2_X. sf-6059407 22474-20028.40 [0206] FIG.34E shows IL-2 expression in terms of % IL-2 postiive cells (left) and IL-2 median fluorescent intensity (MFI; right) following a first stimulation of Her2 CAR T cells that were delivered various combinations of the following: mRNA encoding a dSpCas9-KRAB- DNMT3A/L effector fusion protein, the SpCas9 MED12-targeting gRNA MED12_3, mRNA encoding a dSaCas9-2xVP64 effector fusion protein, and one or both of the SaCas9 IL-2- targeting gRNA IL-2_U and the SaCas9 IL-2-targeting gRNA IL-2_X. FIG.34F shows IL-2+ cell counts following a second stimulation (re-challenge) of the Her2 CAR T cells. [0207] FIG.35 depicts two exemplary dSaCas9 fusion proteins for transcriptional activation: dSaCas9 covalently linked to effector domains NCOA3-FOXO3-NCOA3 (NFN) and VP64 (dSaCas9-NFN-VP64) on the left and dSaCas9 covalently linked to 2 VP64 domains (dSaCas9-2xVP64) on the right. [0208] FIG.36 shows the intracellular expression of IL-2 after first stimulation using an average of two donors as %IL-2 positive cells in the left panel and mean fluorescent levels (MFI; corresponding to average expression levels) in the right panel, in cells that were delivered mRNA encoding dSaCas9-2xVP64, dSaCas9-VP64-NFN, or dSpCas9-2xVP64 mRNA with corresponding IL-2 targeting gRNA(s). No delivery of exemplary fusion protein (CAR only) was used as a negative control. [0209] FIG.37 shows fold IL-2+ Her2 CAR T cell expansion following two (left) or three (right) rounds of stimulation after delivery of various combinations of the following: mRNA encoding a dSpCas9-KRAB-DNMT3A/L effector fusion protein, the SpCas9 MED12-targeting gRNA MED12_3, SpCas9 CISH-targeting gRNA, the SpCas9 TGFBR2-targeting gRNA, mRNA encoding the dSaCas9-NFN-VP64 effector fusion protein, SaCas9 IL-2-targeting gRNAs, and/or non-targeting gRNA (SaNT). No delivery of fusion proteins (CAR only) was also used as a negative control. [0210] FIG.38A and FIG.38B show proliferation of Her2 CAR T cells after a first (top) or second (bottom) stimulation in a first donor (FIG.38A) or second donor (FIG.38B) after delivery of various combinations of the following: mRNA encoding a dSpCas9-KRAB- DNMT3A/L effector fusion protein, SpCas9 MED12-targeting gRNA, SpCas9 CISH-targeting gRNA, SpCas9 TGFBR2-targeting gRNA, mRNA encoding the dSaCas9-NFN-VP64 effector fusion protein, SaCas9 IL-2-targeting gRNAs, and/or non-targeting gRNA (SaNT). No delivery of fusion proteins (CAR alone) and mock delivery (mock) were also used as negative controls. sf-6059407 22474-20028.40 [0211] FIG.39 shows tumor cell killing (i.e., cytotoxic activity) of tumors when incubated with Her CAR T cells derived from a first (top row) or second (bottom row) donor following a first (left), second (middle), or third (right) stimulation after delivery of various combinations of the following: mRNA encoding a dSpCas9-KRAB-DNMT3A/L effector fusion protein, SpCas9 MED12-targeting gRNA, SpCas9 CISH-targeting gRNA, the SpCas9 TGFBR2-targeting gRNA, mRNA encoding the dSaCas9-NFN-VP64 effector fusion protein, SaCas9 IL-2-targeting gRNAs, and/or non-targeting gRNA (SaNT). No delivery of fusion proteins (CAR alone) and mock delivery (mock) were also used as negative controls. [0212] FIG.40 shows viability of T cells 72 hours post-delivery of various combinations of the following: mRNA encoding a dSpCas9-KRAB-DNMT3A/L effector fusion protein, SpCas9 MED12-targeting gRNA, SpCas9 CISH-targeting gRNA, the SpCas9 TGFBR2-targeting gRNA, mRNA encoding the dSaCas9-NFN-VP64 effector fusion protein, SaCas9 IL-2-targeting gRNAs, and/or non-targeting gRNA (SaNT or SpNT). No delivery of fusion proteins (CAR alone) and mock delivery (mock) were also used as negative controls. As additional controls, cells were also delivered : (a) mRNA encoding dSpCas9-2xVP64, and gRNA IL-2_H or (b) mRNA encoding dSaCas9-2xVP64, and gRNAs IL-2_U and IL-2_X, and mRNA encoding dSpCas9-KRAB-DNMT3A/L, and gRNA MED12_3 (IL-2 dSaCas9-2xVP64 + MED12). [0213] FIGS. 41-45 show alignments of a subset of designed engineered zinc finger protein (ZFP) target sites for a given target region: IL-2 gene region 4 (FIG.41), IL-2 region 5 (FIG. 42), IL-2 transcription start site (TSS; FIG.43), MED-12 TSS (FIG.44), and TGFBR2 TSS (FIG.45). Each solid rectangle represents a given target site. Top-performing ZFPs (see Table E17) and guide RNAs (gRNAs) are indicated by a box. [0214] FIG.46 shows the expression of IL-2 following delivery of the top-performing IL-2- targeting ZFP fusion proteins set forth in Table E17, dSpCas9-2xVP64 and gRNA IL2_H (SpCas9), or no delivery (CAR alone). IL-2 expression was represented as % IL-2 positivity following a first stimulation 72 hours post-delivery (top) or cell counts of IL-2-positive CAR T- cells following a second stimulation (bottom). [0215] FIGS. 47A and 47B show the expression of IL-2 and TGFBR2 following delivery of the top-performing TGFBR2 ZFP fusion protein set forth in Table E17, dSpCas9-KRAB- DNMT3A/L and gRNA TGFB2_2 (SpCas9), or no delivery (CAR alone). In addition to the fusion protein sequence listed in Table E17, each TGFBR2 fusion protein also contained a 3x FLAG tag (SEQ ID NO: 287) on its N-terminus. IL-2 expression was represented as % IL-2 sf-6059407 22474-20028.40 positive CAR T cells following a second stimulation (FIG.47A) and expression of TGFBR2 as % knockdown (KD) of TGFBR2 after either 72 hours post-delivery (FIG.47B; left) or 15 days post-delivery (FIG.47B; right), as assessed by qRT-PCR. [0216] FIGS. 48A and 48B show the expression of IL-2 and CD25 following delivery of the top-performing MED12 ZFPs set forth in Table E17, dSpCas9-KRAB-DNMT3A/L and gRNA MED12_3 (SpCas9), or no delivery (CAR alone). In addition to the fusion protein sequence listed in Table E17, each MED12 fusion protein also contained a 3x FLAG tag (SEQ ID NO: 287) on its N-terminus. IL-2 expression is represented as % IL-2 positive CAR T cells following a second stimulation (FIG.48A) and expression of CD25 as % positivity after either 7 days post-delivery (FIG.48B; top) or 15 days post-delivery (FIG.48B; bottom), as assessed by flow cytometery. [0217] FIG.49A-C show overall cell count 72 hours post-delivery of top-performing IL-2- targeting ZFP fusion proteins (FIG.49A), the top-performing TGFBR2-targeting ZFP fusion protein (FIG.49B), and top-performing MED12-targeting ZFP fusions (FIG.49C) as set forth in Table E17. As controls, cells were also delivered a dSpCas9 fusion protein (dSpCas9- 2xVP64 for transcriptional activation of IL-2; dSpCas9-KRAB-DNMT3A/L for transcriptional repression of TGFBR2 or MED12) and a corresponding gRNA or cells did not receive any fusion proteins (CAR alone). [0218] FIG.50A-C shows the expression of IL-2, shown by % IL-2 positive cells, after a first stimulation (FIG.50A) or number of IL-2 positive cells, normalized to CAR alone, in the presence of TGF-beta (TGFb) after a second (FIG.50B) or third (FIG.50C) stimulation using fresh cells following delivery of mRNA encoding (a) dSpCas9-KRAB-DNMT3A/L and TGFBR2-, MED12-, and CISH-targeting gRNAs, (b) dSpCas9-KRAB-DNMT3A/L and TGFBR2- and MED12-targeting gRNAs, (c) dSpCas9-KRAB-DNMT3A/L and CISH- and MED12-targeting gRNAs, (d) dSpCas9-KRAB-DNMT3A/L and TGFBR2-, MED12-, and CISH-targeting gRNAs paired with dSaCas9-NFN-VP64 with IL-2-targeting gRNA (referred to as IL2 in figures), (e) dSpCas9-KRAB-DNMT3A/L and TGFBR2- and MED12-targeting gRNAs paired with dSaCas9-NFN-VP64 with IL-2-targeting gRNA, (f) dSpCas9-KRAB- DNMT3A/L and a TGFBR2-targeting gRNA paired with dSaCas9-NFN-VP64 with IL-2- targeting gRNA, (g) dSpCas9-KRAB-DNMT3A/L and a MED12-targeting gRNA paired with dSaCas9-NFN-VP64 with IL-2-targeting gRNA, (h) dSpCas9-KRAB-DNMT3A/L and TGFBR2-, MED12-, and CISH-targeting gRNAs paired with either IL2_R4_B ZFP fusion sf-6059407 22474-20028.40 protein or IL2_R5_A ZFP fusion protein, (i) TGFBR2_A ZFP fusion protein paired with activation of IL-2 using dSpCas9-2xVP64 and IL-2-targeting gRNA (IL2 dSpCas9-2xVP64), or (j) multiplexed repression of TGFBR2, MED12, and CISH using dSpCas9-KRAB-DNMT3A/L paired with activation of IL-2 using dSaCas9-NFN with corresponding gRNAs (TGFBR2+MED12+CISH+IL2-NFN). As controls, cells were also delivered: (a) dSaCas9-NFN with a non-targeting gRNA SaNT (SEQ ID NO: 439), (b) dSaCas9-NFN-VP64 with a non- targeting gRNA SaNT (SEQ ID NO: 439), (c) no delivery of fusion protein (CAR only), or (d) mock delivery (mock). [0219] FIGS. 51A-C show the level of polyfunctional T cells after a first (FIG.51A), second (FIG.51B), or third simulation (FIG.51C) using fresh cells and after delivery of the same constructs as described for FIG.51A-C. [0220] FIG.52 shows frequency of CAR T cells after each stimulation in cells derived from the first donor (left) or second donor (right) using frozen cells and after delivery of the same constructs as described for FIG.50A-C. [0221] FIG.53 shows a schematic of two dual ZFP fusion protein constructs, Construct 1 and Construct 2, which each contain two ZFP fusion proteins separated by a P2A sequence. One ZFP fusion protein contains a MED12-targeting ZFP fused to DNMT3A-DNMT3L (DNMT3A- 3L) and a KRAB domain, while the second ZFP fusion protein contains a IL-2-targeting ZFP fused to effector domains NCOA3-FOXO3-NCOA3 (NFN). [0222] FIG.54A and FIG.54B shows expression of IL-2, as measured by qPCR, in CAR T cells derived from Donor 1 (FIG.54A) or Donor 2 (FIG.54B) one day post-delivery via electroporation of the indicated constructs expressing both ZFP fusion proteins (Construct 1 or Construct 2), a construct expressing one ZFP fusion protein (IL2_R5_A ZFP FP or MED12_A ZFP FP), a Cas9 fusion protein paired with either a MED12-targeting gRNA (MED12_3) or IL- 2-targeting gRNA (IL2_J), or electroporation of two constructs, each expressing one ZFP fusion fusion, at a concentration of either 75 ug/mL or 37.5 ug/mL each. As a control, a mock electroporation (mock EP) was also performed. [0223] FIG.55 shows expression of MED12, as measured by qPCR, in CAR T cells derived from one of three indicated donors two days post-delivery via electroporation of the indicated constructs expressing both ZFP fusion proteins (Construct 1 or Construct 2), a construct expressing one ZFP fusion protein (IL2_R5_A ZFP FP or MED12_A ZFP FP), a Cas9 fusion protein paired with either a MED12-targeting gRNA (MED12_3) or IL-2-targeting gRNA sf-6059407 22474-20028.40 (IL2_J), or electroporation of two constructs, each expressing one ZFP fusion fusion, at a concentration of either 75 ug/mL or 37.5 ug/mL each. As a control, a mock electroporation (mock EP) was also performed. [0224] FIG.56 shows the amount of IL-2+ CAR T cells (as indicated by the presence of surrogate EGFR) per microliter following a third stimulation and delivery of one of the indicated constructs expressing both ZFP fusion proteins (Construct 1 or Construct 2), a construct expressing one ZFP fusion protein (IL2_R5_A ZFP FP or MED12_A ZFP FP), a Cas9 fusion protein paired with either a MED12-targeting gRNA (MED12_3) or IL-2-targeting gRNA (IL2_J), or electroporation of two constructs, each expressing one ZFP fusion fusion, at a concentration of either 75 ug/mL or 37.5 ug/mL each. As a control, a mock electroporation (mock EP) was also performed. Detailed Description [0225] Provided herein in some embodiments is an epigenetic-modifying DNA-targeting system. In some embodiments, the DNA-targeting system is a multiplexed DNA-targeting system, i.e., targeted to target sites for multiple genes. In some embodiments, the DNA-targeting system comprises synthetic transcription factors that are able to modulate, such as decrease (or downregulate) or increase (or upregulate), transcription of multiple genes in a targeted manner. In some embodiments, the DNA-targeting system comprises at least one first DNA-targeting module (also referred to herein as an activator DNA-targeting module) for increasing transcription of one or more first genes (also referred to herein as activation genes) and at least one second DNA-targeting module (also referred to herein as an repressor DNA-targeting module) for repressing transcription of one or more second genes (also referred to herein as repression genes). In some embodiments, the one or more second genes are all different from the one or more first genes. [0226] In some embodiments, at least one, optionally each, of the first DNA-targeting modules comprises a first fusion protein comprising (i) a first DNA-binding domain for targeting to a target site of one of the one or more first genes and (ii) at least one transcriptional activator domain. In some embodiments, at least one, optionally each, of the second DNA- targeting modules comprises a second fusion protein comprising (i) a second DNA-binding domain for targeting to a target site of one of the one or more second genes and (ii) at least one sf-6059407 22474-20028.40 transcriptional repressor domain. In some embodiments, the first DNA-binding domain or domains are all different from the second DNA-binding domain or domains. [0227] Also provided herein are polynucleotides encoding the DNA-targeting system, as well as vectors and cells containing the provided polynucleotides. Also provided herein are methods of using the epigenetic-modifying DNA-targeting systems, such as for modulating transcription, phenotype, or function of cells, such as T cells, and the resulting modified cells. [0228] The provided embodiments relate to compositions and methods for promoting T cell function, such as one or more T cell effector functions, by epigenetically modifying target sites in multiple target genes. In some embodiments, the methods can be used in connection with T cell therapies, such as in connection with adoptive T cell therapies. In some embodiments, modulating transcription of the multiple genes increases or improves one or more T cell phenotypes or functions. In some embodiments, a T cell effector function is increased, such as the ability to produce cytokines, for example IL-2 or IFN-gamma (IFNg), the ability of T cells to proliferate, the ability of T cells to kill target cells, or the ability of T cells to exhibit a persistent immune response. In particular embodiments, modulation of the multiple genes improves T cell effector functions after or upon T cell stimulation, including following serial stimulation that mimics conditions of repeated antigen encounter as occurs in vivo. [0229] The administration of T cells targeting a specific antigen, also known as Adoptive Cell Therapy (ACT), is a promising approach for treating diseases such as cancer. However, current ACT treatments face challenges, including suboptimal T cell function, expansion, and persistence. Furthermore, the persistence and functionality of the transferred T cells can significantly differ between different T cell subsets and among T cells from different patients. Recent clinical trials for ACT suggest that the ability to persist long term in circulation is dependent on the differentiation stage of the T cells, including the ability to retain a network of transcription factors and metabolic regulators (Pilipow K., et.al., Journal of Clinical Investigation Insight 2018;3(18):e122299). The T cells transferred into the patient are often terminally differentiated and therefore fail to persist long term, ultimately limiting effective anti- tumor response. For instance, while the first CAR T cell therapy was FDA-approved as a cell & gene therapy in 2017, patients whose cancer relapse or do not respond to treatment often suffer from lack of chimeric antigen receptor (CAR) T cell persistence (Mueller et al, Blood (2018)). Moreover, no durable benefit has yet been observed for CAR T cell therapies in solid tumors. [0230] Strategies to mitigate these challenges and enhance the persistence, expansion, and sf-6059407 22474-20028.40 anti-tumor activity of CAR engineered T cells have been tested in preclinical and clinical settings. For instance, strategies for optimizing ex vivo T cell culture conditions, including the addition of cytokines during manufacturing (Besser M.J., Cytotherapy 2009;11(2):206-17), expression of cytokines and/or receptors by the CAR T cells (Krenciute G., Cancer Immunol Res. 201707;5(7):571-581), use of pharmacological inhibitors during expansion to inhibit signaling pathways such as AKT (Urak R.et.al., Journal of Immunotherapy Cancer 2017 Mar 21;5:26) or PI3K (Peterson C.T et.al., Blood Advances 2018 Feb 13;2(3):210-223), immune- depletion, and checkpoint blockade (Cherkassky L. et.al., Journal of clinical investigation 2016 Aug 1;126(8):3130-44), have been so far explored. However, existing strategies have not been entirely satisfactory. In some cases, concerns regarding cytokine-induced toxicity or the emergence of lymphoproliferative diseases as a result of the above-mentioned strategies have raised questions for alternative approaches. [0231] The provided embodiments relate to identification of genomic locations that are epigenetically modified in a T cell to impact or promote T cell effector functions upon T cell stimulation, including those induced in a TCR and/or CAR-induced or dependent manner, such as demonstrated by assessment for cells producing IL-2 and/or IFNg, having the ability to proliferate, or having the ability to kill target cells. In some embodiments, the stimulating conditions or agents include one or more agents, e.g., ligands, which are capable of activating an intracellular signaling domain of a TCR complex. In some aspects, the agent turns on or initiates TCR/CD3 intracellular signaling cascade in a T cell. Such agents can include antibodies, such as those specific for a TCR component and/or costimulatory receptor, e.g., anti-CD3, anti-CD28, for example bound to a solid support such as a bead, and/or one or more cytokines. In some embodiments, the one or more agents are PMA and ionomycin. In some embodiments, the T cell stimulation is an antigen-specific stimulation, in which the cells are stimulated with an agent providing an antigen or epitope thereof that is specific to, or recognized by, an antigen receptor (e.g., CAR) expressed on the T cell. For instance, the stimulating agent may include antigen- expressing target cells. In particular embodiments, the phenotype is or includes the production or secretion of a cytokine, such as IL-2 or IFN-g, in response to a T cell stimulation. The production and/or the secretion of cytokines contributes to immune responses and is involved in different processes, including the induction of anti-viral proteins and the induction of T cell proliferation. Cytokines are not pre-formed factors but are rapidly produced and secreted in response to cellular activation. The production or secretion of cytokines may be measured, sf-6059407 22474-20028.40 detected, and/or quantified by any suitable technique known in the art. [0232] In certain embodiments, the T cell function is the production of one or more cytokines. In particular embodiments, the production of one or more cytokines is measured, detected, and/or quantified by intracellular cytokine staining (ICS). ICS by flow cytometry is a technique well-suited for studying cytokine production at the single-cell level. It detects the production and accumulation of cytokines within the cell (such as within the endoplasmic reticulum) after cell stimulation, allowing for the identification of cell populations that are positive or negative for production of a particular cytokine or for the separation of high producing and low producing cells based on a threshold. ICS can also be used in combination with other flow cytometry protocols for immunephenotyping using cell surface markers or with MHC multimers to access cytokine production in a particular subgroup of cells, making it a flexible and versatile method. Other single-cell techniques for measuring or detecting cytokine production include ELISPOT, limiting dilution, and T cell cloning. [0233] Notably, the target genes, and target sites therein, of the present disclosure were identified by a screening method involving transient delivery in which DNA-binding domain- effector fusion proteins (also called “epi-editors”) were delivered to a T cell transiently (i.e., delivered by a method that results in transient expression and/or presence of the fusion protein in the T cell) followed by primary or serial stimulation of the T cells to assess impact on functional T cell cytokines. It was found herein that the transient delivery of the epigenetic-modifying DNA-targeting systems allowed identification of genomic targets whose modulation substantially impacts T cell function, but without requiring permanent presence of the epigenetic-modifying DNA-targeting systems and/or stable knockdown or knockout of the target gene. This approach is advantageous because it permits identification of target genes and target sites that provide a better safety profile, as their modulation is not reliant on a permanent editor integration, such as by lentiviral transduction. Moreover, the transient screening strategies allow for identification of target genes and target sites therein in which there is a durability of the effect of the epigenetic-modifying DNA-targeting system that is not masked as a result of permanent integration into the genome and expression therefrom. This is in contrast to other screening approaches in which lentiviral delivery of DNA-systems has been employed (Schmidt et al.2022 Science, 375, DOI: 10.1126/science.abj4008; Freimer et al.2022 Nature Genetics, 54:1133-1144). [0234] The provided embodiments can be used to target genes that, when transcriptionally sf-6059407 22474-20028.40 altered by epigenetic modification, can vastly facilitate or promote T cell function, including effector activities required for T cell persistence and function. Such a T cell profile is expected to produce durable effector functions, have better fitness/proliferation benefit, and have the ability to produce pro-proliferation cytokines (e.g., IL-2) and/or cytotoxic cytokines (e.g., IFNg) upon TCR or antigen stimulation. In particular, the provided embodiments provide for epigenetic-modifying DNA-targeting systems (i.e., “epi-editing systems”) and methods that can provide for long-lasting effector function with better fitness. This approach offers substantial clinical solutions to circumvent the problems with T cell persistence, suboptimal functionality, and/or exhaustion. Moreover, the epigenetic modification of the cell does not modify DNA at the sequence level, thereby avoiding safety concerns with gene editing approaches. The ability to epigenetically control the differentiation fate of T cells provides an advantageous approach for increasing the percentage or number of T cells in a population of T cells. [0235] The provided embodiments can also be used to target multiple genes, with one or more first genes targeted for increased transcription and one or more second genes targeted for repressed transcription. In some aspects, T cell function is improved by such combined targeting, relative to any improvements in T cell function achieved by only targeting genes for increased transcription or only targeting genes for decreased transcription. In some embodiments, combined transcriptional activation and repression is affected by using different DNA-binding domains between the first DNA-targeting modules used for increasing transcription and the second DNA-targeting modules used for repressing transcription. In some embodiments, the DNA-binding domains for the first DNA-targeting modules do not bind to the target sites for the one or more second genes, and vice versa. For instance, the DNA-binding domains may be different zinc finger proteins (ZFPs) or transcription activator-like effectors (TALEs) between the first and second DNA-targeting modules. In some embodiments, the DNA-binding domains comprise Cas proteins, and the Cas proteins for the first DNA-targeting modules do not bind to gRNAs targeting the one or more second genes, and vice versa. In some embodiments, the Cas proteins for the first DNA-targeting modules are from a different species than the Cas proteins for the second DNA-targeting modules. In some embodiments, the Cas proteins for the first DNA-targeting modules bind to different protospacer-adjacent motifs (PAMs) than do the Cas proteins for the second DNA-targeting modules. In some aspects, the use of such non-overlapping DNA-binding domains between the first DNA-targeting modules used for increasing transcription and the second DNA-targeting modules used for repressing sf-6059407 22474-20028.40 transcription prevent off-target effects that may lead to, for instance, increased transcription of a gene intended to be repressed, and vice versa. [0236] All publications, including patent documents, scientific articles and databases, referred to in this application are incorporated by reference in their entirety for all purposes to the same extent as if each individual publication were individually incorporated by reference. If a definition set forth herein is contrary to or otherwise inconsistent with a definition set forth in the patents, applications, published applications and other publications that are herein incorporated by reference, the definition set forth herein prevails over the definition that is incorporated herein by reference. [0237] The section headings used herein are for organizational purposes only and are not to be construed as limiting the subject matter described. I. DNA-TARGETING SYSTEMS [0238] In some embodiments, provided herein are epigenetic-modifying DNA-targeting systems capable of specifically targeting a target site for each of multiple genes (i.e., multiple target genes) and modulating transcription of the target genes. In some embodiments, transcriptional modulation of gene expression by the DNA-targeting system promotes or improves function of lymphoid cells. In some embodiments, the DNA-targeting system promotes T cell function, such as one or more T cell effector functions, by epigenetically modifying target sites in the genes. [0239] In some embodiments, the target genes include one or more first genes (also referred to herein as activation genes) for which transcription is activated and one or more second genes (also referred to herein as repression genes) for which transcription is repressed. In some embodiments, the transcriptional modulation is increased transcription for the one or more first genes, and the transcriptional modulation is decreased transcription for the one or more second genes. In some embodiments, the one or more first genes are all different from the one or more second genes. In some embodiments, the target genes include a single first gene. In some embodiments, the target genes include a plurality of first genes, such as at least 2, at least 3, at least 4, at least 5, at least 6, at least 7, at least 8, at least 9, at least 10, at least 11, or at least 12 first genes for which transcription is increased. In some embodiments, the target genes include a single second gene. In some embodiments, the target genes include a plurality of second genes, such as at least 2, at least 3, at least 4, at least 5, at least 6, at least 7, at least 8, at least 9, at least 10, at least 11, or at least 12 second genes. sf-6059407 22474-20028.40 [0240] In some embodiments, the one or more first genes are T cell activator genes. In some embodiments, the one or more first genes, e.g., BATF, CD28, EOMES, IL-2, IL2RB, IRF4, LAT, LCP2, TBX21, and/or VAV1, are positive regulators of T cell function. [0241] In some embodiments, the one or more second genes are T cell repressor genes. In some embodiments, the one or more second genes, e.g., CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and/or RASA2, are negative regulators of T cell function. [0242] In some embodiments, the DNA-targeting system includes at least one first DNA- targeting module (also referred to herein as an activator DNA-targeting module) for increasing transcription of the one or more first genes and at least one second DNA-targeting module (also referred to herein as an repressor DNA-targeting module) for repressing transcription of the one or more second genes. In some embodiments, the DNA-targeting system includes a single first DNA-targeting module. In some embodiments, the DNA-targeting system includes a plurality of first DNA-targeting modules, such as at least 2, at least 3, at least 4, at least 5, at least 6, at least 7, at least 8, at least 9, at least 10, at least 11, or at least 12 first DNA-targeting modules. In some embodiments, the DNA-targeting system includes a single second DNA-targeting module. In some embodiments, the DNA-targeting system includes a plurality of second DNA-targeting module, such as at least 2, at least 3, at least 4, at least 5, at least 6, at least 7, at least 8, at least 9, at least 10, at least 11, or at least 12 second DNA-targeting modules. [0243] In some embodiments, each first DNA-targeting module includes a first fusion protein containing (i) a first DNA-binding domain for targeting to a target site of one of the one or more first genes and (ii) at least one transcriptional activator domain. In some embodiments, the first DNA-binding domain or domains target a plurality of first genes. In some embodiments, the first DNA-binding domain or domains target a single first gene. In some embodiments, the first DNA-binding domain is the same across the at least one first DNA-targeting module. In some embodiments, the first DNA-binding domain is different across the at least one first DNA- targeting module. In some embodiments, the at least one transcriptional activator domain is the same across the at least one first DNA-targeting module. In some embodiments, the at least one transcriptional activator domain is different across the at least one first DNA-targeting module. [0244] In some embodiments, the at least one transcriptional activator domain activates or increases transcription of a first gene as compared to transcription of the first gene in the absence of the DNA-targeting system. In some embodiments, the at least one transcriptional sf-6059407 22474-20028.40 activator domain directly or indirectly leads to increased transcription of the first gene. In some embodiments, the at least one transcriptional activator domain induces, catalyzes, or leads to transcription activation. In some embodiments, the at least one transcriptional activator domain induces transcription activation. In some aspects, the at least one transcriptional activator domain comprises a VP64 domain, a p65 activation domain, a p300 domain, an Rta domain, a CBP domain, a VPR domain, a VPH domain, an HSF1 domain, a TET protein domain, optionally wherein the TET protein is TET1, a SunTag domain, or a domain, portion, variant, or truncation of any of the foregoing. In some embodiments, the at least one transcriptional activator domain is VP64. [0245] In some embodiments, at least one, optionally each, of the first fusion proteins is a dCas9-VP64 fusion protein. In some embodiments, at least one, optionally each, of the first fusion proteins is a dCas9-2xVP64 fusion protein. [0246] In some embodiments, each second DNA-targeting module includes a second fusion protein containing (i) a second DNA-binding domain for targeting to a target site of one of the one or more second genes and (ii) at least one transcriptional repressor domain. In some embodiments, the second DNA-binding domain or domains target a plurality of second genes. In some embodiments, the second DNA-binding domain or domains target a single second gene. In some embodiments, the second DNA-binding domain is the same across the at least one second DNA-targeting module. In some embodiments, the second DNA-binding domain is different across the at least one second DNA-targeting module. In some embodiments, the at least one transcriptional repressor domain is the same across the at least one second DNA-targeting module. In some embodiments, the at least one transcriptional repressor domain is different across the at least one second DNA-targeting module. [0247] In some embodiments, the at least one transcriptional repressor domain inhibits or reduces transcription of a second gene as compared to transcription of the second gene in the absence of the DNA-targeting system. In some embodiments, the at least one transcriptional repressor domain directly or indirectly leads to reduced transcription of the second gene. In some embodiments, the at least one transcriptional repressor domain induces, catalyzes, or leads to transcription repression. In some embodiments, the at least one transcriptional repressor domain induces transcription repression. In some aspects, the at least one transcriptional repressor domain is selected from a KRAB domain, ERF repressor domain, MXI1 domain, SID4X domain, MAD-SID domain, DNMT family protein domain (e.g., DNMT3A or sf-6059407 22474-20028.40 DNMT3B), a fusion of one or more DNMT family proteins or domains thereof (e.g., DNMT3A/L, which comprises a fusion of DNMT3A and DNMT3L domains), LSD1, EZH2, a partially or fully functional fragment or domain of any of the foregoing, and a combination of any of the foregoing. In some embodiments, the at least one transcriptional repressor domain is KRAB. In some embodiments, the at least one transcriptional repressor domain is DNMT3A/L. [0248] In some embodiments, at least one, optionally each, of the second fusion proteins is a dCas9-KRAB fusion protein. In some embodiments, at least one, optionally each, of the second fusion proteins is a dCas9-KRAB-DNMT3A/L fusion protein. [0249] In some embodiments, the first and/or second DNA-binding domain comprises a CRISPR associated (Cas) protein, a zinc finger protein (ZFP), a transcription activator-like effector (TALE), meganuclease, homing endonuclease, I-SceI enzyme, or variants thereof. In some embodiments, the first and/or second DNA-binding domain comprises a catalytically inactive (e.g., nuclease-inactive or nuclease-inactivated) variant of any of the foregoing. In some embodiments, the first and/or second DNA-binding domain comprises a deactivated Cas9 (dCas9) protein that is a catalytically inactivated so that it is inactive for nuclease activity and is not able to cleave the DNA. [0250] In some embodiments, a first and/or second DNA-targeting module is a CRISPR/Cas-based DNA-targeting module. In some embodiments, the first and/or second DNA-binding domain comprises a Cas protein, such as a nuclease-inactive Cas or dCas, e.g., dCas9, and the DNA-targeting module comprising the Cas protein comprises one or more guide RNAs (gRNAs), such as a combination of gRNAs (e.g., two gRNAs or three gRNAs). In some embodiments, the gRNA comprises a spacer sequence that is capable of targeting and/or hybridizing to a target site. In some embodiments, the gRNA is capable of complexing with the Cas protein. In some aspects, the gRNA directs or recruits the Cas protein to the target site. [0251] In some embodiments, at least one, optionally each, of the first DNA-binding domains is a first Cas protein, and the first DNA-targeting module comprising the first Cas protein further comprises a first gRNA for targeting the first DNA-binding domain to a target site of one of the one or more first genes. In some embodiments, at least one, optionally each, of the second DNA-binding domains is a second Cas protein, and the second DNA-targeting module comprising the second Cas protein further comprises a second gRNA for targeting the second DNA-binding domain to a target site of one of the one or more second genes. [0252] In some embodiments, the first DNA-binding domain or domains are all different sf-6059407 22474-20028.40 from the second DNA-binding domain or domains. In some embodiments, the first DNA- binding domain or domains do not bind to the target sites for the one or more second genes, and vice versa. In some embodiments, the first DNA-binding domain or domains are ZFPs, and the second DNA-binding domain or domains are different ZFPs. In some embodiments, the first DNA-binding domain or domains are TALEs, and the second DNA-binding domain or domains are different TALEs. In some embodiments, the first DNA-binding domain or domains are ZFPs, and the second DNA-binding domain or domains are TALEs. In some embodiments, the first DNA-binding domain or domains are TALEs, and the second DNA-binding domain or domains are ZFPs. In some embodiments, the first DNA-binding domain or domains are Cas proteins, and the second DNA-binding domain or domains are ZFPs. In some embodiments, the first DNA-binding domain or domains are ZFPs, and the second DNA-binding domain or domains are Cas proteins. In some embodiments, the first DNA-binding domain or domains are Cas proteins, and the second DNA-binding domain or domains are TALEs. In some embodiments, the first DNA-binding domain or domains are TALEs, and the second DNA- binding domain or domains are Cas proteins. [0253] In some embodiments, the first and second DNA-binding domains are both Cas poteins. In some embodiments, the first Cas protein or proteins for the first DNA-targeting modules do not bind to gRNAs targeting the one or more second genes, and vice versa. In some embodiments, the first Cas protein or proteins for the first DNA-targeting modules are from a different species than the second Cas protein or proteins for the second DNA-targeting modules. In some embodiments, the first Cas protein or proteins for the first DNA-targeting modules bind to different protospacer-adjacent motifs (PAMs) than do the second Cas protein or proteins for the second DNA-targeting modules. [0254] Exemplary components and features of the DNA-targeting systems are provided below in the following subsections. A. Target Genes and Target Sites For Promoting Lymphoid Activation and Function [0255] In some embodiments, modulation of the target genes promotes T cell activation or function. In some embodiments, increased transcription of the one or more first genes and repressed transcription of the one or more second genes promotes T cell activation or function. [0256] In some embodiments, the target site for a gene is a target site in the gene or a regulatory DNA element thereof. In some embodiments, the target site for each gene is a target sf-6059407 22474-20028.40 site in the gene. In some embodiments, the target site for each gene is a target site in a regulatory DNA element of the gene. In some embodiments, the target sites across genes are a mix of target sites in a gene and target sites in a regulatory DNA element of a gene. In some embodiments, the target genes are genes in a lymphoid cell, such as a T cell. [0257] In some embodiments, a regulatory DNA element is a sequence to which a gene regulatory protein may bind and affect transcription of the gene. In some embodiments, the regulatory DNA element is a cis, trans, distal, proximal, upstream, or downstream regulatory DNA element of a gene. In some embodiments, the regulatory DNA element is a promoter or enhancer of the gene. In some embodiments, the target site is located within a promoter, enhancer, exon, intron, untranslated region (UTR), 5’ UTR, or 3’ UTR of the gene. In some embodiments, the regulatory DNA element is a promoter. In some embodiments, a promoter is a nucleotide sequence to which RNA polymerase binds to begin transcription of the gene. In some embodiments, a promoter is a nucleotide sequence located within about 100bp, about 500bp, or about 1000bp of a transcriptional start site of the gene. In some embodiments, a promoter is within 500bp of a transcriptional start site of the gene. In some embodiments the target site is located within a sequence of unknown or known function that is suspected of being able to control expression of a gene. 1. Lymphoid Cells and Modulated Effector Functions [0258] In some embodiments, the provided DNA-targeting systems provide for transcriptional modulation to repress or increase expression of multiple target genes. In some embodiments, the target genes regulate a cellular phenotype, e.g., of a T cell. In some embodiments, modulated expression of the target genes promotes increased T cell effector function upon T cell stimulation. In some embodiments, the increased T cell effector function is increased compared to a T cell in which expression of the genes has not been modulated, e.g., not modulated with a provided DNA-targeting system. Methods for modulating T cell function or functions of other lymphoid cells by provided DNA-targeting systems are further described below and in Section IV. [0259] In some embodiments, the DNA-targeting system is transiently delivered to the cell. In some embodiments, delivery of the DNA-targeting system, for example by transient delivery, promotes increased T cell effector function upon T cell stimulation. In some embodiments, the T cell effector function is increased in comparison to a comparable T cell to which the DNA- targeting system has not been delivered. sf-6059407 22474-20028.40 [0260] In some aspects, transient delivery refers to any method of delivery that results in expression and/or presence of one or more components of the DNA-targeting system in the cell for a limited duration. For example, delivery of mRNA (such as by electroporation) encoding the fusion protein of the DNA-targeting system to a cell can result in transient expression of the fusion protein in the cell, for example until the mRNA is degraded. In other examples, the DNA- targeting system can be expressed from one or more nucleic acids encoding the DNA-targeting system, wherein the nucleic acids encoding the DNA-targeting system are not incorporated into the genome of the cell, and are eventually degraded and/or removed from the cell such that expression of the DNA-targeting system does not persist. In other examples, one or more components of the DNA-targeting system, such as a fusion protein and optionally a gRNA can be synthesized in vitro and delivered directly to the cell (e.g. by electroporation) without the need for an expression vector, resulting in transient presence of the DNA-targeting system, for example until the fusion protein and/or gRNA are degraded. In some aspects, transient delivery differs from non-transient methods of delivery that result in stable expression, such as methods involving incorporation of an expression vector for a DNA-targeting system or component thereof into the genome of the cell. [0261] In some embodiments, delivery of the DNA-targeting system to the cell (e.g. T cell), such as by transient delivery, promotes a phenotype in the cell (e.g. T cell). In some embodiments, the phenotype is increased activation or function in the cell (e.g. T cell). In some embodiments, delivery of the DNA-targeting system to the cell (e.g. T cell), such as by transient delivery, promotes increased activation or function in the cell (e.g. T cell). In some embodiments, the phenotype is increased T cell effector function upon T cell stimulation. In some embodiments, the T cell effector function is increased compared to a T cell that has not been delivered the epigenetic-modifying DNA-targeting system. In some embodiments, the T cell effector function is characterized by an activity selected from the group consisting of IL-2 production, IFN-gamma production, TNF-alpha production, T cell proliferation or a combination of any of the foregoing. [0262] In some embodiments, the provided DNA-targeting systems promote or increase an improved T cell effector function as may occur after stimulation in vitro, ex vivo, or in vivo. In some embodiments, the T cell stimulation is a polyclonal T cell stimulation. In some embodiments, the T cell stimulation is with an anti-CD3 and anti-CD28 activation reagent. In some embodiments, the T cell stimulation is an antigen-specific activity that is mediated or sf-6059407 22474-20028.40 induced by specific binding of an antigen to an antigen receptor on the surface of the T cell. In some embodiments, the T cell expresses a chimeric antigen receptor (CAR) or engineered T cell receptor (eTCR) directed against an antigen, and the T cell stimulation is an antigen-specific stimulation of the CAR or eTCR. In some embodiments, the T cell stimulation is with antigen- expressing target cells. In some embodiments, the T cell stimulation occurs when the T cell contacts a cell expressing the antigen. In some embodiments, the T cell stimulation is a restimulation after at least one prior T cell stimulation of the T cells. In some embodiments, the T cells are stimulated and then are transiently delivered a provided DNA-targeting system prior to assessment of a T cell effector function or phenotype. [0263] In certain embodiments, the cell composition that contains T cells is stimulated with an anti-CD3/anti-CD28 activation reagent for an amount of time, and an effector function is measured at one or more time points during or after the incubation. In some embodiments, such an activation reagent has anti-CD3/anti-CD28 coated on a support, such as magnetic beads or other matrix. Exemplary activation reagents include Dynabeads™ and T cell TransAct™. In some embodiments, the T cells are incubated with the activation reagent for 3 hours to 72 hours, such as 12 hours to 48 hours, for example 12 hours, 18 hours, 24 hours, 36 hours, or 48 hours, or any value between any of the foregoing. In some embodiments, cells can be assessed directly for an effector function, such as production of cytokines or ability to proliferate. In some embodiments, the supernatant of the culture can be collected, and the amount of a soluble factor, e.g., a cytokine, is detected. In some embodiments, the T cells can be collected and re-exposed to the activation reagent to monitor cytolytic activity. In some embodiments, cells can be restimulated one or more times, such as by serial stimulation methods, and serially assessed for effector functions after each stimulation. [0264] In certain embodiments, the antigen-specific activity is measured by incubating the cell composition that contains T cells expressing the antigen receptor, e.g., a CAR, with antigen- expressing cells for an amount of time, and an effector function is measured at one or more time points during or after the incubation. In some embodiments, the T cells are incubated with the antigen specific agent, such as antigen-expressing cells, for 3 hours to 96 hours, such as 12 hours to 72 hours, for example 12 hours, 24 hours, 48 hours, or 72 hours, or any value between any of the foregoing. In some embodiments, cells can be assessed directly for an effector function, such as production of cytokines or ability to proliferate. In some embodiments, the supernatant of the culture can be collected, and the amount of a soluble factor, e.g., a cytokine, is detected. n sf-6059407 22474-20028.40 some embodiments, the T cells can be collected and re-exposed to antigen-expressing target cells to monitor cell killing (cytolytic activity) of target cells. In some embodiments, cells can be restimulated one or more times, such as by serial stimulation methods, and serially assessed for effector functions after each stimulation. In some embodiments, the T cells with the engineered antigen receptor (e.g., a CAR) are incubated with a constant number of the antigen-expressing cells, such as at an effector to target (E:T) ratio of 1:4 to 4:1, such as at a ratio of 1:4, 1:3, 1:2, or 1:1. [0265] In some embodiments, the cell (e.g., T cell) exhibits increased cytokine production. In some embodiments, the increased cytokine production occurs upon T cell stimulation. In some embodiments, T cell effector function is characterized by cytokine production. In some embodiments, the cytokine production is increased by at least about 1.1 fold, 1.2 fold, 1.3 fold, 1.4 fold, 1.5 fold, 2 fold, 5 fold, 10 fold, 50 fold, or 100 fold in comparison to a cell that has not been delivered the epigenetic-modifying DNA-targeting system. In some embodiments, the cytokine production is production of IL-2, IFN-gamma, TNF-alpha, or a combination thereof. In some embodiments, the T cell effector function is characterized by IL-2 production. In some embodiments, the cell (e.g., T cell) exhibits increased IL-2 production. In some embodiments, the T cell effector function is characterized by IFN-gamma production. In some embodiments, the cell (e.g., T cell) exhibits increased IFN-gamma production. In some embodiments, the T cell effector function is characterized by IL-2 production and IFN-gamma production. In some embodiments, the cell (e.g., T cell) exhibits increased IL-2 production and increased IFN- gamma production. In some embodiments, the T cell effector function is characterized by polyfunctional production of IL-2, IFN-gamma, and TNF-alpha. In some embodiments, the cell (e.g., T cell) exhibits increased IL-2, IFN-gamma, and TNF-alpha production. [0266] Suitable techniques for the measurement of the production or secretion of a soluble factor, such as a cytokine, are known in the art. Production and/or secretion of a soluble factor can be measured by determining the concentration or amount of the extracellular amount of the factor, or determining the amount of transcriptional activity of the gene that encodes the factor. Suitable techniques include assays such as an immunoassay, an aptamer-based assay, a histological or cytological assay, an mRNA expression level assay, an enzyme linked immunosorbent assay (ELISA), immunoblotting, immunoprecipitation, radioimmunoassay (RIA), immunostaining, flow cytometry assay, surface plasmon resonance (SPR), chemiluminescence assay, lateral flow immunoassay, inhibition assay or avidity assay, protein sf-6059407 22474-20028.40 microarrays, high-performance liquid chromatography (HPLC), Meso Scale Discovery (MSD) electrochemiluminescence, and bead based multiplex immunoassays (MIA). In some embodiments, the suitable technique may employ a detectable binding reagent that specifically binds the soluble factor. [0267] In some embodiments, cytokine production is measured as a percentage of cells being positive for the cytokine, for example as measured by intracellular cytokine staining (ICS) and flow cytometry. Intracellular cytokine staining (ICS) by flow cytometry is a technique well- suited for studying cytokine production at the single-cell level. It detects the production and accumulation of cytokines within the endoplasmic reticulum after cell stimulation, allowing for the identification of cell populations that are positive or negative for production of a particular cytokine or for the separation of high producing and low producing cells based on a threshold. ICS can also be used in combination with other flow cytometry protocols for immunephenotyping using cell surface markers or with MHC multimers to access cytokine production in a particular subgroup of cells, making it an extremely flexible and versatile method. Other single-cell techniques for measuring or detecting cytokine production include ELISPOT, limiting dilution, and T cell cloning. [0268] In some embodiments, the cytokine production is measured as the amount of cytokine secreted from the cell, for example as measured by ELISA (enzyme-linked immunosorbent assay). ELISA is a plate-based assay technique designed for detecting and quantifying substances such as peptides, cytokines, antibodies, and hormones. In an ELISA, the soluble factor, such as a cytokine, is immobilized to a solid surface and then complexed with an antibody that is linked to an enzyme. Detection is accomplished by assessing the conjugated enzyme activity via incubation with a substrate to produce a detectable signal. [0269] In some embodiments, the T cell effector function is characterized by activity that further comprises T cell proliferation. In some embodiments, the cell (e.g., T cell) exhibits increased proliferation. In some embodiments, the increased proliferation occurs upon T cell stimulation. In some embodimnets, the proliferation is increased by at least about 1.1 fold, 1.2 fold, 1.3 fold, 1.4 fold, 1.5 fold, 2 fold, 5 fold, 10 fold, 50 fold, or 100 fold in comparison to a cell that has not been delivered the epigenetic-modifying DNA-targeting system. In some embodiments, the proliferation is measured as the increase in cell numbers before and after stimulation. In some embodiments, the increased proliferation is measured as the number of cells after stimulation in a cell population delivered with the epigenetic-modifying DNA- sf-6059407 22474-20028.40 targeting system compared to the number of cells after stimulation in a cell population not delivered the epigenetic-modifying DNA-targeting system. In some embodiments, the cell (e.g., T cell) does not exhibit increased proliferation. [0270] In some embodiments, the T cell effector function is characterized by activity that further comprises killing of target cells. In some embodiments, the cell (e.g., T cell) exhibits increased killing of target cells. In some embodiments, the increased killing of target cells occurs upon T cell stimulation. In some embodiments, the stimulation is performed by contacting the cells (e.g., T cells) with target cells. In some embodiments, T cells are incubated with antigen-expressing target cells at ratios between 4:1 and 1:4, inclusive, such as at ratios of 1:4, 1:3, 1:2, or 1:1. In some embodiments, the killing of target cells is increased by at least about 1.1 fold, 1.2 fold, 1.3 fold, 1.4 fold, 1.5 fold, 2 fold, 5 fold, 10 fold, 50 fold, or 100 fold in comparison to a cell that has not been delivered the epigenetic-modifying DNA-targeting system. In some embodiments, the killing is measured as the ability of the cells to kill target cells when contacted by the target cells. Killing of target cells can be measured by any suitable assay, for example as described herein in the Examples. In some embodiments, the killing is measured in an in vitro assay, wherein cells delivered with the epigenetic-modifying DNA- targeting system are co-cultured with the target cells, and the number of target cells are measured over time. In some embodiments, reduced numbers and/or proliferation of the target cells are indicative of target cell killing. The cytolytic activity can be measured by directly or indirectly measuring the target cell number over time. For example, the target cells may be incubated with a detectable marker prior to being incubated with antigen receptor (e.g., CAR)- expressing cells, such a marker that is detectable then the target cell is lysed, or a detectable marker that is detectable in viable target cells. These readouts provide direct or indirect of target cell number and/or target cell death and can be measured at different time points during the assay. A reduction of target cell number and/or an increase of target cell death can indicate the cytolytic activity of the cells. Suitable methods for performing cytolytic assays are known in the art and include chromium-51 release assays, non-radioactive chromium assays, and flow cytometric assays that use fluorescent dyes such as carboxyfluorescein succinimidyl ester (CFSE), PKH-2, and PKH-26. [0271] In some embodiments, the T cell effector function is characterized by activity that further comprises T cell persistence. In some embodiments, the cell (e.g., T cell) exhibits increased persistence (e.g., T cell persistence). In some embodiments, persistence relates to the sf-6059407 22474-20028.40 ability of cells to remain present and/or maintain an immune response in the presence of target cells. In some embodiments, persistence can be measured in vitro or in vivo, for example after administration of cells to a subject. Persistence can be measured by any suitable method, for example as described in Section IV. [0272] In certain embodiments, the ability of T cells to persist can be measured as a pharmacokinetic property of the cell composition following its administration to a subject. In some embodiments, the pharmacokinetic parameter can include exposure, number, concentration, persistence, and proliferation. In some cases, pharmacokinetics can be assessed by measuring such parameters as the maximum (peak) plasma concentration
, the peak time (i.e., when maximum plasma concentration (Cmax) occurs; Tmax), the minimum plasma concentration (i.e., the minimum plasma concentration between doses of a therapeutic agent, e.g., CAR+ T cells; Cmin), the elimination half-life (T1/2), and area under the curve (i.e., the area under the curve generated by plotting time versus plasma concentration of the therapeutic agent CAR+ T cells; AUC), following administration. The parameters of the administered engineered T cells can be measured in samples of blood from the subject. For example, nucleic acid-based methods, such as quantitative PCR (qPCR) or flow cytometry-based methods, or other assays, such as an immunoassay, ELISA, or chromatography/mass spectrometry-based assays, can be used. [0273] In some aspects, nucleic acid-based methods, such as quantitative PCR (qPCR), is used to assess the quantity of cells expressing the antigen receptor (e.g., CAR-expressing cells administered for T cell based therapy) in the blood, serum, organ, or tissue sample (e.g., disease site, e.g., tumor sample) of the subject. In some aspects, persistence is quantified as copies of DNA or plasmid encoding the receptor, e.g., CAR, per microgram of DNA, or as the number of antigen receptor-expressing, e.g., CAR-expressing, cells per microliter of the sample, e.g., of blood or serum, or per total number of peripheral blood mononuclear cells (PBMCs), white blood cells, or T cells per microliter of the sample. In some embodiments, the primers or probe used for qPCR or other nucleic acid-based methods are specific for binding, recognizing, and/or amplifying nucleic acids encoding the antigen receptor and/or other components or elements of the plasmid and/or vector, including regulatory elements, e.g., promoters, transcriptional, and/or post-transcriptional regulatory elements or response elements, or markers, e.g., surrogate markers. In some embodiments, the primers can be specific for regulatory elements, such as the woodchuck hepatitis virus post-transcriptional regulatory element (WPRE). sf-6059407 22474-20028.40 [0274] In some embodiments, any of the phenotypes described herein, such as increased IL- 2 production, increased IFN-gamma production, increased IL-2 production and increased IFN- gamma production, increased IL-2, IFN-gamma and TNF-alpha production, increased proliferation or proliferation that is not increased, increased killing of target cells, and/or increased persistence, are observed after stimulation (e.g., T cell stimulation). [0275] In some embodiments, the phenotype, such as any phenotype described herein, including increased T cell effector function, occurs 48 hours or more after the transient delivery of the epigenetic-modifying DNA-targeting system to the T cell. In some embodiments, the phenotype, such as increased T cell effector function, occurs up to 6 days, up to 9 days, up to 12 days, up to 15 days, up to 21 days, up to 28 days, up to 35 days, up to 42 days, up to 49 days, up to 56 days, up to 63 days, or up to 71 days after the transient delivery of the epigenetic- modifying DNA-targeting system to the T cell. [0276] In some aspects, the phenotype is one that is characterized by a cell surface phenotype of the cells. In some embodiments, the phenotype comprises expression of one or more cell-surface markers selected from IL-2+, TNFa+, IFNg+, and any combination thereof. In some embodiments, the phenotype is a phenotype in a T cell, such as a CD3+ T cell, which may be a CD4+ T cell or CD8+ T cell. Thus, in some embodiments, the phenotype comprises expression of one or more cell-surface markers selected from CD3+, CD4+, CD8+, IL-2+, TNFa+, IFNg+, and any combination thereof. In some aspects, the phenotype comprises expression of IL-2+. In some embodiments, the phenotype comprises expression of IL-2- and IFNg+. [0277] It is understood that embodiments of the provided epigenetic-modifying DNA- targeting systems are not limited to modulating expression of target genes and promoting phenotypes in T cells, but may also be used to modulate any one or more of the target genes as described herein in any lymphoid cell. In addition to T cells, lymphoid cells can include NK cells, NKT cells, and any cells that have been differentiated from stem cells into such lymphoid cells and/or have been differentiated from progenitor cells, such as common lymphoid progenitors (CLPs). In some embodiments, the lymphoid cells are differentiated from stem cells, such as hematopoietic stem or progenitor cells, or progenitor cells. In some embodiments, the lymphoid cells are trans-differentiated from a non-pluripotent cell of non-hematopoietic lineage. [0278] In some embodiments, the lymphoid cell for modulation is an isolated or enriched population of lymphoid immune cells, such as a population isolated or enriched in T, NK, and/or sf-6059407 22474-20028.40 NKT cells. In some embodiments, the cells for modulation are isolated or enriched T cells. In some embodiments, the cells for modulation are isolated or enriched NK cells. In some embodiments, the cells for modulation are isolated or enriched NK T cells. [0279] In some embodiments, isolated or enriched populations or subpopulations of immune cells comprising T, NK, and/or NKT cells for modulation can be obtained from a unit of blood using any number of techniques known to the skilled artisan, such as Ficoll™ separation. In one embodiment, T, NK, or NKT cells from the circulating blood of an individual are obtained by apheresis and separated from other nucleated white blood cells, red blood cells, and platelets, such as by Ficoll™ separation or affinity-based selection. In some embodiments, the cells are primary cells. In some embodiments, the primary cells are isolated or enriched from a peripheral blood sample from a subject, such as a human subject. [0280] In some embodiments, the lymphoid cells for modulation are differentiated in vitro from a stem cell or progenitor cell. In some embodiments, the lymphoid cells, such as T, NK, or NKT cells or lineages thereof, can be differentiated from a stem cell, a hematopoietic stem or progenitor cell (HSC), or a progenitor cell. The progenitor cell can be a CD34+ hemogenic endothelium cell, a multipotent progenitor cell, a T cell progenitor, an NK cell progenitor, or an NKT cell progenitor. In some embodiments, the progenitor cells is a lymphoid progenitor cells, such as a common lymphoid progenitor cell, early thymic progenitor cells, pre-T cell progenitor cells, pre-NK progenitor cell, T progenitor cell, NK progenitor cell, or NKT progenitor cell. The stem cell can be a pluripotent stem cell, such as induced pluripotent stem cells (iPSCs) and embryonic stem cells (ESCs). The iPSC is a non-naturally occurring reprogrammed pluripotent cell. Once the cells of a subject have been reprogrammed to a pluripotent state, the cells can then be programmed or differentiated to a desired cell type or subtypes, such as T, NK, or NKT cells. [0281] In some embodiments, the iPSC is differentiated to a T, NK, or NKT cells by a multi- stage differentiation platform wherein cells from various stages of development can be induced to assume a hematopoietic phenotype, ranging from mesodermal stem cells to fully differentiated T, NK, or NKT cells (See e.g. U.S. Pat. No.10,626,372). [0282] In some embodiments, the population or subpopulation of lymphoid cells is trans- differentiated in vitro from a non-pluripotent cell of non-hematopoietic fate to a hematopoietic lineage cell or from a non-pluripotent cell of a first hematopoietic cell type to a different hematopoietic cell type, which can be a T, NK, or NKT progenitor cell or a fully differentiated specific type of immune cell, such as an T, NK, or NKT cell (see, e.g., U.S. Pat. No.9,376,664 sf-6059407 22474-20028.40 and U.S. application Ser. No.15/072,769, the disclosure of which is incorporated herein in their entirety). In some embodiments, the non-pluripotent cell of non-hematopoietic fate is a somatic cell, such as a skin fibroblast, an adipose tissue-derived cell, and a human umbilical vein endothelial cell (HUVEC). Somatic cells useful for trans-differentiation may be immortalized somatic cells. [0283] Various strategies are being pursued to induce pluripotency, or increase potency, in cells (Takahashi, K., and Yamanaka, S., Cell 126, 663-676 (2006); Takahashi et al., Cell 131, 861-872 (2007); Yu et al., Science 318, 1917-1920 (2007); Zhou et al., Cell Stem Cell 4, 381- 384 (2009); Kim et al., Cell Stem Cell 4, 472-476 (2009); Yamanaka et al., 2009; Saha, K., Jaenisch, R., Cell Stem Cell 5, 584-595 (2009)), and improve the efficiency of reprogramming (Shi et al., Cell Stem Cell 2, 525-528 (2008a); Shi et al., Cell Stem Cell 3, 568-574 (2008b); Huangfu et al., Nat Biotechnol 26, 795-797 (2008a); Huangfu et al., Nat Biotechnol 26, 1269- 1275 (2008b); Silva et al., Plos Bio 6, e253. Doi: 10.1371/journal. Pbio.0060253 (2008); Lyssiotis et al., PNAS 106, 8912-8917 (2009); Ichida et al., Cell Stem Cell 5, 491-503 (2009); Maherali, N., Hochedlinger, K., Curr Biol 19, 1718-1723 (2009b); Esteban et al., Cell Stem Cell 6, 71-79 (2010); and Feng et al., Cell Stem Cell 4, 301-312 (2009)), the disclosures of which are hereby incorporated by reference in their entireties. [0284] It is understood that a cell that is positive (+) for a particular cell surface marker is a cell that expresses the marker on its surface at a level that is detectable. Likewise, it is understood that a cell that is negative (-) for a particular cell surface marker is a cell that expresses the marker on its surface at a level that is not detectable. Antibodies and other binding entities can be used to detect expression levels of marker proteins to identify or detect a given cell surface marker. Suitable antibodies may include polyclonal, monoclonal, fragments (such as Fab fragments), single chain antibodies, and other forms of specific binding molecules. Antibody reagents for cell surface markers are readily known to a skilled artisan. A number of well-known methods for assessing expression level of surface markers or proteins may be used, such as detection by affinity-based methods, e.g., immunoaffinity-based methods, e.g., in the context of surface markers, such as by flow cytometry. In some embodiments, the label is a fluorophore, and the method for detection or identification of cell surface markers on cells (e.g., T cells) is by flow cytometry. In some embodiments, different labels are used for each of the different markers by multicolor flow cytometry. In some embodiments, surface expression can be determined by flow cytometry, for example by staining with an antibody that specifically sf-6059407 22474-20028.40 binds to the marker and detecting the binding of the antibody to the marker. [0285] In some embodiments, a cell (e.g., T cell) is positive (pos or +) for a particular marker if there is detectable presence on or in the cell of a particular marker, which can be an intracellular marker or a surface marker. In some embodiments, surface expression is positive if staining by flow cytometry is detectable at a level substantially above the staining detected carrying out the same procedures with an isotype-matched control under otherwise identical conditions and/or at a level substantially similar to, or in some cases higher than, a cell known to be positive for the marker and/or at a level higher than that for a cell known to be negative for the marker. [0286] In some embodiments, a cell (e.g. T cell) is negative (neg or -) for a particular marker if there is an absence of detectable presence on or in the cell of a particular marker, which can be an intracellular marker or a surface marker. In some embodiments, surface expression is negative if staining is not detectable by flow cytometry at a level substantially above the staining detected carrying out the same procedures with an isotype-matched control under otherwise identical conditions and/or at a level substantially lower than a cell known to be positive for the marker and/or at a level substantially similar to a cell known to be negative for the marker. [0287] In some aspects, the phenotype can be characterized by one or more functions of the cells. In some aspects, the phenotype is characterized by polyfunctional activity of the T cells to produce more than one T cell stimulatory cytokine, such as determined in a polyfunctional cytokine secretion assay following stimulation of the T cells with a stimulatory agent. In some embodiments, the T cell is polyfunctional for producing two or more cytokines. In some embodiments, a T cell is polyfunctional for producing two or more cytokines selected from among interferon-gamma (IFN-gamma), interleukin 2 (IL-2), and TNF-alpha. In some embodiments, a polyfunctional T cell produces IFN-gamma, IL-2, and TNF-alpha. In some embodiments, the stimulatory agent is a non-specific or non-antigen-dependent T cell stimulatory agent. In some embodiments, the non-specific or non-antigen dependent T cell stimulatory agent is a polyclonal stimulatory agent. In some embodiments, the non-specific or non-antigen dependent stimulatory agent comprises PMA/ionomycin, anti-CD3/anti-CD28, phytohemagglutinin (PHA), or concanavalin A (ConA). In some embodiments, the non-specific or non-antigen dependent T cell stimulatory agent contains PMA/ionomycin. [0288] In particular embodiments, the production of one or more cytokines is measured, detected, and/or quantified by intracellular cytokine staining. Intracellular cytokine staining sf-6059407 22474-20028.40 (ICS) by flow cytometry is a technique well-suited for studying cytokine production at the single-cell level. It detects the production and accumulation of cytokines within the endoplasmic reticulum after cell stimulation, allowing for the identification of cell populations that are positive or negative for production of a particular cytokine or for the separation of high producing and low producing cells based on a threshold. In some embodiments, the stimulation can be performed using nonspecific stimulation, e.g., is not an antigen-specific stimulation. For example, PMA/ionomycin can be used for nonspecific cell stimulation. ICS can also be used in combination with other flow cytometry protocols for immunephenotyping using cell surface markers or with MHC multimers to access cytokine production in a particular subgroup of cells, making it an extremely flexible and versatile method. Other single-cell techniques for measuring or detecting cytokine production include ELISPOT, limiting dilution, and T cell cloning. In some embodiments, the assays to assay polyfunctional cytokine secretion of multiple cytokines can include multiplexed assays or other assays to assess polyfunctionality (see, e.g., Xue et al., (2017) Journal for ImmunoTherapy of Cancer 5:85). 2. Genes and Target Sites for Increasing Transcription [0289] In some embodiments, the target genes include one or more first genes for which transcription is to be increased. In some embodiments, the target genes include a single first gene. In some embodiments, the target genes include a plurality of first genes, such as at least 2, at least 3, at least 4, at least 5, at least 6, at least 7, at least 8, at least 9, at least 10, at least 11, or at least 12 first genes for which transcription is increased. [0290] In some embodiments, the one or more first genes are T cell activator genes. In some embodiments, the one or more first genes are positive regulators of T cell function. In some embodiments, the one or more first genes are selected from BATF, CD28, EOMES, IL-2, IL2RB , IRF4, LAT, LCP2, TBX21, and VAV1. In some embodiments, the one or more first genes is or compise IL-2. [0291] In some embodiments, the DNA-targeting system comprises a plurality of first DNA- targeting modules. In some embodiments, each first DNA-targeting module targets a target site. In some embodiments, the plurality of first DNA-targeting modules target a plurality of first genes, wherein the plurality of first genes are independently selected from the group consisting of BATF, CD28, EOMES, IL-2, IL2RB, IRF4, LAT, LCP2, TBX21, and VAV1. In some embodiments, the plurality of first DNA-targeting modules target 2 or 3 genes selected from the sf-6059407 22474-20028.40 group consisting of BATF, CD28, EOMES, IL-2, IL2RB, IRF4, LAT, LCP2, TBX21, and VAV1. [0292] In some embodiments, the DNA-targeting system targets a combination of first genes set forth in Table 1. In some embodiments, the plurality of first DNA-targeting modules target a combination of first genes set forth in Table 1. In some embodiments, transcription of each of the first genes of the combination is increased by the DNA-targeting system. Table 1. Combinations of first genes targeted by a multiplexed epigenetic-modifying DNA- targeting system for increasing transcription of first genes


sf-6059407 22474-20028.40


[0293] In some embodiments, the DNA-targeting system targets a target site for BATF, CD28, EOMES, IL-2, IL2RB, IRF4, LAT, LCP2, TBX21, and/or VAV1. In some embodiments, the target site comprises a sequence selected from any one of SEQ ID NOS:7-9, 78, 144-156, 170, 172-177, 184-191, and 451-453 or a contiguous portion thereof of at least 14 nucleotides, or a complementary sequence of any of the foregoing. In some embodiments, the target site is a sf-6059407 22474-20028.40 contiguous portion of any one of SEQ ID NOS:7-9, 78, 144-156, 170, 172-177, 184-191, and 451-453 that is 15, 16, 17, 18 or 19 nucleotides in length, or a complementary sequence of any of the foregoing. In some embodiments, the target site is a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to all or a contiguous portion of a target site sequence described herein above. In some embodiments, the target site is the sequence set forth in any one of SEQ ID NOS:7-9, 78, 144-156, 170, 172-177, 184-191, and 451-453. In some embodiments, the target site comprises a sequence selected from any one of SEQ ID NOS:7-9, 78, 144-156, 170, 172-177, and 184-191, or a contiguous portion thereof of at least 14 nucleotides, or a complementary sequence of any of the foregoing. In some embodiments, the target site is a contiguous portion of any one of SEQ ID NOS:7-9, 78, 144-156, 170, 172-177, and 184-191 that is 15, 16, 17, 18 or 19 nucleotides in length, or a complementary sequence of any of the foregoing. In some embodiments, the target site is a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to all or a contiguous portion of a target site sequence described herein above. In some embodiments, the target site is the sequence set forth in any one of SEQ ID NOS:7-9, 78, 144-156, 170, 172-177, and 184-191. In some embodiments, the target site is a sequence set forth in Table 5. [0294] In some embodiments, the DNA-targeting system targets a combination of first genes, such as any combination shown in Table 1. In some embodiments, the DNA-targeting system targets a combination of first genes selected from: BATF and IL-2; BATF and VAV1; CD28 and BATF; CD28 and EOMES; CD28 and IL-2; CD28 and LCP2; CD28 and TBX21; CD28 and VAV1; EOMES and BATF; EOMES and LCP2; EOMES and TBX21; EOMES and VAV1; LCP2 and BATF; LCP2 and IL-2; LCP2 and TBX21; LCP2 and VAV1; TBX21 and BATF; TBX21 and IL-2; TBX21 and TBX21; TBX21 and VAV1; and VAV1 and IL-2. [0295] In some embodiments, the DNA-targeting system targets IL-2 and VAV1. In some embodiments, the DNA-targeting system targets a target site for IL-2 comprising the sequence set forth in SEQ ID NO:78, and a target site for VAV1 comprising the sequence set forth in SEQ ID NO:170. [0296] In some embodiments, the DNA-targeting system targets IL2RB and VAV1. [0297] In some embodiments, the DNA-targeting system targets a combination of target sites for a combination of first genes for transcriptional activation, as shown in Table 2. Table 2. First gene and target site combinations for transcriptional activation sf-6059407 22474-20028.40

sf-6059407 22474-20028.40

[0298] In some embodiments, the DNA-targeting system targets a target site for BATF comprising the sequence set forth in SEQ ID NO:172, and a target site for IL-2 comprising the sequence set forth in SEQ ID NO:78. In some embodiments, the DNA-targeting system targets a target site for BATF comprising the sequence set forth in SEQ ID NO:172, and a target site for VAV1 comprising the sequence set forth in SEQ ID NO:170. In some embodiments, the DNA- targeting system targets a target site for CD28 comprising the sequence set forth in SEQ ID NO:144, and a target site for BATF comprising the sequence set forth in SEQ ID NO:172. In some embodiments, the DNA-targeting system targets a target site for CD28 comprising the sequence set forth in SEQ ID NO:144, and a target site for EOMES comprising the sequence set forth in SEQ ID NO:149. In some embodiments, the DNA-targeting system targets a target site for CD28 comprising the sequence set forth in SEQ ID NO:144, and a target site for IL-2 comprising the sequence set forth in SEQ ID NO:78. In some embodiments, the DNA-targeting system targets a target site for CD28 comprising the sequence set forth in SEQ ID NO:144, and a target site for LCP2 comprising the sequence set forth in SEQ ID NO:151. In some sf-6059407 22474-20028.40 embodiments, the DNA-targeting system targets a target site for CD28 comprising the sequence set forth in SEQ ID NO:144, and a target site for TBX21 comprising the sequence set forth in SEQ ID NO:155. In some embodiments, the DNA-targeting system targets a target site for CD28 comprising the sequence set forth in SEQ ID NO:144, and a target site for VAV1 comprising the sequence set forth in SEQ ID NO:170. In some embodiments, the DNA-targeting system targets a target site for EOMES comprising the sequence set forth in SEQ ID NO:149, and a target site for BATF comprising the sequence set forth in SEQ ID NO:172. In some embodiments, the DNA-targeting system targets a target site for EOMES comprising the sequence set forth in SEQ ID NO:149, and a target site for LCP2 comprising the sequence set forth in SEQ ID NO:151. In some embodiments, the DNA-targeting system targets a target site for EOMES comprising the sequence set forth in SEQ ID NO:149, and a target site for TBX21 comprising the sequence set forth in SEQ ID NO:155. In some embodiments, the DNA-targeting system targets a target site for EOMES comprising the sequence set forth in SEQ ID NO:149, and a target site for VAV1 comprising the sequence set forth in SEQ ID NO:170. In some embodiments, the DNA-targeting system targets a target site for LCP2 comprising the sequence set forth in SEQ ID NO:151, and a target site for BATF comprising the sequence set forth in SEQ ID NO:172. In some embodiments, the DNA-targeting system targets a target site for LCP2 comprising the sequence set forth in SEQ ID NO:151, and a target site for IL-2 comprising the sequence set forth in SEQ ID NO:78. In some embodiments, the DNA-targeting system targets a target site for LCP2 comprising the sequence set forth in SEQ ID NO:151, and a target site for TBX21 comprising the sequence set forth in SEQ ID NO:155. In some embodiments, the DNA-targeting system targets a target site for LCP2 comprising the sequence set forth in SEQ ID NO:151, and a target site for VAV1 comprising the sequence set forth in SEQ ID NO:170. In some embodiments, the DNA-targeting system targets a target site for TBX21 comprising the sequence set forth in SEQ ID NO:155, and a target site for BATF comprising the sequence set forth in SEQ ID NO:172. In some embodiments, the DNA-targeting system targets a target site for TBX21 comprising the sequence set forth in SEQ ID NO:155, and a target site for IL-2 comprising the sequence set forth in SEQ ID NO:78. In some embodiments, the DNA-targeting system targets a target site for TBX21 comprising the sequence set forth in SEQ ID NO:155, and a target site for TBX21 comprising the sequence set forth in SEQ ID NO:155. In some embodiments, the DNA-targeting system targets a target site for TBX21 comprising the sequence set forth in SEQ ID NO:155, and a target site for VAV1 comprising the sf-6059407 22474-20028.40 sequence set forth in SEQ ID NO:170. In some embodiments, the DNA-targeting system targets a target site for VAV1 comprising the sequence set forth in SEQ ID NO:170, and a target site for IL-2 comprising the sequence set forth in SEQ ID NO:78. [0299] In some embodiments, delivery of the DNA-targeting system increases expression (e.g. transcription) of one or more first genes. In some embodiments, the increase in gene expression in a cell (e.g. T cell) is about a log2 fold change of greater than 1.0. For instance, the log2 fold change is greater than at or about 1.5, at or about 2.0, at or about 2.5, at or about 3.0, at or about 4.0, at or about 5.0, at or about 6.0, at or about 7.0, at or about 8.0, at or about 9.0, at or about 10.0, or any value between any of the foregoing compared to the level of the first gene in a control cell. 3. Genes and Target Sites for Decreasing Transcription [0300] In some embodiments, the target genes include one or more second genes for which transcription is to be repressed. In some embodiments, the target genes include a single second gene. In some embodiments, the target genes include a plurality of second genes, such as at least 2, at least 3, at least 4, at least 5, at least 6, at least 7, at least 8, at least 9, at least 10, at least 11, or at least 12 first genes for which transcription is repressed. [0301] In some embodiments, the one or more second genes are T cell repressor genes. In some embodiments, the one or more second genes are negative regulators of T cell function. In some embodiments, the one or more second genes are selected from CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and RASA2. In some embodiments, the one or more second genes is or compise MED12. [0302] In some embodiments, the DNA-targeting system comprises a plurality of second DNA-targeting modules. In some embodiments, each second DNA-targeting module targets a target site. In some embodiments, the plurality of second DNA-targeting modules target a plurality of second genes, wherein the plurality of second genes are independently selected from the group consisting of CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and RASA2. In some embodiments, the plurality of second DNA-targeting modules target 2 or 3 genes selected from the group consisting of CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and RASA2. [0303] In some embodiments, the DNA-targeting system targets a combination of second genes set forth in Table 3. In some embodiments, the plurality of second DNA-targeting sf-6059407 22474-20028.40 modules target a combination of second genes set forth in Table 3. In some embodiments, transcription of each of the second genes of the combination is repressed by the DNA-targeting system. Table 3. Combinations of second genes targeted by a multiplexed epigenetic-modifying DNA-targeting system for decreasing transcription of second genes


sf-6059407 22474-20028.40
sf-6059407 22474-20028.40
sf-6059407 22474-20028.40
sf-6059407 22474-20028.40
sf-6059407 22474-20028.40
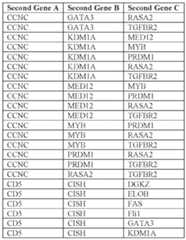

[0304] In some embodiments, the DNA-targeting system targets a target site for CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and/or RASA2. In some embodiments, the target site comprises a sequence selected from any one of SEQ ID NOS: 1-6, 10-33, 80-90, 102-112, 200-211, 292-295, 300-302, 306- 308, and 454-457 or a contiguous portion thereof of at least 14 nucleotides, or a complementary sequence of any of the foregoing. In some embodiments, the target site is a contiguous portion of any one of SEQ ID NOS:1-6, 10-33, 80-90, 102-112, 200-211, 292-295, 300-302, 306-308, and 454-457 that is 15, 16, 17, 18 or 19 nucleotides in length, or a complementary sequence of any of the foregoing. In some embodiments, the target site is a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to all or a contiguous portion of a target site sequence described herein above. In some embodiments, the target site is the sequence set forth in any one of SEQ ID NOS:1-6, 10-33, 80-90, 102-112, 200-211, 292-295, 300-302, 306-308, and 454-457. In some embodiments, the target site comprises a sequence selected from any one of SEQ ID NOS: 1-6, 10-33, 80-90, 102-112, 200-211, 292-295, 300-302, and 306-308, or a contiguous portion thereof of at least 14 nucleotides, or a complementary sequence of any of the foregoing. In some embodiments, the target site is a contiguous portion of any one of SEQ ID NOS:1-6, 10-33, 80- sf-6059407 22474-20028.40 90, 102-112, 200-211, 292-295, 300-302, and 306-308 that is 15, 16, 17, 18 or 19 nucleotides in length, or a complementary sequence of any of the foregoing. In some embodiments, the target site is a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to all or a contiguous portion of a target site sequence described herein above. In some embodiments, the target site is the sequence set forth in any one of SEQ ID NOS:1-6, 10-33, 80-90, 102-112, 200-211, 292-295, 300-302, and 306-308. In some embodiments, the target site is a sequence set forth in Table 6. [0305] In some embodiments, the DNA-targeting system targets a combination of second genes, such as any combination shown in Table 3. In some embodiments, the DNA-targeting system targets a combination of second genes selected from: CBLB and CCNC; CBLB and CD5; CBLB and CISH; CBLB and DGKZ; CBLB and ELOB; CBLB and FAS; CBLB and Fli1; CBLB and GATA3; CBLB and KDM1A; CBLB and MED12; CBLB and MYB; CBLB and PRDM1; CBLB and RASA2; CD5 and CISH; CD5 and MYB; CISH and DGKZ; CISH and MYB; CISH and RASA2; GATA3 and CD5; GATA3 and CISH; GATA3 and MYB; MED12 and CBLB; MED12 and CD5; MED12 and CISH; MED12 and DGKZ; MED12 and ELOB; MED12 and GATA3; MED12 and MYB; MED12 and PRDM1; MED12 and RASA2; MYB and RASA2; MED12 and TGFBR2; PRDM1 and CISH; PRDM1 and GATA3; PRDM1 and MYB; PRDM1 and RASA2; CD5, CISH, and MYB; GATA3, CBLB, and MYB; GATA3, CD5, and MYB; and PRDM1, GATA3, and CISH. [0306] In some embodiments, the DNA-targeting system targets CBLB and MYB. In some embodiments, the DNA-targeting system targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and a target site for MYB comprising the sequence set forth in SEQ ID NO:18. [0307] In some embodiments, the DNA-targeting system targets CBLB and MED12. In some embodiments, the DNA-targeting system targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and a target site for MED12 comprising the sequence set forth in SEQ ID NO:81. [0308] In some embodiments, the DNA-targeting system targets CBLB and CCNC. In some embodiments, the DNA-targeting system targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and a target site for CCNC comprising the sequence set forth in SEQ ID NO:104. sf-6059407 22474-20028.40 [0309] In some embodiments, the DNA-targeting system targets MED12 and TGFBR2. In some embodiments, the DNA-targeting system targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and a target site for TGFBR2 comprising the sequence set forth in SEQ ID NO:301. [0310] In some embodiments, the DNA-targeting system targets MED12, TGFBR2 and CISH. In some embodiments, the DNA-targeting system targets a target site for MED12 compirsing the sequence set forth in SEQ ID NO:81, a target site for TGFBR2 comprising the sequence set forth in SEQ ID NO:301, and a target site for CISH comprising the sequence set forth in SEQ ID NO:28 or 30. [0311] In some embodiments, the DNA-targeting system targets a combination of target sites for a combination of second genes for transcriptional repression, as shown in Table 4. Table 4. Second gene and target site combinations for transcriptional repression

sf-6059407 22474-20028.40
sf-6059407 22474-20028.40
sf-6059407 22474-20028.40

[0312] In some embodiments, the DNA-targeting system targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and a target site for CCNC comprising the sequence set forth in SEQ ID NO:104. In some embodiments, the DNA-targeting system targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and a target site for CD5 comprising the sequence set forth in SEQ ID NO:3. In some embodiments, the DNA- targeting system targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and a target site for CISH comprising the sequence set forth in SEQ ID NO:30. In some embodiments, the DNA-targeting system targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and a target site for DGKZ comprising the sequence set forth in SEQ ID NO:13. In some embodiments, the DNA-targeting system targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and a target site for ELOB comprising the sequence set forth in SEQ ID NO:24. In some embodiments, the DNA-targeting system targets a sf-6059407 22474-20028.40 target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and a target site for FAS comprising the sequence set forth in SEQ ID NO:204. In some embodiments, the DNA- targeting system targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and a target site for Fli1 comprising the sequence set forth in SEQ ID NO:208. In some embodiments, the DNA-targeting system targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and a target site for GATA3 comprising the sequence set forth in SEQ ID NO:26. In some embodiments, the DNA-targeting system targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and a target site for KDM1A comprising the sequence set forth in SEQ ID NO:4. In some embodiments, the DNA-targeting system targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and a target site for MED12 comprising the sequence set forth in SEQ ID NO:81. In some embodiments, the DNA-targeting system targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and a target site for MYB comprising the sequence set forth in SEQ ID NO:18. In some embodiments, the DNA-targeting system targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and a target site for PRDM1 comprising the sequence set forth in SEQ ID NO:32. In some embodiments, the DNA-targeting system targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and a target site for RASA2 comprising the sequence set forth in SEQ ID NO:19. In some embodiments, the DNA-targeting system targets a target site for CD5 comprising the sequence set forth in SEQ ID NO:3, and a target site for CISH comprising the sequence set forth in SEQ ID NO:30. In some embodiments, the DNA-targeting system targets a target site for CD5 comprising the sequence set forth in SEQ ID NO:3, and a target site for MYB comprising the sequence set forth in SEQ ID NO:18. In some embodiments, the DNA-targeting system targets a target site for CISH comprising the sequence set forth in SEQ ID NO:30, and a target site for DGKZ comprising the sequence set forth in SEQ ID NO:13. In some embodiments, the DNA-targeting system targets a target site for CISH comprising the sequence set forth in SEQ ID NO:30, and a target site for MYB comprising the sequence set forth in SEQ ID NO:18. In some embodiments, the DNA-targeting system targets a target site for CISH comprising the sequence set forth in SEQ ID NO:30, and a target site for RASA2 comprising the sequence set forth in SEQ ID NO:19. In some embodiments, the DNA-targeting system targets a target site for GATA3 comprising the sequence set forth in SEQ ID NO:26, and a target site for CD5 comprising the sequence set forth in SEQ ID NO:3. In some embodiments, the DNA-targeting system targets a target site for sf-6059407 22474-20028.40 GATA3 comprising the sequence set forth in SEQ ID NO:26, and a target site for CISH comprising the sequence set forth in SEQ ID NO:30. In some embodiments, the DNA-targeting system targets a target site for GATA3 comprising the sequence set forth in SEQ ID NO:26, and a target site for MYB comprising the sequence set forth in SEQ ID NO:18. In some embodiments, the DNA-targeting system targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and a target site for CBLB comprising the sequence set forth in SEQ ID NO:11. In some embodiments, the DNA-targeting system targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and a target site for CD5 comprising the sequence set forth in SEQ ID NO:3. In some embodiments, the DNA-targeting system targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and a target site for CISH comprising the sequence set forth in SEQ ID NO:30. In some embodiments, the DNA-targeting system targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and a target site for DGKZ comprising the sequence set forth in SEQ ID NO:13. In some embodiments, the DNA-targeting system targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and a target site for ELOB comprising the sequence set forth in SEQ ID NO:24. In some embodiments, the DNA-targeting system targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and a target site for GATA3 comprising the sequence set forth in SEQ ID NO:26. In some embodiments, the DNA-targeting system targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and a target site for MYB comprising the sequence set forth in SEQ ID NO:18. In some embodiments, the DNA-targeting system targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and a target site for PRDM1 comprising the sequence set forth in SEQ ID NO:32. In some embodiments, the DNA-targeting system targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and a target site for RASA2 comprising the sequence set forth in SEQ ID NO:19. In some embodiments, the DNA-targeting system targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and a target site for TGFBR2 comprising the sequence set forth in SEQ ID NO:301. In some embodiments, the DNA-targeting system targets a target site for MYB comprising the sequence set forth in SEQ ID NO:18, and a target site for RASA2 comprising the sequence set forth in SEQ ID NO:19. In some embodiments, the DNA-targeting system targets a target site for PRDM1 comprising the sequence set forth in SEQ ID NO:32, and a target site for CISH comprising the sequence set forth in SEQ ID NO:30. In some sf-6059407 22474-20028.40 embodiments, the DNA-targeting system targets a target site for PRDM1 comprising the sequence set forth in SEQ ID NO:32, and a target site for GATA3 comprising the sequence set forth in SEQ ID NO:26. In some embodiments, the DNA-targeting system targets a target site for PRDM1 comprising the sequence set forth in SEQ ID NO:32, and a target site for MYB comprising the sequence set forth in SEQ ID NO:18. In some embodiments, the DNA-targeting system targets a target site for PRDM1 comprising the sequence set forth in SEQ ID NO:32, and a target site for RASA2 comprising the sequence set forth in SEQ ID NO:19. In some embodiments, the DNA-targeting system targets a target site for CD5 comprising the sequence set forth in SEQ ID NO:3, a target site for CISH comprising the sequence set forth in SEQ ID NO:30, and a target site for MYB comprising the sequence set forth in SEQ ID NO:18. In some embodiments, the DNA-targeting system targets a target site for GATA3 comprising the sequence set forth in SEQ ID NO:26, a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and a target site for MYB comprising the sequence set forth in SEQ ID NO:18. In some embodiments, the DNA-targeting system targets a target site for GATA3 comprising the sequence set forth in SEQ ID NO:26, a target site for CD5 comprising the sequence set forth in SEQ ID NO:3, and a target site for MYB comprising the sequence set forth in SEQ ID NO:18. In some embodiments, the DNA-targeting system targets a target site for PRDM1 comprising the sequence set forth in SEQ ID NO:32, a target site for GATA3 comprising the sequence set forth in SEQ ID NO:26, and a target site for CISH comprising the sequence set forth in SEQ ID NO:30. In some embodiments, the DNA-targeting system targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, a target site for TGFBR2 comprising the sequence set forth in SEQ ID NO:301, and a target site for CISH comprising the sequence set forth in SEQ ID NO: 28 or 30. [0313] In some embodiments, delivery of the DNA-targeting system reduces (e.g., decreases or represses) transcription of one or more second genes. In some embodiments, the reduction in gene expression in a cell (e.g., T cell) is about a log2 fold change of less than -1.0. For instance, the log2 fold change is less than at or about -1.5, at or about -2.0, at or about -2.5, at or about - 3.0, at or about -4.0, at or about -5.0, at or about -6.0, at or about -7.0, at or about -8.0, at or about -9.0, at or about -10.0, or any value between any of the foregoing compared to the level of the second gene in a control cell. sf-6059407 22474-20028.40 B. CRISPR/Cas-Based DNA-Targeting Systems and DNA-Binding Domains [0314] Provided herein are multiplexed epigenetic-targeting DNA-targeting systems based on CRISPR/Cas systems, i.e., CRISPR/Cas-based DNA-targeting systems, that are able to bind to a target site for each of multiple target genes. In some embodiments, the CRISPR/Cas DNA- binding domain is nuclease inactive, such as includes a dCas (e.g., dCas9) so that the system binds to the target site for a target gene without mediating nucleic acid cleavage at the target site. The CRISPR/Cas-based DNA-targeting systems may be used to modulate expression of target genes in a cell, such as a T cell. In some embodiments, the CRISPR/Cas-based DNA- targeting system can include any known Cas enzyme, and generally a nuclease-inactive or dCas. In some embodiments, the dCas protein is any suitable dCas protein. In some embodiments, the dCas protein is a dCas9 protein, such dSpCas9 or dSaCas9. In some embodiments, the CRISPR/Cas-based DNA-targeting system includes a fusion protein of a nuclease-inactive Cas protein and an effector domain, and at least one gRNA. In some embodiments, the effector domain increases transcription of the one or more first genes (e.g., the effector domain is a transcriptional activator domain). In some embodiments, the effector domain reduces transcription of the one or more second genes (e.g., the effector domain is a transcriptional repressor domain). [0315] In some embodments, the DNA-targeting system comprises at least one first DNA- targeting module for increasing transcription of the one or more first genes and at least one second DNA-targeting module for repressing transcription of the one or more second genes. In some embodiments, at least one, optionally each, of the first DNA-targeting modules comprises (a) a first fusion protein comprising (i) a first Cas protein and (ii) at least one transcriptional activator domain and (b) a first gRNA for targeting the first Cas protein domain to a target site of one of the one or more first genes. In some embodiments, at least one, optionally each, of the second DNA-targeting modules comprises (a) a second fusion protein comprising (i) a second Cas protein and (ii) at least one transcriptional repressor domain and (b) a second gRNA for targeting the second Cas protein domain to a target site of one of the one or more second genes. [0316] In some embodiments, the first Cas protein is the same across the at least one first DNA-targeting module. In some embodiments, the first Cas protein is different across the at least one first DNA-targeting module. In some embodiments, the first gRNA is the same across the at least one first DNA-targeting module. In some embodiments, the first gRNA is different across the at least one first DNA-targeting module. sf-6059407 22474-20028.40 [0317] In some embodiments, the second Cas protein is the same across the at least one second DNA-targeting module. In some embodiments, the second Cas protein is different across the at least one second DNA-targeting module. In some embodiments, the second gRNA is the same across the at least one second DNA-targeting module. In some embodiments, the second gRNA is different across the at least one second DNA-targeting module. [0318] In some embodiments, the first Cas protein or proteins are all different from the second Cas protein or proteins. In some embodiments, the first Cas protein or proteins do not bind to the second gRNA or gRNAs, and the second Cas protein or proteins do not bind to the first gRNA or gRNAs. In some embodiments, the first Cas protein or proteins are from a different Staphylococcus species than the second Cas protein or proteins. In some embodiments, the first Cas protein or proteins do not bind to the protospacer-adjacent motifs (PAMs) that the second Cas protein or proteins bind to. [0319] In some embodiments, at least one, optionally each, of the first DNA-targeting modules comprises (a) a first fusion protein comprising (i) a dSpCas9 protein and (ii) at least one transcriptional activator domain and (b) a dSpCas9 gRNA for targeting the dSpCas9 Cas protein to a target site of one of the one or more first genes. In some embodiments, at least one, optionally each, of the second DNA-targeting modules comprises (a) a second fusion protein comprising (i) a dSaCas9 protein and (ii) at least one transcriptional repressor domain and (b) a dSaCas9 gRNA for targeting the dSaCas9 Cas protein to a target site of one of the one or more second genes. [0320] In some embodiments, at least one, optionally each, of the first DNA-targeting modules comprises (a) a first fusion protein comprising (i) a dSaCas9 protein and (ii) at least one transcriptional activator domain and (b) a dSaCas9 gRNA for targeting the dSaCas9 Cas protein to a target site of one of the one or more first genes. In some embodiments, at least one, optionally each, of the second DNA-targeting modules comprises (a) a second fusion protein comprising (i) a dSpCas9 protein and (ii) at least one transcriptional repressor domain and (b) a dSpCas9 gRNA for targeting the dSpCas9 Cas protein to a target site of one of the one or more second genes. [0321] In some embodiments, at least one, optionally each, of the first DNA-targeting modules comprises (a) a first fusion protein comprising (i) a first Cas protein that binds to a first PAM 5’-NGG-3’ (SEQ ID NO: 224), wherein N is any nucleotide, and (ii) at least one transcriptional activator domain and (b) a first gRNA for targeting the first Cas protein to a sf-6059407 22474-20028.40 target site of one of the one or more first genes. In some embodiments, at least one, optionally each, of the second DNA-targeting modules comprises (a) a second fusion protein comprising (i) a second Cas protein that binds to a second PAM 5’- NNGRRT-3’ (SEQ ID NO: 225), wherein N is any nucleotide, and R is G or A, and (ii) at least one transcriptional repressor domain and (b) a second gRNA for targeting the second Cas protein to a target site of one of the one or more second genes. In some embodiments, the first Cas protein does not bind to the second PAM, and the second Cas protein does not bind to the first PAM. [0322] In some embodiments, at least one, optionally each, of the first DNA-targeting modules comprises (a) a first fusion protein comprising (i) a first Cas protein that binds to a first PAM 5’- NNGRRT-3’ (SEQ ID NO: 225), wherein N is any nucleotide, and R is G or A, and (ii) at least one transcriptional activator domain and (b) a first gRNA for targeting the first Cas protein to a target site of one of the one or more first genes. In some embodiments, at least one, optionally each, of the second DNA-targeting modules comprises (a) a second fusion protein comprising (i) a second Cas protein that binds to a second PAM 5’-NGG-3’ (SEQ ID NO: 224), wherein N is any nucleotide, and (ii) at least one transcriptional repressor domain and (b) a second gRNA for targeting the second Cas protein to a target site of one of the one or more second genes. In some embodiments, the first Cas protein does not bind to the second PAM, and the second Cas protein does not bind to the first PAM. [0323] The CRISPR system (also known as CRISPR/Cas system, or CRISPR-Cas system) refers to a conserved microbial nuclease system, found in the genomes of bacteria and archaea, that provides a form of acquired immunity against invading phages and plasmids. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) refers to loci containing multiple repeating DNA elements that are separated by non-repeating DNA sequences called spacers. Spacers are short sequences of foreign DNA that are incorporated into the genome between CRISPR repeats, serving as a “memory” of past exposures. Spacers encode the DNA-targeting portion of RNA molecules that confer specificity for nucleic acid cleavage by the CRISPR system. CRISPR loci contain or are adjacent to one or more CRISPR-associated (Cas) genes, which can act as RNA-guided nucleases for mediating the cleavage, as well as non-protein coding DNA elements that encode RNA molecules capable of programming the specificity of the CRISPR-mediated nucleic acid cleavage. [0324] In Type II CRISPR/Cas systems with the Cas protein Cas9, two RNA molecules and the Cas9 protein form a ribonucleoprotein (RNP) complex to direct Cas9 nuclease activity. The sf-6059407 22474-20028.40 CRISPR RNA (crRNA) contains a spacer sequence that is complementary to a target nucleic acid sequence (target site) and that encodes the sequence specificity of the complex. The trans- activating crRNA (tracrRNA) base-pairs to a portion of the crRNA and forms a structure that complexes with the Cas9 protein, forming a Cas/RNA RNP complex. [0325] Naturally occurring CRISPR/Cas systems, such as those with Cas9, have been engineered to allow efficient programming of Cas/RNA RNPs to target desired sequences in cells of interest, both for gene-editing and modulation of gene expression. The tracrRNA and crRNA have been engineered to form a single chimeric guide RNA molecule, commonly referred to as a guide RNA (gRNA), for example as described in WO 2013/176772, WO 2014/093661, WO 2014/093655, Jinek, M. et al. Science 337(6096):816-21 (2012), or Cong, L. et al. Science 339(6121):819-23 (2013). The spacer sequence of the gRNA can be chosen by a user to target the Cas/gRNA RNP complex to a desired locus, e.g. a desired target site in the target gene. [0326] Cas proteins have also been engineered to be catalytically inactivated or nuclease inactive to allow targeting of Cas/gRNA RNPs without inducing cleavage at the target site. Mutations in Cas proteins can reduce or abolish nuclease activity of the Cas protein, rendering the Cas protein catalytically inactive. Cas proteins with reduced or abolished nuclease activity are referred to as deactivated Cas (dCas), or nuclease-inactive Cas (iCas) proteins, as referred to interchangeably herein. An exemplary deactivated Cas9 (dCas9) derived from S. pyogenes contains silencing mutations of the RuvC and HNH nuclease domains (D10A and H840A), for example as described in WO 2013/176772, WO 2014/093661, Jinek, M. et al. Science 337(6096):816-21 (2012), and Qi, L. et al. Cell 152(5):1173-83 (2013). Exemplary dCas variants derived from the Cas12 system (i.e. Cpf1) are described, for example in WO 2017/189308 and Zetsche, B. et al. Cell 163(3):759-71 (2015). Conserved domains that mediate nucleic acid cleavage, such as RuvC and HNH endonuclease domains, are readily identifiable in Cas orthologues, and can be mutated to produce inactive variants, for example as described in Zetsche, B. et al. Cell 163(3):759-71 (2015). [0327] dCas-fusion proteins with transcriptional and/or epigenetic regulators have been used as a versatile platform for ectopically regulating gene expression in target cells. These include fusion of a Cas with an effector domain, such as a transcriptional activator or transcriptional repressor. For example, fusing dCas9 with a transcriptional activator such as VP64 (a polypeptide composed of four tandem copies of VP16, a 16 amino acid transactivation domain sf-6059407 22474-20028.40 of the Herpes simplex virus) can result in robust induction of gene expression. Alternatively, fusing dCas9 with a transcriptional repressor such as KRAB (Krüppel associated box) can result in robust repression of gene expression. A variety of dCas-fusion proteins with effector domains can be engineered for regulation of gene expression, for example as described in WO 2014/197748, WO 2016/130600, WO 2017/180915, WO 2021/226555, WO 2013/176772, WO 2014/152432, WO 2014/093661, WO 2021/247570, Adli, M. Nat. Commun.9, 1911 (2018), Perez-Pinera, P. et al. Nat. Methods 10, 973–976 (2013), Mali, P. et al. Nat. Biotechnol.31, 833–838 (2013), Maeder, M. L. et al. Nat. Methods 10, 977–979 (2013), Gilbert, L. A. et al. Cell 154(2):442-451 (2013), and Nuñez, J.K. et al. Cell 184(9):2503-2519 (2021). 1. CRISPR/Cas-Based DNA-Binding Domains [0328] In some aspects, the first and/or second DNA-binding domain comprises a CRISPR- associated (Cas) protein. In particular embodiments here, the Cas protein is nuclease-inactive (i.e., is a dCas protein). [0329] In some embodiments, the Cas protein is derived from a Class 1 CRISPR system (i.e., multiple Cas protein system), such as a Type I, Type III, or Type IV CRISPR system. In some embodiments, the Cas protein is derived from a Class 2 CRISPR system (i.e., single Cas protein system), such as a Type II, Type V, or Type VI CRISPR system. In some embodiments, the Cas protein is from a Type V CRISPR system. In some embodiments, the Cas protein is a Cas12 protein (i.e., Cpf1), for example as described in WO 2017/189308 and Zetsche, B. et al. Cell.163(3):759-71 (2015). In some embodiments, the Cas protein is derived from a Type II CRISPR system. In some embodiments, the Cas protein is a Cas9 protein, for example as described in WO 2013/176772, WO 2014/152432, WO 2014/093661, WO 2014/093655, Jinek, M. et al. Science 337(6096):816-21 (2012), Mali, P. et al. Science 339(6121):823-6 (2013), Cong, L. et al. Science 339(6121):819-23 (2013), Perez-Pinera, P. et al. Nat. Methods 10, 973– 976 (2013), or Mali, P. et al. Nat. Biotechnol.31, 833–838 (2013). Various CRISPR/Cas systems and associated Cas proteins for use in gene editing and regulation have been described, for example in Moon, S.B. et al. Exp. Mol. Med.51, 1–11 (2019), Zhang, F. Q. Rev. Biophys. 52, E6 (2019), and Makarova K.S. et al. Methods Mol. Biol.1311:47-75 (2015). [0330] In some embodiments, the dCas9 protein comprises a sequence derived from a naturally occurring Cas9 molecule. In some embodiments, the dCas9 protein comprises a sequence derived from a naturally occurring Cas9 molecule of S. pyogenes, S. thermophilus, S. aureus, C. jejuni, N. meningitidis, F. novicida, S. canis, or S. auricularis. In some embodiments, sf-6059407 22474-20028.40 the dCas9 protein comprises a sequence derived from a naturally occurring Cas9 molecule of S. aureus. In some embodiments, the dCas9 protein comprises a sequence derived from a naturally occurring Cas9 molecule of S. pyogenes. [0331] Non-limiting examples of Cas9 orthologs from other bacterial strains include Cas proteins identified in Acaryochloris marina MBIC11017; Acetohalobium arabaticum DSM 5501; Acidithiobacillus caldus; Acidithiobacillus ferrooxidans ATCC 23270; Alicyclobacillus acidocaldarius LAA1; Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446; Allochromatium vinosum DSM 180; Ammonifex degensii KC4; Anabaena variabilis ATCC 29413; Arthrospira maxima CS-328; Arthrospira platensis str. Paraca; Arthrospira sp. PCC 8005; Bacillus pseudomycoides DSM 12442; Bacillus selenitireducens MLS10; Burkholderiales bacterium 1_1_47; Caldicelulosiruptor becscii DSM 6725; Candidatus Desulforudis audaxviator MP104C; Caldicellulosiruptor hydrothermalis 108; Clostridium phage c-st; Clostridium botulinum A3 str. Loch Maree; Clostridium botulinum Ba4 str.657; Clostridium difficile QCD- 63q42; Crocosphaera watsonii WH 8501; Cyanothece sp. ATCC 51142; Cyanothece sp. CCY0110; Cyanothece sp. PCC 7424; Cyanothece sp. PCC 7822; Exiguobacterium sibiricum 255-15; Finegoldia magna ATCC 29328; Ktedonobacter racemifer DSM 44963; Lactobacillus delbrueckii subsp. bulgaricus PB2003/044-T3-4; Lactobacillus salivarius ATCC 11741; Listeria innocua; Lyngbya sp. PCC 8106; Marinobacter sp. ELB17; Methanohalobium evestigatum Z- 7303; Microcystis phage Ma-LMM01; Microcystis aeruginosa NIES-843; Microscilla marina ATCC 23134; Microcoleus chthonoplastes PCC 7420; Neisseria meningitidis; Nitrosococcus halophilus Nc4; Nocardiopsis dassonvillei subsp. dassonvillei DSM 43111; Nodularia spumigena CCY9414; Nostoc sp. PCC 7120; Oscillatoria sp. PCC 6506; Pelotomaculum_thermopropionicum SI; Petrotoga mobilis SJ95; Polaromonas naphthalenivorans CJ2; Polaromonas sp. JS666; Pseudoalteromonas haloplanktis TAC125; Streptomyces pristinaespiralis ATCC 25486; Streptomyces pristinaespiralis ATCC 25486; Streptococcus thermophilus; Streptomyces viridochromogenes DSM 40736; Streptosporangium roseum DSM 43021; Synechococcus sp. PCC 7335; and Thermosipho africanus TCF52B (Chylinski et al., RNA Biol., 2013; 10(5): 726-737). [0332] In some aspects, the Cas protein is a variant that lacks nuclease activity (i.e., is a dCas protein). In some embodiments, the Cas protein is mutated so that nuclease activity is reduced or eliminated. Such Cas proteins are referred to as deactivated Cas or dead Cas (dCas) or nuclease-inactive Cas (iCas) proteins, as referred to interchangeably herein. In some sf-6059407 22474-20028.40 embodiments, the variant Cas protein is a variant Cas9 protein that lacks nuclease activity or that is a deactivated Cas9 (dCas9 or iCas9) protein. [0333] In some embodiments, the Cas9 protein is derived from a Staphylococcus aureus Cas9 (SaCas9) protein or a variant thereof. In some embodiments, the variant Cas9 is a Staphylococcus aureus dCas9 protein (dSaCas9) that comprises at least one amino acid mutation selected from D10A and N580A, with reference to numbering of positions of SEQ ID NO:124. In some embodiments, the variant Cas9 protein comprises the sequence set forth in SEQ ID NO:125, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. [0334] In some embodiments, the Cas9 protein is derived from a Streptococcus pyogenes Cas9 (SpCas9) protein or a variant thereof. In some embodiments, the variant Cas9 is a Streptococcus pyogenes dCas9 (dSpCas9) protein that comprises at least one amino acid mutation selected from D10A and H840A, with reference to numbering of positions of SEQ ID NO:126. In some embodiments, the variant Cas9 protein comprises the sequence set forth in SEQ ID NO:127, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 2. Guide RNAs (gRNAs) [0335] In some embodiments, the Cas protein (e.g., dCas9) is provided in combination or as a complex with one or more guide RNA (gRNA). In some aspects, the gRNA is a nucleic acid that promotes the specific targeting or homing of the gRNA/Cas RNP complex to the target site of a target gene. In some embodiments, a target site of a gRNA may be referred to as a protospacer. [0336] In some embodiments, the gRNA is capable of complexing with the Cas protein. In some embodiments, the gRNA comprises a gRNA spacer sequence (i.e., a spacer sequence or a guide sequence) that is capable of hybridizing to the target site, or that is complementary to the target site. In some embodiments, the gRNA comprises a scaffold sequence that complexes with or binds to the Cas protein. [0337] In some embodiments, the gRNA is a chimeric gRNA. In general, gRNAs can be unimolecular (i.e., composed of a single RNA molecule), or modular (comprising more than one, and typically two, separate RNA molecules). Modular gRNAs can be engineered to be unimolecular, wherein sequences from the separate modular RNA molecules are comprised in a single gRNA molecule, sometimes referred to as a chimeric gRNA, synthetic gRNA, or single sf-6059407 22474-20028.40 gRNA. In some embodiments, the chimeric gRNA is a fusion of two non-coding RNA sequences: a crRNA sequence and a tracrRNA sequence, for example as described in WO 2013/176772, or Jinek, M. et al. Science 337(6096):816-21 (2012). In some embodiments, the chimeric gRNA mimics the naturally occurring crRNA:tracrRNA duplex involved in the Type II Effector system, wherein the naturally occurring crRNA:tracrRNA duplex acts as a guide for the Cas9 protein. [0338] In some aspects, the spacer sequence of a gRNA is a polynucleotide sequence comprising at least a portion that has sufficient complementarity with the target site to hybridize with the target site in the target gene and direct sequence-specific binding of a Cas/gRNA complex to the sequence of the target site. Full complementarity is not necessarily required, provided there is sufficient complementarity to cause hybridization. In some embodiments, the gRNA comprises a spacer sequence that is complementary, e.g., at least 80%, 85%, 90%, 95%, 98%, 99%, or 100% (e.g., fully complementary), to the target site. The strand of the target nucleic acid comprising the target site sequence may be referred to as the “complementary strand” of the target nucleic acid. [0339] In some aspects, a gRNA targets a target site in double-stranded DNA. Thus, in some aspects, the sequence of the target site may be defined by the sequence that the gRNA spacer hybridizes to, or by the sequence complementary to the sequence that the gRNA spacer hybridizes to. In some aspects, the sequence of the target site may be defined by the sequence that the gRNA spacer displaces in order to hybridize to the DNA. In some embodiments, the sequence of the target site is the sequence that the gRNA hybridizes to. [0340] In some embodiments, the gRNA spacer sequence is between about 14 nucleotides (nt) and about 26 nt, or between 16 nt and 22 nt in length. In some embodiments, the gRNA spacer sequence is 14 nt, 15 nt, 16 nt, 17 nt,18 nt, 19 nt, 20 nt, 21 nt or 22 nt, 23 nt, 24 nt, 25 nt, or 26 nt in length. In some embodiments, the gRNA spacer sequence is 18 nt, 19 nt, 20 nt, 21 nt or 22 nt in length. In some embodiments, the gRNA spacer sequence is 20 nt in length. [0341] A target site of a gRNA may be referred to as a protospacer. In some aspects, the spacer is designed to target a protospacer with a specific protospacer-adjacent motif (PAM), i.e., a sequence immediately adjacent to the protospacer that contributes to and/or is required for Cas binding specificity. Different CRISPR/Cas systems have different PAM requirements for targeting. For example, in some embodiments, S. pyogenes Cas9 uses the PAM 5’-NGG-3’ (SEQ ID NO: 224), where N is any nucleotide. In some embodiments, S. aureus Cas9 uses the sf-6059407 22474-20028.40 PAM 5’- NNGRRT-3’ (SEQ ID NO: 225), where N is any nucleotide, and R is G or A. In some embodiments, N. meningitidis Cas9 uses the PAM 5′-NNNNGATT-3’ (SEQ ID NO: 226), where N is any nucleotide. In some embodiments, C. jejuni Cas9 uses the PAM 5′- NNNNRYAC-3′ (SEQ ID NO: 227), where N is any nucleotide, R is G or A, and Y is C or T. In some embodiments, S. thermophilus uses the PAM 5’-NNAGAAW-3’ (SEQ ID NO: 228), where N is any nucleotide and W is A or T. In some embodiments, F. Novicida Cas9 uses the PAM 5’-NGG-3’ (SEQ ID NO: 224), where N is any nucleotide. In some embodiments, T. denticola Cas9 uses the PAM 5’-NAAAAC-3’ (SEQ ID NO: 229), where N is any nucleotide. In some embodiments, Cas12a (also known as Cpf1) from various species, uses the PAM 5’- TTTV-3’ (SEQ ID NO: 230). In some embodiments, Cas proteins may use or be engineered to use different PAMs from those listed above. For example, mutated SpCas9 proteins may use the PAMs 5’-NGG-3’ (SEQ ID NO: 224), 5’-NGAN-3’ (SEQ ID NO: 231), 5’-NGNG-3’ (SEQ ID NO: 232), 5’-NGAG-3’ (SEQ ID NO: 233), or 5’-NGCG-3’ (SEQ ID NO: 234). In some embodiments, the protospacer-adjacent motif (PAM) of a gRNA for complexing with S. pyogenes Cas9 is NGG, as set forth in SEQ ID NO: 224. In some embodiments, the PAM of a gRNA for complexing with S. aureus Cas9 is NNGRRT, as set forth in SEQ ID NO: 225. [0342] A spacer sequence may be selected to reduce the degree of secondary structure within the spacer sequence. Secondary structure may be determined by any suitable polynucleotide folding algorithm. [0343] In some embodiments, the gRNA (including the guide sequence) will comprise the base uracil (U), whereas DNA encoding the gRNA molecule will comprise the base thymine (T). While not wishing to be bound by theory, in some embodiments, it is believed that the complementarity of the guide sequence with the target sequence contributes to specificity of the interaction of the gRNA molecule/Cas molecule complex with a target nucleic acid. It is understood that in a guide sequence and target sequence pair, the uracil bases in the guide sequence will pair with the adenine bases in the target sequence. [0344] In some embodiments, one, more than one, or all of the nucleotides of a gRNA can have a modification, e.g., to render the gRNA less susceptible to degradation and/or improve bio-compatibility. By way of non-limiting example, the backbone of the gRNA can be modified with a phosphorothioate, or other modification(s). In some cases, a nucleotide of the gRNA can comprise a 2’ modification, e.g., a 2-acetylation, e.g., a 2’ methylation, or other modification(s). [0345] In some embodiments, the gRNA further comprises a scaffold sequence. In some sf-6059407 22474-20028.40 aspects, the scaffold sequence (in some cases including a crRNA sequence and/or a tracrRNA sequence) will be different depending on the Cas protein. In some aspects, different CRISPR/Cas systems have different gRNA scaffold sequences for associating with Cas protein. [0346] In some embodiments, the scaffold sequence is an SpCas9 scaffold sequence. In some embodiments, the scaffold sequence comprises the sequence set forth in SEQ ID NO:69 (GUUUAAGAGCUAUGCUGGAAACAGCAUAGCAAGUUUAAAUAAGGCUAGUCCGU UAUCAACUUGAAAAAGUGGCACCGAGUCGGUGC), or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to all or a portion thereof. In some embodiments, the scaffold sequence is set forth in SEQ ID NO: 69. [0347] In some embodiments, the scaffold sequence is an SaCas9 scaffold sequence. In some embodiments, the scaffold sequence comprises the sequence set forth in SEQ ID NO:387 (GUUUUAGUACUCUGGAAACAGAAUCUACUAAAACAAGGCAAAAUGCCGUGUUU AUCUCGUCAACUUGUUGGCGAGA), or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to all or a portion thereof. In some embodiments, the scaffold sequence is set forth in SEQ ID NO: 387. [0348] In some embodiments, the scaffold sequence is an Acidaminococcus sp. Cas12a scaffold sequence. In some embodiments, the scaffold sequence comprises a sequence set forth in SEQ ID NO:443, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to SEQ ID NO:443. In some embodiments, the scaffold sequence is set forth in SEQ ID NO: 443. [0349] In some embodiments, the scaffold sequence is a CasPhi-2 scaffold sequence. In some embodiments, the scaffold sequence comprises a sequence set forth in SEQ ID NO:444, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to SEQ ID NO:444. In some embodiments, the scaffold sequence is set forth in SEQ ID NO: 444. [0350] In some embodiments, the scaffold sequence is a Un1Cas12f1 scaffold sequence. In some embodiments, the scaffold sequence comprises a sequence set forth in SEQ ID NO:445, a sequence “GGAAUGAAC” as set forth in SEQ ID NO:446, or a sequence “UUUUAUUUU” as set forth in SEQ ID NO:447, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to SEQ ID sf-6059407 22474-20028.40 NO:445, 446, or 447. In some embodiments, the scaffold sequence comprises a sequence set forth in SEQ ID NO:445, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to SEQ ID NO:445. In some embodiments, the scaffold sequence is set forth in SEQ ID NO: 445. In some embodiments, the scaffold sequence comprises a sequence set forth in SEQ ID NO:446, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to SEQ ID NO:446. In some embodiments, the scaffold sequence is set forth in SEQ ID NO: 446. In some embodiments, the scaffold sequence comprises a sequence set forth in SEQ ID NO:447, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to SEQ ID NO:447. In some embodiments, the scaffold sequence is set forth in SEQ ID NO: 447. [0351] In some embodiments, the scaffold sequence is a C. jejuni Cas9 scaffold sequence. In some embodiments, the scaffold sequence comprises a sequence set forth in SEQ ID NO:448, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to SEQ ID NO:448. In some embodiments, the scaffold sequence is set forth in SEQ ID NO: 448. [0352] In some embodiments, the scaffold sequence is a Cas12k scaffold sequence. In some embodiments, the scaffold sequence comprises a sequence set forth in SEQ ID NO:449, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to SEQ ID NO:449. In some embodiments, the scaffold sequence is set forth in SEQ ID NO: 449. [0353] In some embodiments, the scaffold sequence is a CasMini scaffold sequence. In some embodiments, the scaffold sequence comprises a sequence set forth in SEQ ID NO:450, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to SEQ ID NO:450. In some embodiments, the scaffold sequence is set forth in SEQ ID NO: 450. [0354] Methods for designing gRNAs and exemplary targeting domains can include those described in, e.g., International PCT Pub. Nos. WO 2014/197748, WO 2016/130600, WO 2017/180915, WO 2021/226555, WO 2013/176772, WO 2014/152432, WO 2014/093661, WO 2014/093655, WO 2015/089427, WO 2016/049258, WO 2016/123578, WO 2021/076744, WO 2014/191128, WO 2015/161276, WO 2017/193107, and WO 2017/093969. sf-6059407 22474-20028.40 a. gRNAs for Transcriptional Activation [0355] In some embodiments, a first gRNA targets a target site for a first gene for transcriptional activation. In some embodiments, the first gRNA targets a target site for a first gene, such as a first gene in a T cell, wherein the first gene is selected from BATF, CD28, EOMES, IL-2, IL2RB, IRF4, LAT, LCP2, TBX21, and VAV1. In some embodiments, the first gRNA targets IL-2. [0356] In some embodiments, the first gRNA is a Cas9 gRNA. In some embodiments, the first gRNA is a Cas12 gRNA. [0357] In some embodiments, the first gRNA is an SpCas9 gRNA. In some embodiments, the first gRNA is an SaCas9 gRNA. [0358] In some embodiments, the first gRNA targets a target site that comprises a sequence selected from any one of SEQ ID NOS:7-9, 78, 144-156, 170, 172-177, 184-191, and 388-412 as shown in Table 5, a contiguous portion thereof of at least 14 nucleotides, a complementary sequence of any of the foregoing, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. In some embodiments, the target site is a contiguous portion of any one of SEQ ID NOS:7-9, 78, 144-156, 170, 172-177, 184-191, and 388-412 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the target site is set forth in any one of SEQ ID NOS: 7-9, 78, 144-156, 170, 172-177, 184-191, and 388-412. [0359] In some embodiments, the first gRNA comprises a spacer sequence selected from any one of SEQ ID NOS:41-43, 79, 157-169, 171, 178-183, 192-199, and 413-437, as shown in Table 5, or a contiguous portion thereof of at least 14 nt, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. In some embodiments, the spacer sequence of the first gRNA is a contiguous portion of any one of SEQ ID NOS:41-43, 79, 157-169, 171, 178-183, 192-199, and 413-437 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the spacer sequence of the first gRNA is set forth in any one of SEQ ID NOS:41- 43, 79, 157-169, 171, 178-183, 192-199, and 413-437. [0360] In some embodiments, the first gRNA is an SpCas9 gRNA that targets a target site that comprises a sequence selected from any one of SEQ ID NOS:7-9, 78, 144-156, 170, 172- 177, 184-191, and 388-403 as shown in Table 5, a contiguous portion thereof of at least 14 nucleotides, a complementary sequence of any of the foregoing, or a sequence having at or at sf-6059407 22474-20028.40 least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. In some embodiments, the target site is a contiguous portion of any one of SEQ ID NOS:7-9, 78, 144-156, 170, 172-177, 184-191, and 388-403 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the target site is set forth in any one of SEQ ID NOS: 7-9, 78, 144-156, 170, 172-177, 184-191, and 388- 403. [0361] In some embodiments, the first gRNA is an SpCas9 gRNA that comprises a spacer sequence selected from any one of SEQ ID NOS:41-43, 79, 157-169, 171, 178-183, 192-199, and 413-428, as shown in Table 5, or a contiguous portion thereof of at least 14 nt, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. In some embodiments, the spacer sequence of the first gRNA is a contiguous portion of any one of SEQ ID NOS:41-43, 79, 157-169, 171, 178-183, 192-199, and 413-428 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the spacer sequence of the first gRNA is set forth in any one of SEQ ID NOS:41-43, 79, 157-169, 171, 178-183, 192-199, and 413-428. [0362] In some embodiments, the first gRNA is an SaCas9 gRNA that targets a target site that comprises a sequence selected from any one of SEQ ID NOS:404-412 as shown in Table 5, a contiguous portion thereof of at least 14 nucleotides, a complementary sequence of any of the foregoing, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. In some embodiments, the target site is a contiguous portion of any one of SEQ ID NOS:404-412 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the target site is set forth in any one of SEQ ID NOS: 404-412. [0363] In some embodiments, the first gRNA is an SpCas9 gRNA that comprises a spacer sequence selected from any one of SEQ ID NOS:429-437, as shown in Table 5, or a contiguous portion thereof of at least 14 nt, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. In some embodiments, the spacer sequence of the first gRNA is a contiguous portion of any one of SEQ ID NOS:429-437 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the spacer sequence of the first gRNA is set forth in any one of SEQ ID NOS:429-437. [0364] In some embodiments, the first gRNA targets IL-2. sf-6059407 22474-20028.40 [0365] In some embodiments, the first gRNA targets an IL-2 target site that comprises a sequence selected from any one of SEQ ID NOS:78 and 388-412 as shown in Table 5, a contiguous portion thereof of at least 14 nucleotides, a complementary sequence of any of the foregoing, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. In some embodiments, the IL-2 target site is a contiguous portion of any one of SEQ ID NOS:78 and 388-412 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the IL- 2 target site is set forth in any one of SEQ ID NOS: 78 and 388-412. [0366] In some embodiments, the first gRNA comprises an IL-2-targeting spacer sequence selected from any one of SEQ ID NOS:79 and 413-437, as shown in Table 5, or a contiguous portion thereof of at least 14 nt, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. In some embodiments, the IL-2 targeting spacer sequence of the first gRNA is a contiguous portion of any one of SEQ ID NOS:79 and 413-437 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the IL-2 targeting spacer sequence of the first gRNA is set forth in any one of SEQ ID NOS:79 and 413-437. [0367] In some embodiments, the first gRNA is an SpCas9 gRNA that targets an IL-2 target site that comprises a sequence selected from any one of SEQ ID NOS:78 and 388-403 as shown in Table 5, a contiguous portion thereof of at least 14 nucleotides, a complementary sequence of any of the foregoing, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. In some embodiments, the IL-2 target site is a contiguous portion of any one of SEQ ID NOS:78 and 388-403 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the IL- 2 target site is set forth in any one of SEQ ID NOS:78 and 388-403. [0368] In some embodiments, the first gRNA is an SpCas9 gRNA that comprises an IL-2 targeting spacer sequence selected from any one of SEQ ID NOS:79 and 413-428, as shown in Table 5, or a contiguous portion thereof of at least 14 nt, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. In some embodiments, the IL-2 targeting spacer sequence of the first gRNA is a contiguous portion of any one of SEQ ID NOS:79 and 413-428 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the IL-2 targeting spacer sequence of the first gRNA is set forth in any one of SEQ ID NOS:79 and 413-428. sf-6059407 22474-20028.40 [0369] In some embodiments, the first gRNA is an SaCas9 gRNA that targets an IL-2 target site that comprises a sequence selected from any one of SEQ ID NOS:404-412 as shown in Table 5, a contiguous portion thereof of at least 14 nucleotides, a complementary sequence of any of the foregoing, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. In some embodiments, the IL-2 target site is a contiguous portion of any one of SEQ ID NOS:404-412 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the IL-2 target site is set forth in any one of SEQ ID NOS: 404-412. [0370] In some embodiments, the first gRNA is an SpCas9 gRNA that comprises an IL-2- targeting spacer sequence selected from any one of SEQ ID NOS:429-437, as shown in Table 5, or a contiguous portion thereof of at least 14 nt, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. In some embodiments, the IL-2 targeting spacer sequence of the first gRNA is a contiguous portion of any one of SEQ ID NOS:429-437 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the IL-2 targeting spacer sequence of the first gRNA is set forth in any one of SEQ ID NOS:429-437. [0371] In some embodiments, a combination of first gRNAs that each target a target site for a first gene for transcriptional activation is used. In some embodiments, a combination of first gRNAs comprising at least two first gRNAs targeting at least two different first genes for transcriptional activation is used. In some embodiments, the first gRNAs target a combination of first genes selected from the combinations of first genes listed in Table 1. [0372] In some embodiments, the combination of first gRNAs comprises a plurality of first gRNAs targeted to at least two different first genes. In some embodiments, one first gRNA targets BATF, CD28, EOMES, IL-2, IL2RB, IRF4, LAT, LCP2, TBX21, or VAV1, and the other first gRNA targets a different gene from BATF, CD28, EOMES, IL-2, IL2RB, IRF4, LAT, LCP2, TBX21, and VAV1. In some embodiments, one first gRNA targets IL-2, and the other first gRNA targets BATF, CD28, EOMES, IL-2, IL2RB, IRF4, LAT, LCP2, TBX21, or VAV1. In some embodiments, one first gRNA targets VAV1, and the other first gRNA targets BATF, CD28, EOMES, IL-2, IL2RB, IRF4, LAT, LCP2, TBX21, or VAV1. [0373] In some embodiments, one first gRNA targets IL-2 and the other first gRNA targets VAV1. In some embodiments, one first gRNA targets a target site for IL-2 having the sequence sf-6059407 22474-20028.40 set forth in SEQ ID NO:78 and the other first gRNA targets a target site for VAV1 having the sequence set forth in SEQ ID NO:170. [0374] In some embodiments, the combination of first gRNAs comprises at least three first gRNAs each targeting a different first gene selected from BATF, CD28, EOMES, IL-2, IL2RB, IRF4, LAT, LCP2, TBX21, and VAV1. [0375] In some embodiments, the combination of first gRNAs targets a combination of target sites for a combination of first genes for transcriptional activation, for instance as shown in Table 2. [0376] In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for BATF comprising the sequence set forth in SEQ ID NO:172, and another first gRNA that targets a target site for IL-2 comprising the sequence set forth in SEQ ID NO:78. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for BATF comprising the sequence set forth in SEQ ID NO:172, and another first gRNA that targets a target site for VAV1 comprising the sequence set forth in SEQ ID NO:170. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for CD28 comprising the sequence set forth in SEQ ID NO:144, and another first gRNA that targets a target site for BATF comprising the sequence set forth in SEQ ID NO:172. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for CD28 comprising the sequence set forth in SEQ ID NO:144, and another first gRNA that targets a target site for EOMES comprising the sequence set forth in SEQ ID NO:149. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for CD28 comprising the sequence set forth in SEQ ID NO:144, and another first gRNA that targets a target site for IL-2 comprising the sequence set forth in SEQ ID NO:78. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for CD28 comprising the sequence set forth in SEQ ID NO:144, and another first gRNA that targets a target site for LCP2 comprising the sequence set forth in SEQ ID NO:151. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for CD28 comprising the sequence set forth in SEQ ID NO:144, and another first gRNA that targets a target site for TBX21 comprising the sequence set forth in SEQ ID NO:155. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for CD28 comprising the sequence set forth in SEQ ID NO:144, and another first gRNA that targets a target site for VAV1 comprising the sequence set forth in sf-6059407 22474-20028.40 SEQ ID NO:170. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for EOMES comprising the sequence set forth in SEQ ID NO:149, and another first gRNA that targets a target site for BATF comprising the sequence set forth in SEQ ID NO:172. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for EOMES comprising the sequence set forth in SEQ ID NO:149, and another first gRNA that targets a target site for LCP2 comprising the sequence set forth in SEQ ID NO:151. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for EOMES comprising the sequence set forth in SEQ ID NO:149, and another first gRNA that targets a target site for TBX21 comprising the sequence set forth in SEQ ID NO:155. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for EOMES comprising the sequence set forth in SEQ ID NO:149, and another first gRNA that targets a target site for VAV1 comprising the sequence set forth in SEQ ID NO:170. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for LCP2 comprising the sequence set forth in SEQ ID NO:151, and another first gRNA that targets a target site for BATF comprising the sequence set forth in SEQ ID NO:172. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for LCP2 comprising the sequence set forth in SEQ ID NO:151, and another first gRNA that targets a target site for IL-2 comprising the sequence set forth in SEQ ID NO:78. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for LCP2 comprising the sequence set forth in SEQ ID NO:151, and another first gRNA that targets a target site for TBX21 comprising the sequence set forth in SEQ ID NO:155. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for LCP2 comprising the sequence set forth in SEQ ID NO:151, and another first gRNA that targets a target site for VAV1 comprising the sequence set forth in SEQ ID NO:170. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for TBX21 comprising the sequence set forth in SEQ ID NO:155, and another first gRNA that targets a target site for BATF comprising the sequence set forth in SEQ ID NO:172. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for TBX21 comprising the sequence set forth in SEQ ID NO:155, and another first gRNA that targets a target site for IL-2 comprising the sequence set forth in SEQ ID NO:78. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for TBX21 comprising the sequence set forth in SEQ ID sf-6059407 22474-20028.40 NO:155, and another first gRNA that targets a target site for TBX21 comprising the sequence set forth in SEQ ID NO:155. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for TBX21 comprising the sequence set forth in SEQ ID NO:155, and another first gRNA that targets a target site for VAV1 comprising the sequence set forth in SEQ ID NO:170. In some embodiments, the combination of first gRNAs comprises one first gRNA that targets a target site for VAV1 comprising the sequence set forth in SEQ ID NO:170, and another first gRNA that targets a target site for IL-2 comprising the sequence set forth in SEQ ID NO:78. Table 5. First genes, target sites, and first gRNAs for transcriptional activation

sf-6059407 22474-20028.40
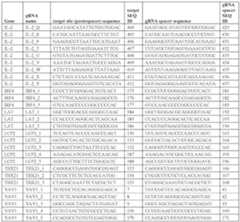
b. gRNAs for Transcriptional Repression [0377] In some embodiments, a second gRNA targets a target site for a second gene for transcriptional repression. In some embodiments, the second gRNA targets a target site for a second gene, such as a second gene in a T cell, wherein the second gene is selected from the list consisting of: CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and RASA2. In some embodiments, the second gRNA targets MED12. [0378] In some embodiments, the second gRNA is a Cas9 gRNA. In some embodiments, the sf-6059407 22474-20028.40 second gRNA is a Cas12 gRNA. [0379] In some embodiments, the second gRNA is an SpCas9 gRNA. In some embodiments, the second gRNA is an SaCas9 gRNA. [0380] In some embodiments, the second gRNA targets a target site that comprises a sequence selected from any one of SEQ ID NOS:1-6, 10-33, 80-90, 102-112, 200-211, 292-295, 300-302, and 306-308, as shown in Table 6, a contiguous portion thereof of at least 14 nucleotides, a complementary sequence of any of the foregoing, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. In some embodiments, the target site is a contiguous portion of any one of SEQ ID NOS: 1-6, 10-33, 80-90, 102-112, and 200-211 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the target site is set forth in any one of SEQ ID NOS: 1-6, 10-33, 80-90, 102-112, 200-211, 292-295, 300-302, and 306-308. [0381] In some embodiments, the second gRNA comprises a spacer sequence selected from any one of SEQ ID NOS:35-40, 44-67, 91-101, 113-123, 212-223, 296-299, 303-305, and 309- 311, as shown in Table 6, or a contiguous portion thereof of at least 14 nt, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. In some embodiments, the spacer sequence of the second gRNA is a contiguous portion of any one of SEQ ID NOS:35-40, 44-67, 91-101, 113-123, 212-223, 296-299, 303-305, and 309-311 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the spacer sequence of the second gRNA is set forth in any one of SEQ ID NOS:35-40, 44-67, 91-101, 113-123, 212-223, 296-299, 303-305, and 309-311. [0382] In some embodiments, the second gRNA is an SpCas9 gRNA that targets a target site that comprises a sequence selected from any one of SEQ ID NOS:1-6, 10-33, 80-90, 102-112, 200-211, 292-295, 300-302, and 306-308, as shown in Table 6, a contiguous portion thereof of at least 14 nucleotides, a complementary sequence of any of the foregoing, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. In some embodiments, the target site is a contiguous portion of any one of SEQ ID NOS: 1-6, 10-33, 80-90, 102-112, and 200-211 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the target site is set forth in any one of SEQ ID NOS: 1-6, 10-33, 80-90, 102-112, 200-211, 292-295, 300-302, and 306-308. sf-6059407 22474-20028.40 [0383] In some embodiments, the second gRNA is an SpCas9 gRNA that comprises a spacer sequence selected from any one of SEQ ID NOS:35-40, 44-67, 91-101, 113-123, 212-223, 296- 299, 303-305, and 309-311, as shown in Table 6, or a contiguous portion thereof of at least 14 nt, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. In some embodiments, the spacer sequence of the second gRNA is a contiguous portion of any one of SEQ ID NOS:35-40, 44-67, 91-101, 113-123, 212-223, 296-299, 303-305, and 309-311 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the spacer sequence of the second gRNA is set forth in any one of SEQ ID NOS:35-40, 44-67, 91-101, 113-123, 212-223, 296-299, 303-305, and 309-311. [0384] In some embodiments, the second gRNA targets MED12. [0385] In some embodiments, the second gRNA targets a MED12 target site that comprises a sequence selected from any one of SEQ ID NOS:80-90, as shown in Table 6, a contiguous portion thereof of at least 14 nucleotides, a complementary sequence of any of the foregoing, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. In some embodiments, the MED12 target site is a contiguous portion of any one of SEQ ID NOS: 80-90 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the MED12 target site is set forth in any one of SEQ ID NOS: 80-90. [0386] In some embodiments, the second gRNA comprises a MED12-targeting spacer sequence selected from any one of SEQ ID NOS:91-101, as shown in Table 6, or a contiguous portion thereof of at least 14 nt, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. In some embodiments, the MED12-targeting spacer sequence of the second gRNA is a contiguous portion of any one of SEQ ID NOS:91-101 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the MED12-targeting spacer sequence of the second gRNA is set forth in any one of SEQ ID NOS:91-101. [0387] In some embodiments, the second gRNA is an SpCas9 gRNA that targets a MED12 target site that comprises a sequence selected from any one of SEQ ID NOS:80-90, as shown in Table 6, a contiguous portion thereof of at least 14 nucleotides, a complementary sequence of any of the foregoing, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. sf-6059407 22474-20028.40 In some embodiments, the MED12 target site is a contiguous portion of any one of SEQ ID NOS:80-90 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the MED12 target site is set forth in any one of SEQ ID NOS:80-90. [0388] In some embodiments, the second gRNA is an SpCas9 gRNA that comprises a MED12-targeting spacer sequence selected from any one of SEQ ID NOS:91-101, as shown in Table 6, or a contiguous portion thereof of at least 14 nt, or a sequence having at or at least 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, 99.5%, 99.9%, or 100% sequence identity to any of the foregoing. In some embodiments, the MED12-targeting spacer sequence of the second gRNA is a contiguous portion of any one of SEQ ID NOS:91-101 that is 14, 15, 16, 17, 18 or 19 nucleotides in length. In some embodiments, the MED12-targeting spacer sequence of the second gRNA is set forth in any one of SEQ ID NOS:91-101. [0389] In some embodiments, a combination of second gRNAs that each target a target site for a second gene for transcriptional repression is used. In some embodiments, a combination of second gRNAs comprising at least two second gRNAs targeting at least two different second genes for transcriptional repression is used. In some embodiments, the second gRNAs target a combination of second genes selected from the combinations of second genes listed in Table 3. [0390] In some embodiments, the combination of second gRNAs comprises a plurality of second gRNAs targeted to at least two different second genes. In some embodiments, one second gRNA targets CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, or RASA2, and the other second gRNA targets a different gene from CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and RASA2. [0391] In some embodiments, one second gRNA targets CBLB, and the other second gRNA targets CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, or RASA2. In some embodiments, one second gRNA targets CCNC, and the other second gRNA targets CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, or RASA2. In some embodiments, one second gRNA targets MED12, and the other second gRNA targets CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, or RASA2. In some embodiments, one second gRNA targets MYB, and the other second gRNA targets CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, or RASA2. sf-6059407 22474-20028.40 [0392] In some embodiments, one second gRNA targets CBLB and the other second gRNA targets MYB. In some embodiments, one second gRNA targets a target site for CBLB having the sequence set forth in SEQ ID NO:11 and the other second gRNA targets a target site for MYB having the sequence set forth in SEQ ID NO:18. [0393] In some embodiments, one second gRNA targets CBLB and the other second gRNA targets CCNC. In some embodiments, one second gRNA targets a target site for CBLB having the sequence set forth in SEQ ID NO:11 and the other second gRNA targets a target site for CCNC having the sequence set forth in SEQ ID NO:104. [0394] In some embodiments, one second gRNA targets CBLB and the other second gRNA targets MED12. In some embodiments, one second gRNA targets a target site for CBLB having the sequence set forth in SEQ ID NO:11 and the other second gRNA targets a target site for MED12 having the sequence set forth in SEQ ID NO:81. [0395] In some embodiments, one second gRNA targets MED12 and the other second gRNA targets TGFBR2. In some embodiments, one second gRNA targets a target site for MED12 having the sequence set forth in SEQ ID NO:81 and the other second gRNA targets a target site for TGFBR2 having the sequence set forth in SEQ ID NO:301. [0396] In some embodiments, one second gRNA targets MED12 and the other second gRNA targets CISH. In some embodiments, one second gRNA targets a target site for MED12 having the sequence set forth in SEQ ID NO:81 and the other second gRNA targets a target site for CISH having the sequence set forth in SEQ ID NO:28 or 30. [0397] In some embodiments, the combination of second gRNAs comprises at least three second gRNAs each targeting a different second gene selected from CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and RASA2. [0398] In some embodiments, the combination of second gRNAs targets a combination of target sites for a combination of second genes for transcriptional repression, for intance as shown in Table 4. [0399] In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and another second gRNA that targets a target site for CCNC comprising the sequence set forth in SEQ ID NO:104. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and another second gRNA that targets a target site for CD5 comprising the sequence set sf-6059407 22474-20028.40 forth in SEQ ID NO:3. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and another second gRNA that targets a target site for CISH comprising the sequence set forth in SEQ ID NO:30. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and another second gRNA that targets a target site for DGKZ comprising the sequence set forth in SEQ ID NO:13. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and another second gRNA that targets a target site for ELOB comprising the sequence set forth in SEQ ID NO:24. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and another second gRNA that targets a target site for FAS comprising the sequence set forth in SEQ ID NO:204. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and another second gRNA that targets a target site for Fli1 comprising the sequence set forth in SEQ ID NO:208. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and another second gRNA that targets a target site for GATA3 comprising the sequence set forth in SEQ ID NO:26. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and another second gRNA that targets a target site for KDM1A comprising the sequence set forth in SEQ ID NO:4. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and another second gRNA that targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and another second gRNA that targets a target site for MYB comprising the sequence set forth in SEQ ID NO:18. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and another second gRNA that targets a target site for PRDM1 comprising the sequence set forth in SEQ ID NO:32. In some embodiments, the combination of second gRNAs comprises one sf-6059407 22474-20028.40 second gRNA that targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and another second gRNA that targets a target site for RASA2 comprising the sequence set forth in SEQ ID NO:19. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for CD5 comprising the sequence set forth in SEQ ID NO:3, and another second gRNA that targets a target site for CISH comprising the sequence set forth in SEQ ID NO:30. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for CD5 comprising the sequence set forth in SEQ ID NO:3, and another second gRNA that targets a target site for MYB comprising the sequence set forth in SEQ ID NO:18. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for CISH comprising the sequence set forth in SEQ ID NO:30, and another second gRNA that targets a target site for DGKZ comprising the sequence set forth in SEQ ID NO:13. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for CISH comprising the sequence set forth in SEQ ID NO:30, and another second gRNA that targets a target site for MYB comprising the sequence set forth in SEQ ID NO:18. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for CISH comprising the sequence set forth in SEQ ID NO:30, and another second gRNA that targets a target site for RASA2 comprising the sequence set forth in SEQ ID NO:19. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for GATA3 comprising the sequence set forth in SEQ ID NO:26, and another second gRNA that targets a target site for CD5 comprising the sequence set forth in SEQ ID NO:3. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for GATA3 comprising the sequence set forth in SEQ ID NO:26, and another second gRNA that targets a target site for CISH comprising the sequence set forth in SEQ ID NO:30. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for GATA3 comprising the sequence set forth in SEQ ID NO:26, and another second gRNA that targets a target site for MYB comprising the sequence set forth in SEQ ID NO:18. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and another second gRNA that targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and another sf-6059407 22474-20028.40 second gRNA that targets a target site for CD5 comprising the sequence set forth in SEQ ID NO:3. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and another second gRNA that targets a target site for CISH comprising the sequence set forth in SEQ ID NO:30. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and another second gRNA that targets a target site for DGKZ comprising the sequence set forth in SEQ ID NO:13. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and another second gRNA that targets a target site for ELOB comprising the sequence set forth in SEQ ID NO:24. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and another second gRNA that targets a target site for GATA3 comprising the sequence set forth in SEQ ID NO:26. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and another second gRNA that targets a target site for MYB comprising the sequence set forth in SEQ ID NO:18. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and another second gRNA that targets a target site for PRDM1 comprising the sequence set forth in SEQ ID NO:32. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and another second gRNA that targets a target site for RASA2 comprising the sequence set forth in SEQ ID NO:19. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, and another second gRNA that targets a target site for TGFBR2 comprising the sequence set forth in SEQ ID NO:301. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for MYB comprising the sequence set forth in SEQ ID NO:18, and another second gRNA that targets a target site for RASA2 comprising the sequence set forth in SEQ ID NO:19. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for PRDM1 comprising the sequence set forth in SEQ ID NO:32, and another second gRNA that targets a target site for CISH comprising the sequence set sf-6059407 22474-20028.40 forth in SEQ ID NO:30. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for PRDM1 comprising the sequence set forth in SEQ ID NO:32, and another second gRNA that targets a target site for GATA3 comprising the sequence set forth in SEQ ID NO:26. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for PRDM1 comprising the sequence set forth in SEQ ID NO:32, and another second gRNA that targets a target site for MYB comprising the sequence set forth in SEQ ID NO:18. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for PRDM1 comprising the sequence set forth in SEQ ID NO:32, and another second gRNA that targets a target site for RASA2 comprising the sequence set forth in SEQ ID NO:19. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for CD5 comprising the sequence set forth in SEQ ID NO:3, another second gRNA that targets a target site for CISH comprising the sequence set forth in SEQ ID NO:30, and a third gRNA that targets a target site for MYB comprising the sequence set forth in SEQ ID NO:18. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for GATA3 comprising the sequence set forth in SEQ ID NO:26, another second gRNA that targets a target site for CBLB comprising the sequence set forth in SEQ ID NO:11, and a third gRNA that targets a target site for MYB comprising the sequence set forth in SEQ ID NO:18. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for GATA3 comprising the sequence set forth in SEQ ID NO:26, another second gRNA that targets a target site for CD5 comprising the sequence set forth in SEQ ID NO:3, and a third gRNA that targets a target site for MYB comprising the sequence set forth in SEQ ID NO:18. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for PRDM1 comprising the sequence set forth in SEQ ID NO:32, another second gRNA that targets a target site for GATA3 comprising the sequence set forth in SEQ ID NO:26, and a third gRNA that targets a target site for CISH comprising the sequence set forth in SEQ ID NO:30. In some embodiments, the combination of second gRNAs comprises one second gRNA that targets a target site for MED12 comprising the sequence set forth in SEQ ID NO:81, another second gRNA that targets a target site for TGFBR2 comprising the sequence set forth in SEQ ID NO:301, and a third gRNA that targets a target site for CISH comprising the sequence set forth in SEQ ID NO:28 or 30. sf-6059407 22474-20028.40 Table 6. Second genes, target sites, and second gRNAs for transcriptional repression

sf-6059407 22474-20028.40

sf-6059407 22474-20028.40 C. Other DNA-Binding Domains and DNA-Targeting Systems [0400] In some embodiments, the first and/or second DNA-binding domain comprises a zinc finger protein (ZFP); a transcription activator-like effector (TALE); a meganuclease; a homing endonuclease; or an I-SceI enzyme or a variant thereof. In some embodiments, the first and/or second DNA-binding domain comprises a catalytically inactive variant of any of the foregoing. In some embodiments, the first and/or second DNA-binding domain is an engineered zinc finger protein (eZFP) or a TALE. In some embodiments, the first and/or second DNA-binding domain is an engineered zinc finger protein (eZFP). In some embodiments, the first and/or second DNA- binding domain is a TALE. [0401] In some embodiments, a ZFP, a zinc finger DNA binding protein, or zinc finger DNA binding domain is a protein, or a domain within a larger protein, that binds DNA in a sequence-specific manner through one or more zinc fingers, which are regions of amino acid sequence within the binding domain whose structure is stabilized through coordination of a zinc ion. The term zinc finger DNA binding protein is often abbreviated as zinc finger protein or ZFP. Among the ZFPs are artificial or engineered ZFPs (eZFPs) comprising ZFP domains targeting specific DNA sequences, typically 9-18 nucleotides long, generated by assembly of individual fingers. ZFPs include those in which a single finger domain is approximately 30 amino acids in length and contains an alpha helix containing two invariant histidine residues coordinated through zinc with two cysteines of a single beta turn, and having two, three, four, five, or six fingers. Generally, sequence-specificity of a ZFP may be altered by making amino acid substitutions at the four helix positions (−1, 2, 3, and 6) on a zinc finger recognition helix. Thus, for example, the ZFP or ZFP-containing molecule is non-naturally occurring, e.g., is an eZFP that is engineered to bind to a target site of choice. [0402] In some embodiments, zinc fingers are custom-designed (i.e., designed by the user) or obtained from a commercial source. Various methods for designing zinc finger proteins are available. For example, methods for designing zinc finger proteins to bind to a target DNA sequence of interest are described, for example, in Liu, Q. et al., PNAS, 94(11):5525-30 (1997); Wright, D.A. et al., Nat. Protoc., 1(3):1637-52 (2006); Gersbach, C.A. et al., Acc. Chem. Res., 47(8):2309-18 (2014); Bhakta M.S. et al., Methods Mol. Biol., 649:3-30 (2010); and Gaj et al., Trends Biotechnol, 31(7):397-405 (2013). In addition, various web-based tools for designing zinc finger proteins to bind to a DNA target sequence of interest are publicly available. See, for example, the Zinc Finger Tools design web site from Scripps available on the world wide web at sf-6059407 22474-20028.40 scripps.edu/barbas/zfdesign/zfdesignhome.php. Various commercial services for designing zinc finger proteins to bind to a DNA target sequence of interest are also available. See, for example, the commercially available services or kits offered by Creative Biolabs (world wide web at creative-biolabs.com/Design-and-Synthesis-of-Artificial-Zinc-Finger-Proteins.html), the Zinc Finger Consortium Modular Assembly Kit available from Addgene (world wide web at addgene.org/kits/zfc-modular-assembly/), or the CompoZr Custom ZFN Service from Sigma Aldrich (world wide web at sigmaaldrich.com/life-science/zinc-finger-nuclease- technology/custom-zfn.html). [0403] Transcription activator-like effectors (TALEs) are proteins naturally found in Xanthomonas bacteria. TALEs comprise a plurality of repeated amino acid sequences, each repeat having binding specificity for one base in a target sequence. Each repeat comprises a pair of variable residues in position 12 and 13 (repeat variable diresidue; RVD) that determine the nucleotide specificity of the repeat. In some embodiments, RVDs associated with recognition of the different nucleotides are HD for recognizing C, NG for recognizing T, NI for recognizing A, NN for recognizing G or A, NS for recognizing A, C, G or T, HG for recognizing T, IG for recognizing T, NK for recognizing G, HA for recognizing C, ND for recognizing C, HI for recognizing C, HN for recognizing G, NA for recognizing G, SN for recognizing G or A and YG for recognizing T, TL for recognizing A, VT for recognizing A or G and SW for recognizing A. In some embodiments, RVDs can be mutated towards other amino acid residues in order to modulate their specificity towards nucleotides A, T, C and G and in particular to enhance this specificity. Binding domains with similar modular base-per-base nucleic acid binding properties can also be derived from different bacterial species. These alternative modular proteins may exhibit more sequence variability than TALE repeats. [0404] In some embodiments, a “TALE DNA binding domain” or “TALE” is a polypeptide comprising one or more TALE repeat domains/units. The repeat domains, each comprising a repeat variable diresidue (RVD), are involved in binding of the TALE to its cognate target DNA sequence. A single “repeat unit” (also referred to as a “repeat”) is typically 33-35 amino acids in length and exhibits at least some sequence homology with other TALE repeat sequences within a naturally occurring TALE protein. TALE proteins may be designed to bind to a target site using canonical or non-canonical RVDs within the repeat units. See, e.g., U.S. Pat. Nos. 8,586,526 and 9,458,205. sf-6059407 22474-20028.40 [0405] Zinc finger and TALE DNA-binding domains can be engineered to bind to a predetermined nucleotide sequence, for example via engineering (altering one or more amino acids) of the recognition helix region of a naturally occurring zinc finger protein, by engineering of the amino acids in a TALE repeat involved in DNA binding (the repeat variable diresidue or RVD region), or by systematic ordering of modular DNA-binding domains, such as TALE repeats or ZFP domains. Therefore, engineered zinc finger proteins or TALE proteins are proteins that are non-naturally occurring. Non-limiting examples of methods for engineering zinc finger proteins and TALEs are design and selection. A designed protein is a protein not occurring in nature whose design/composition results principally from rational criteria. Rational criteria for design include application of substitution rules and computerized algorithms for processing information in a database storing information of existing ZFP or TALE designs (canonical and non-canonical RVDs) and binding data. See, for example, U.S. Pat. Nos. 9,458,205; 8,586,526; 6,140,081; 6,453,242; and 6,534,261; see also WO 98/53058; WO 98/53059; WO 98/53060; WO 02/016536 and WO 03/016496. 1. Exemplary ZFPs [0406] In some embodiments, the first and/or second DNA-binding domain comprises a zinc finger protein (ZFP). In some aspects, provided herein are exemplary ZFPs that are capable of binding to, or bind to, a target site, such as any provided herein in Section I.A.2 and I.A.3. In some aspects, the exemplary ZFPs can facilitate specific targeting of effector domains for transcriptional activation of a target gene provided in Section I, e.g., the IL-2 gene, for gene- specific transcriptional activation of IL-2. In some aspects, the exemplary ZFPs can facilitate specific targeting of effector domains for transcription repression of a target gene provided in Section I, e.g., the MED12 or TGFBR2 genes, for gene-specific transcriptional repression of MED12 or TGFBR2. Thus, in some aspects, the exemplary ZFPs facilitates promotion of lymphoid activation and function. [0407] In some embodiments, the target site for an ZFP provided herein is within in the IL-2 gene. In some embodiments, the target site within the IL-2 gene is within region 4, region 5, or the region surrounding the transcriptional start site (TSS). In some embodiments, the target site within the IL-2 gene is within region 4 or region 5. In some embodiments, the target site for an ZFP provided herein is within in the MED12 gene. In some embodiments, the target site within the MED12 gene is the region surrounding the MED12 TSS. In some embodiments, the target sf-6059407 22474-20028.40 site for an ZFP provided herein is within in the TGFBR2 gene. In some embodiments, the target site within the TGFBR2 gene is the region surrounding the TGFBR2 TSS. [0408] In some embodiments, the target site for an ZFP provided herein comprises the nucleotide sequence set forth in any one of SEQ ID NOS:451-457, a contiguous portion thereof of at least 12 nt, or a complementary sequence of any of the foregoing. In some embodiments, the target site for an ZFP provided herein comprises the nucleotide sequence set forth in any one of SEQ ID NOS:451-457. In some embodiments, the target site is comprised in double-stranded DNA, such as genomic DNA. In some embodiments, the target site is double-stranded DNA, such as genomic DNA. In some embodiments, the ZFP is capable of binding to the target site. In some embodiments, the ZFP binds to the target site. In some embodiments, the binding is target- specific. For example, in some embodiments, a ZFP binds to the target site, and not to other sites comprising different sequences. For example, in some embodiments, an individual ZFP disclosed herein binds to the target site set forth in SEQ ID NO:451, and does not bind to a different target site, such as the target site set forth in SEQ ID NO:457. In some embodiments, the target site for a ZFP provided herein comprises a sequence set forth in Table 7. Table 7. ZFP target sequences

[0409] In some embodiments, the target site for a ZFP provided herein comprises the nucleotide sequence set forth in SEQ ID NO:451, a contiguous portion thereof of at least 12 nt, or a complementary sequence of any of the foregoing. In some embodiments, the target site for a ZFP provided herein comprises the sequence set forth in SEQ ID NO: 451. [0410] In some embodiments, the target site for a ZFP provided herein comprises the nucleotide sequence set forth in SEQ ID NO:452, a contiguous portion thereof of at least 12 nt, or a complementary sequence of any of the foregoing. In some embodiments, the target site for a ZFP provided herein comprises the sequence set forth in SEQ ID NO: 452. [0411] In some embodiments, the target site for a ZFP provided herein comprises the nucleotide sequence set forth in SEQ ID NO:453, a contiguous portion thereof of at least 12 nt, sf-6059407 22474-20028.40 or a complementary sequence of any of the foregoing. In some embodiments, the target site for a ZFP provided herein comprises the sequence set forth in SEQ ID NO: 453. [0412] In some embodiments, the target site for an ZFP provided herein comprises the nucleotide sequence set forth in SEQ ID NO:454, a contiguous portion thereof of at least 12 nt, or a complementary sequence of any of the foregoing. In some embodiments, the target site for a ZFP provided herein comprises the sequence set forth in SEQ ID NO: 454. [0413] In some embodiments, the target site for a ZFP provided herein comprises the nucleotide sequence set forth in SEQ ID NO:455, a contiguous portion thereof of at least 12 nt, or a complementary sequence of any of the foregoing. In some embodiments, the target site for a ZFP provided herein comprises the sequence set forth in SEQ ID NO: 455. [0414] In some embodiments, the target site for a ZFP provided herein comprises the nucleotide sequence set forth in SEQ ID NO:456, a contiguous portion thereof of at least 12 nt, or a complementary sequence of any of the foregoing. In some embodiments, the target site for a ZFP provided herein comprises the sequence set forth in SEQ ID NO: 456. [0415] In some embodiments, the target site for a ZFP provided herein comprises the nucleotide sequence set forth in SEQ ID NO:457, a contiguous portion thereof of at least 12 nt, or a complementary sequence of any of the foregoing. In some embodiments, the target site for a ZFP provided herein comprises the sequence set forth in SEQ ID NO: 457. [0416] In some embodiments, characteristics of ZFPs targeting specific target sites provided herein are shown in Table 8. In some embodiments, the ZFP comprises six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, each comprising a corresponding recognition region F1-F6, as shown in Table 8. In some embodiments, the recognition regions F1-F6 facilitate specific binding to the indicated target site sequence in Table 8. In some embodiments, the ZFP comprises an amino acid sequence comprising the recognition regions, as shown in Table 8. In some embodiments, the ZFP can be encoded by a DNA sequence as shown in Table 8. Table 8. Exemplary ZFP engineered zinc finger proteins sf-6059407 22474-20028.40
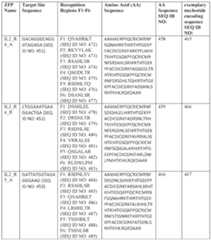
sf-6059407 22474-20028.40
sf-6059407 22474-20028.40

[0417] In some embodiments, provided herein is an ZFP, such as IL2_R4_A as described herein. In some embodiments, the ZFP targets a target site comprising the nucleotide sequence set forth in SEQ ID NO:451, a contiguous portion thereof of at least 12 nt, or a complementary sequence of any of the foregoing. In some embodiments, the ZFP targets a target site comprising the nucleotide sequence set forth in SEQ ID NO:451. In some embodiments, the target site is double-stranded DNA. In some embodiments, the ZFP comprises six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, each comprising a corresponding zinc finger recognition region F1 through F6, and the amino acid sequence of each zinc finger recognition region is as follows: F1: QNAHRKT (SEQ ID NO: 472), F2: RKYYLAK (SEQ ID NO: 473), F3: RSAHLSR (SEQ ID NO: 474), F4: QSGDLTR (SEQ ID NO: 475), F5: RSDHLTQ (SEQ ID NO: 476), and F6: DSANLSR (SEQ ID NO: 477). In some embodiments, the ZFP comprises the amino acid sequence set forth in SEQ ID NO:458, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the ZFP comprises the amino acid sequence set forth in SEQ ID NO:458. In some embodiments, the ZFP is encoded by the nucleotide sequence set forth in SEQ ID NO:465, or a nucleotide sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the ZFP is encoded by the nucleotide sequence set forth in SEQ ID NO:465. [0418] In some embodiments, provided herein is an ZFP, such as IL2_R4_B as described herein. In some embodiments, the ZFP targets a target site comprising the nucleotide sequence sf-6059407 22474-20028.40 set forth in SEQ ID NO:452, a contiguous portion thereof of at least 12 nt, or a complementary sequence of any of the foregoing. In some embodiments, the ZFP targets a target site comprising the nucleotide sequence set forth in SEQ ID NO:452. In some embodiments, the target site is double-stranded DNA. In some embodiments, the ZFP comprises six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, each comprising a corresponding zinc finger recognition region F1 through F6, and the amino acid sequence of each zinc finger recognition region is as follows: F1: DSSHLEL (SEQ ID NO: 478), F2: DRSNLTR (SEQ ID NO: 479), F3: RSDNLSE (SEQ ID NO: 480), F4: VRRALSS (SEQ ID NO: 481), F5: QSGALAR (SEQ ID NO: 482), and F6: RLDWLPM (SEQ ID NO: 483). In some embodiments, the ZFP comprises the amino acid sequence set forth in SEQ ID NO:459, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the ZFP comprises the amino acid sequence set forth in SEQ ID NO:459. In some embodiments, the ZFP is encoded by the nucleotide sequence set forth in SEQ ID NO:466, or a nucleotide sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the ZFP is encoded by the nucleotide sequence set forth in SEQ ID NO:466. [0419] In some embodiments, provided herein is an ZFP, such as IL2_R5_A as described herein. In some embodiments, the ZFP targets a target site comprising the nucleotide sequence set forth in SEQ ID NO:453, a contiguous portion thereof of at least 12 nt, or a complementary sequence of any of the foregoing. In some embodiments, the ZFP targets a target site comprising the nucleotide sequence set forth in SEQ ID NO:453. In some embodiments, the target site is double-stranded DNA. In some embodiments, the ZFP comprises six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, each comprising a corresponding zinc finger recognition region F1 through F6, and the amino acid sequence of each zinc finger recognition region is as follows: F1: RSDNLSV (SEQ ID NO: 484), F2: RSAHLSR (SEQ ID NO: 485), F3: QNAHRKT (SEQ ID NO: 486), F4: LRHHLTR (SEQ ID NO: 487), F5: TSSNRKT (SEQ ID NO: 488), and F6: TSSNLSR (SEQ ID NO: 489). In some embodiments, the ZFP comprises the amino acid sequence set forth in SEQ ID NO:460, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the ZFP comprises the amino acid sequence set forth in SEQ ID NO:460. In some embodiments, the ZFP is encoded by the nucleotide sequence set forth in SEQ ID NO:467, or a nucleotide sequence that has at least 90%, sf-6059407 22474-20028.40 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the ZFP is encoded by the nucleotide sequence set forth in SEQ ID NO:467. [0420] In some embodiments, provided herein is an ZFP, such as MED12_A as described herein. In some embodiments, the ZFP targets a target site comprising the nucleotide sequence set forth in SEQ ID NO:454, a contiguous portion thereof of at least 12 nt, or a complementary sequence of any of the foregoing. In some embodiments, the ZFP targets a target site comprising the nucleotide sequence set forth in SEQ ID NO:454. In some embodiments, the target site is double-stranded DNA. In some embodiments, the ZFP comprises six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, each comprising a corresponding zinc finger recognition region F1 through F6, and the amino acid sequence of each zinc finger recognition region is as follows: F1: DRSHLTR (SEQ ID NO: 490), F2: DRSYRNT (SEQ ID NO: 491), F3: QRRSLPH (SEQ ID NO: 492), F4: RSADLSR (SEQ ID NO: 493), F5: RSDTLSE (SEQ ID NO: 494), and F6: NRRGRWS (SEQ ID NO: 495). In some embodiments, the ZFP comprises the amino acid sequence set forth in SEQ ID NO:461, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the ZFP comprises the amino acid sequence set forth in SEQ ID NO:461. In some embodiments, the ZFP is encoded by the nucleotide sequence set forth in SEQ ID NO:468, or a nucleotide sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the ZFP is encoded by the nucleotide sequence set forth in SEQ ID NO:468. [0421] In some embodiments, provided herein is an ZFP, such as MED12_B as described herein. In some embodiments, the ZFP targets a target site comprising the nucleotide sequence set forth in SEQ ID NO:455, a contiguous portion thereof of at least 12 nt, or a complementary sequence of any of the foregoing. In some embodiments, the ZFP targets a target site comprising the nucleotide sequence set forth in SEQ ID NO:455. In some embodiments, the target site is double-stranded DNA. In some embodiments, the ZFP comprises six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, each comprising a corresponding zinc finger recognition region F1 through F6, and the amino acid sequence of each zinc finger recognition region is as follows: F1: RSANLAR (SEQ ID NO: 496), F2: DRSALAR (SEQ ID NO: 497), F3: RSDALST (SEQ ID NO: 498), F4: QSATRTK (SEQ ID NO: 499), F5: RSDTLSE (SEQ ID NO: 500), and F6: FRYARQY (SEQ ID NO: 501). In some embodiments, the ZFP comprises the amino acid sequence set forth in SEQ ID NO:462, or a portion thereof, or sf-6059407 22474-20028.40 an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the ZFP comprises the amino acid sequence set forth in SEQ ID NO:462. In some embodiments, the ZFP is encoded by the nucleotide sequence set forth in SEQ ID NO:469, or a nucleotide sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the ZFP is encoded by the nucleotide sequence set forth in SEQ ID NO:469. [0422] In some embodiments, provided herein is an ZFP, such as MED12_C as described herein. In some embodiments, the ZFP targets a target site comprising the nucleotide sequence set forth in SEQ ID NO:456, a contiguous portion thereof of at least 12 nt, or a complementary sequence of any of the foregoing. In some embodiments, the ZFP targets a target site comprising the nucleotide sequence set forth in SEQ ID NO:456. In some embodiments, the target site is double-stranded DNA. In some embodiments, the ZFP comprises six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, each comprising a corresponding zinc finger recognition region F1 through F6, and the amino acid sequence of each zinc finger recognition region is as follows: F1: DQSNLRA (SEQ ID NO: 502), F2: QNAHRKT (SEQ ID NO: 503), F3: TSGSLSR (SEQ ID NO: 504), F4: DRSNLSS (SEQ ID NO: 505), F5: RSAHLSR (SEQ ID NO: 506), and F6: RSDHLTQ (SEQ ID NO: 507). In some embodiments, the ZFP comprises the amino acid sequence set forth in SEQ ID NO:463, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the ZFP comprises the amino acid sequence set forth in SEQ ID NO:463. In some embodiments, the ZFP is encoded by the nucleotide sequence set forth in SEQ ID NO:470, or a nucleotide sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the ZFP is encoded by the nucleotide sequence set forth in SEQ ID NO:470. [0423] In some embodiments, provided herein is an ZFP, such as TGFBR2_A as described herein. In some embodiments, the ZFP targets a target site comprising the nucleotide sequence set forth in SEQ ID NO:457, a contiguous portion thereof of at least 12 nt, or a complementary sequence of any of the foregoing. In some embodiments, the ZFP targets a target site comprising the nucleotide sequence set forth in SEQ ID NO:457. In some embodiments, the target site is double-stranded DNA. In some embodiments, the ZFP comprises six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, each comprising a corresponding zinc finger recognition region F1 through F6, and the amino acid sequence of each zinc finger sf-6059407 22474-20028.40 recognition region is as follows: F1: RSDHLSA (SEQ ID NO: 508), F2: QSSDLRR (SEQ ID NO: 509), F3: HHNNRTH (SEQ ID NO: 510), F4: RNASRTR (SEQ ID NO: 511), F5: RSDHLSA (SEQ ID NO: 512), and F6: RSANLTR (SEQ ID NO: 513). In some embodiments, the ZFP comprises the amino acid sequence set forth in SEQ ID NO:464, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the ZFP comprises the amino acid sequence set forth in SEQ ID NO:464. In some embodiments, the ZFP is encoded by the nucleotide sequence set forth in SEQ ID NO:471, or a nucleotide sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the ZFP is encoded by the nucleotide sequence set forth in SEQ ID NO:471. [0424] In some embodiments, an ZFP, such as any provided herein, can be paired with any of the CRISPR/Cas-based DNA-binding domain and gRNA described herein in Section 1.B. In some embodiments, for transcriptional activation, an exemplary ZFP targets IL-2 and a gRNA targets BATF, CD28, EOMES, IL-2, IL2RB, IRF4, LAT, LCP2, TBX21, and VAV1. In some embodiments, for transcriptional activation, an exemplary ZFP targets IL-2 and a gRNA targets IL-2, wherein each DNA-binding domain does not target the same target site within IL-2. [0425] In some embodiments, for transcriptional repression, an exemplary ZFP targets MED12 or TGFBR2 and a gRNA targets CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and RASA2. In some embodiments, for transcriptional repression, an exemplary ZFP targets MED12 and a gRNA targets CISH or TGFBR2. In some embodiments, for transcriptional repression, an exemplary ZFP targets MED12 and a gRNA targets CISH. In some embodiments, for transcriptional repression, an exemplary ZFP targets MED12 and a gRNA targets TGFBR2. In some embodiments, for transcriptional repression, an exemplary ZFP targets TGFBR2 and a gRNA targets CISH. In some embodiments, for transcriptional repression, an exemplary ZFP targets TGFBR2 and a gRNA targets MED12. [0426] In some embodiments, a first DNA-binding domain for targeting to a target site of one of the one or more activation genes is an ZFP, such as an IL-2-targeting ZFP, e.g., IL2_R4_A, IL2_R4_B, and IL2_R5_A, and a second DNA-binding domain for targeting to a target site of the one or more repression genes is an ZFP, such as an MED12- or TGFBR2 ZFP, e.g., MED12_A-C or TGFBR2_A. In some embodiments, a first DNA-binding domain for targeting to a target site of one of the one or more activation genes is a ZFP, such as an IL-2- sf-6059407 22474-20028.40 targeting ZFP, e.g., IL2_R4_A, IL2_R4_B, and IL2_R5_A, and a second DNA-binding domain for targeting to a target site of the one or more repression genes is a Cas protein and a gRNA for targeting the Cas protein to a target site of one of more repression genes, e.g., MED12, TGFBR2, and/or CISH. In some embodiments, a first DNA-binding domain for targeting to a target site of one of the one or more activation genes is a Cas protein and a gRNA for targeting the Cas protein to a target site of one of more activation genes, e.g., IL-2, and a second DNA- binding domain for targeting to a target site of the one or more repression genes is an ZFP, such as an MED12- or TGFBR2 ZFP, e.g., MED12_A-C or TGFBR2_A. D. Effector Domains [0427] In some aspects, the DNA-targeting systems provided herein further include effector domains that include one or more transcriptional activator domains and one or more transcriptional repressor domains. In some embodiments, the effector domains mediate targeted epigenetic modification to increase or decrease transcription of the target genes. In some embodiments, the at least one transcriptional activator domain is the same across the at least one first DNA-targeting module. In some embodiments, the at least one transcriptional activator domain is different across the at least one first DNA-targeting module. In some embodiments, the at least one transcriptional repressor domain is the same across the at least one second DNA- targeting module. In some embodiments, the at least one transcriptional repressor domain is different across the at least one second DNA-targeting module. 1. Transcriptional Activator Domains [0428] In some aspects, the transcriptional activator domain activates, induces, catalyzes, or leads to increased transcription of a first gene when ectopically recruited to the first gene or DNA regulatory element thereof. In some embodiments, the transcriptional activator domain activates, induces, catalyzes, or leads to transcription activation, transcription co-activation, transcription elongation, transcription de-repression, transcription factor release, polymerization, histone modification, histone acetylation, histone deacetylation, nucleosome remodeling, chromatin remodeling, reversal of heterochromatin formation, proteolysis, ubiquitination, deubiquitination, phosphorylation, dephosphorylation, DNA methylation, DNA demethylation, histone methylation, histone demethylation, or DNA base oxidation. In some embodiments, the transcriptional activator domain activates, induces, catalyzes or leads to transcription activation, transcription co-activation, or transcription elongation. In some embodiments, the transcriptional sf-6059407 22474-20028.40 activator domain induces transcription activation. In some embodiments, the transcriptional activator domain has one of the aforementioned activities itself (i.e., acts directly). In some embodiments, the transcriptional activator domain recruits and/or interacts with a polypeptide domain that has one of the aforementioned activities (i.e., acts indirectly). [0429] Gene expression of endogenous mammalian genes, such as human genes, can be achieved by targeting a fusion protein comprising a DNA-binding domain, such as a dCas9, and an effector domain, such as a transcriptional activator domain, to mammalian genes or regulatory DNA elements thereof (e.g., a promoter or enhancer) via one or more gRNAs. Any of a variety of effector domains for transcriptional activation (e.g., transcriptional activator domains) are known and can be used in accord with the provided embodiments. Transcriptional activator domains, as well as activation of target genes by Cas fusion proteins (with a variety of Cas molecules) and the transcriptional activator domains, are described, for example, in WO 2014/197748, WO 2016/130600 , WO 2017/180915, WO 2021/226555 , WO 2021/226077, WO 2013/176772 , WO 2014/152432, WO 2014/093661, WO2024/015881, Adli, M. Nat. Commun. 9, 1911 (2018), Perez-Pinera, P. et al. Nat. Methods 10, 973–976 (2013), Mali, P. et al. Nat. Biotechnol.31, 833–838 (2013), and Maeder, M. L. et al. Nat. Methods 10, 977–979 (2013). [0430] In some embodiments, a transcriptional activator domain comprises a domain of a protein selected from among VP64, p65, Rta, p300, CBP, VPR, VPH, HSF1, a TET protein (e.g. TET1), a partially or fully functional fragment or domain thereof, and a combination of any of the foregoing. In some embodiments, a transcriptional activator domain further comprises at least one domain of a protein selected from among FOXO3 and NCOA3, that exhibits transcriptional activation, is capable of inducing or activating transcription from a gene, is a functional transcriptional activation domain, and/or has a function of transcription activation. In some embodiments, a transcriptional activator domain further comprises at least one domain selected from among FOXO3 and NCOA3. [0431] In some embodiments, the transcriptional activator domain comprises a VP64 domain. For example, dCas9-VP64 can be targeted to a target site by one or more first gRNAs to activate a first gene. VP64 is a polypeptide composed of four tandem copies of VP16, a 16 amino acid transactivation domain of the Herpes simplex virus. VP64 domains, including in dCas fusion proteins, have been described, for example, in WO 2014/197748, WO 2013/176772, WO 2014/152432, and WO 2014/093661. In some embodiments, the transcriptional activator domain comprises at least one VP16 domain, or a VP16 tetramer (“VP64”) or a variant thereof. sf-6059407 22474-20028.40 An exemplary VP64 domain is set forth in SEQ ID NO:142. In some embodiments, the transcriptional activator domain comprises SEQ ID NO:142, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:142, or a portion thereof. In some embodiments, the transcriptional activator domain is set forth in SEQ ID NO:142. [0432] In some embodiments, the transcriptional activator domain comprises a p65 activation domain (p65AD). p65AD is the principal transactivation domain of the 65kDa polypeptide of the nuclear form of the NF-KB transcription factor. An exemplary sequence of human transcription factor p65 is available at the Uniprot database under accession number Q04206. p65 domains, including in dCas fusion proteins, have been described, for example, in WO 2017/180915 and Chavez, A. et al. Nat. Methods 12, 326–328 (2015). An exemplary p65 activation domain is set forth in SEQ ID NO:248. In some embodiments, the transcriptional activator domain comprises SEQ ID NO:248, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:248, or a portion thereof. In some embodiments, the transcriptional activator domain is set forth in SEQ ID NO:248. [0433] In some embodiments, the transcriptional activator domain comprises an R transactivator (Rta) domain. Rta is an immediate-early protein of Epstein-Barr virus (EBV) and is a transcriptional activator that induces lytic gene expression and triggers virus reactivation. The Rta domain, including in dCas fusion proteins, has been described, for example in WO 2017/180915 and Chavez, A. et al. Nat. Methods 12, 326–328 (2015). An exemplary Rta domain is set forth in SEQ ID NO:249. In some embodiments, the transcriptional activator domain comprises SEQ ID NO:249, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:249, or a portion thereof. In some embodiments, the transcriptional activator domain is set forth in SEQ ID NO:249. [0434] In some embodiments, the transcriptional activator domain comprises a CREB- binding protein (CBP) domain or a p300 domain. In some aspects, CBP refers to the CREB- binding protein encoded by the human CREBBP gene. CBP is a coactivator that interacts with cAMP-response element binding protein (CREB). In some aspects, p300 refers to the Histone acetyltransferase p300 protein encoded by the human EP300 gene, and is a coactivator closely related to CBP. CBP and p300 each interact with a variety of transcriptional activators to affect sf-6059407 22474-20028.40 gene transcription (Gerritsen, M.E. et al. PNAS 94(7):2927-2932 (1997)). In some embodiments, the transcriptional activator domain comprises a p300 domain. p300 domains (such as the catalytic core of p300), including in dCas fusion proteins for gene activation, have been described, for example, in WO 2016/130600, WO 2017/180915, and Hilton, I.B. et al., Nat. Biotechnol.33(5):510-517 (2015). An exemplary human CBP sequence is set forth in SEQ ID NO:250. An exemplary human p300 sequence is set forth in SEQ ID NO:251. An exemplary p300 domain is set forth in SEQ ID NO:252. In some embodiments, the transcriptional activator domain comprises any one of SEQ ID NOS:250-252, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any one of SEQ ID NOS:250-252, or a portion thereof. In some embodiments, the transcriptional activator domain comprises SEQ ID NO:252, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:252, or a portion thereof. In some embodiments, the transcriptional activator domain is set forth in SEQ ID NO:252. [0435] In some embodiments, the transcriptional activator domain comprises a HSF1 domain. In some aspects, HSF1 refers to the Heat shock factor protein 1 protein encoded by the human HSF1 gene. HSF1, including in dCas fusion proteins for gene activation, has been described, for example, in WO 2021/226555, WO 2015/089427, and Konermann et al. Nature 517(7536):583-8 (2015). An exemplary human HSF1 sequence is set forth in SEQ ID NO:254. An exemplary HSF1 domain sequence is set forth in SEQ ID NO:253. In some embodiments, the transcriptional activator domain comprises SEQ ID NO:253 or SEQ ID NO:254, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:253 or SEQ ID NO:254, or a portion thereof. In some embodiments, the transcriptional activator domain comprises SEQ ID NO:253, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:253, or a portion thereof. In some embodiments, the transcriptional activator domain is set forth in SEQ ID NO:253. [0436] In some embodiments, the transcriptional activator domain comprises the tripartite activator VP64-p65-Rta (also known as VPR). VPR comprises three transcription activation domains (VP64, p65, and Rta) fused by short amino acid linkers and can effectively upregulate target gene expression. VPR, including in dCas fusion proteins for gene activation, has been described, for example, in WO 2021/226555 and Chavez, A. et al. Nat. Methods 12, 326–328 sf-6059407 22474-20028.40 (2015). An exemplary VPR polypeptide is set forth in SEQ ID NO:255. In some embodiments, the transcriptional activator domain comprises SEQ ID NO:255, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:255, or a portion thereof. In some embodiments, the transcriptional activator domain is set forth in SEQ ID NO:255. [0437] In some embodiments, the transcriptional activator domain comprises VPH. VPH is a tripartite activator polypeptide comprising VP64, mouse p65, and HSF1. VPH, including in dCas fusion proteins for gene activation, has been described, for example, in WO 2021/226555. An exemplary VPH polypeptide is set forth in SEQ ID NO:256. In some embodiments, the transcriptional activator domain comprises SEQ ID NO:256, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:256, or a portion thereof. In some embodiments, the transcriptional activator domain is set forth in SEQ ID NO:256. [0438] In some embodiments, the transcriptional activator effector domain has demethylase activity. The effector domain may include an enzyme that remove methyl (CH3-) groups from nucleic acids, proteins (in particular histones), and other molecules. The effector domain may covert the methyl group to hydroxymethylcytosine in a mechanism for demethylating DNA. Alternatively, the transcriptional activator domain can convert the methyl group to hydroxymethylcytosine in a mechanism for demethylating DNA. The effector domain can catalyze this reaction. For example, the transcriptional activator domain that catalyzes this reaction may comprise a domain from a TET protein, for example TET1 (Ten-eleven translocation methylcytosine dioxygenase 1). In some aspects, TET1 refers to the Methylcytosine dioxygenase TET1 protein encoded by the human TET1 gene. TET1 catalyzes the conversion of the modified genomic base 5-methylcytosine (5mC) into 5- hydroxymethylcytosine (5hmC) and plays a key role in active DNA demethylation. TET1, including in dCas fusion proteins for gene activation, has been described, for example, in WO 2021/226555. An exemplary human TET1 sequence is set forth in SEQ ID NO:257. An exemplary TET1 catalytic domain is set forth in SEQ ID NO:258. In some embodiments, the transcriptional activator domain comprises SEQ ID NO:257 or SEQ ID NO:258, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:257 or SEQ ID NO:258, or a portion thereof. In some embodiments, the transcriptional activator domain comprises SEQ ID NO:258, sf-6059407 22474-20028.40 or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:258, or a portion thereof. In some embodiments, the transcriptional activator domain is set forth in SEQ ID NO:258. [0439] In some embodiments, the transcriptional activator domain may comprise a SunTag domain. SunTag is a repeating peptide array which can recruit multiple copies of an antibody- fusion protein that binds the repeating peptide. The antibody-fusion protein may comprise an additional transcriptional activator domain, such as a VP64, to induce increased transcription of the target gene. SunTag, including in dCas fusion proteins for gene activation, has been described, for example, in WO 2016/011070 and Tanenbaum, M. et al. Cell.159(3):635–646 (2014). An exemplary SunTag effector domain includes a repeating GCN4 peptide having the amino acid sequence LLPKNYHLENEVARLKKLVGER (SEQ ID NO:246) separated by linkers having the amino acid sequence GGSGG (SEQ ID NO:247). In some embodiments, the transcriptional activator domain comprises the sequence set forth in SEQ ID NO:246, a domain thereof, a portion thereof, or a variant thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. In some embodiments, the SunTag effector domain recruits an antibody-fusion protein that comprises a transcriptional activator effector domain (e.g., VP64) and binds the GCN4 peptide, thereby activating transcription at the target site and acting as a transcriptional activator domain. [0440] In some embodiments, a transcriptional activation domain comprises a FOXO3 domain, i.e. a domain from FOXO3. In some aspects, FOXO3 refers to the Forkhead box protein O3 encoded by the human FOXO3 gene. FOXO3 functions as a transcriptional activator that recognizes and binds to specific DNA sequences. An exemplary human FOXO3 sequence is set forth in SEQ ID NO:531. An exemplary FOXO3 domain sequence is set forth in SEQ ID NO:532 and SEQ ID NO:533. In some embodiments, the transcriptional activation domain comprises a sequence set forth in any of SEQ ID NOS:531-533 or a domain or a portion thereof, such as a contiguous portion thereof of at least 10, 15, 20, 22, 25, 30, 35, 37, 40, 42, 45, 47, 49, 50, 55, 57, 60, 61, 62, 65, 70, 72, 75, 76, or 80 amino acids, such as at least 20 amino acids, or a variant thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to a sequence set forth in any of SEQ ID NOS: 531- 533 or a domain or a portion thereof, such as a contiguous portion thereof of at least 10, 15, 20, 22, 25, 30, 35, 37, 40, 42, 45, 47, 49, 50, 55, 57, 60, 61, 62, 65, 70, 72, 75, 76, or 80 amino acids, such as at least 20 amino acids, or a variant thereof. In some embodiments, the sf-6059407 22474-20028.40 transcriptional activation domain is or comprises an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:532. In some embodiments, the transcriptional activation domain comprises a contiguous portion of SEQ ID NO:531 that is at least 80 amino acids in length. In some embodiments, the transcriptional activation domain comprises SEQ ID NO:532. In some embodiments, the transcriptional activation domain is set forth in SEQ ID NO:532. An exemplary nucleotide sequence encoding the transcriptional activation domain set forth in SEQ ID NO:532 is set forth in SEQ ID NO:534. In some embodiments, the transcriptional activation domain is or comprises an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:533. In some embodiments, the transcriptional activation domain comprises a contiguous portion of SEQ ID NO:531 that is at least 42 amino acids in length. In some embodiments, the transcriptional activation domain comprises SEQ ID NO:533. In some embodiments, the transcriptional activation domain is set forth in SEQ ID NO:533. [0441] In some embodiments, a transcriptional activation domain comprises a NCOA3 domain, i.e. a domain from NCOA3. In some aspects, NCOA3 refers to the Nuclear receptor coactivator 3 protein encoded by the human NCOA3 gene. NCOA3 functions as a transcriptional coactivator for steroid receptors and nuclear receptors. An exemplary human NCOA3 sequence is set forth in SEQ ID NO:535. An exemplary NCOA3 domain sequence is set forth in SEQ ID NO:536 and SEQ ID NO:537. In some embodiments, the transcriptional activation domain comprises a sequence set forth in any of SEQ ID NOS:535-538 or a domain or a portion thereof, such as a contiguous portion thereof of at least 10, 15, 20, 22, 25, 30, 35, 37, 40, 42, 45, 47, 49, 50, 55, 57, 60, 61, 62, 65, 70, 72, 75, 76, or 80 amino acids, such as at least 20 amino acids, or a variant thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to a sequence set forth in any of SEQ ID NOS: 535-538 or a domain or a portion thereof, such as a contiguous portion thereof of at least 10, 15, 20, 22, 25, 30, 35, 37, 40, 42, 45, 47, 49, 50, 55, 57, 60, 61, 62, 65, 70, 72, 75, 76, or 80 amino acids, such as at least 20 amino acids, or a variant thereof. In some embodiments, the transcriptional activation domain is or comprises an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:536. In some embodiments, the transcriptional activation domain comprises a contiguous portion of SEQ ID NO:535 that is at least 80 amino acids in length. In some embodiments, the transcriptional activation domain comprises SEQ ID NO:536. In some sf-6059407 22474-20028.40 embodiments, the transcriptional activation domain is set forth in SEQ ID NO:536. An exemplary nucleotide sequence encoding the transcriptional activation domain set forth in SEQ ID NO:536 is set forth in SEQ ID NO:530. In some embodiments, the transcriptional activation domain is or comprises an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:537. In some embodiments, the transcriptional activation domain comprises a contiguous portion of SEQ ID NO:535 that is at least 49 amino acids in length. In some embodiments, the transcriptional activation domain comprises SEQ ID NO:537. In some embodiments, the transcriptional activation domain is set forth in SEQ ID NO:537. [0442] In some embodiments, the transcriptional activation domain comprises a fusion of NCOA3 and FOXO3 domains as described herein, e.g. the NCOA3 domain set forth in SEQ ID NO: 537 and the FOXO3 domain set forth in SEQ ID NO: 533. In some embodiments, the transcriptional activation domain comprises a fusion of two NCOA3 domains and one FOXO3. The fusion protein contains these domains can be arranged in any order. In some embodiments, the transcriptional activation domain is arranged, from N terminus to C terminus, as follows: a first NCOA3 domain, a FOXO3 domain, and a second NCOA3 domain, also referred to as a NCOA3-FOXO3-NCOA3 domain (NFN). In some embodiments, the NFN domain is or comprises an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO: 528. In some embodiments, the NFN domain comprises SEQ ID NO: 528. The NFN domain is set forth in SEQ ID NO: 528. In some embodiments, the domains are either directly linked to each other, or they are linked via a linker, such as a peptide linker. [0443] In some embodiments, the fusion protein comprises a transcriptional activation domain that is a NFN domain. In some embodiments, the fusion protein further comprises an additional transcriptional activation domain that is a VP64 domain. In some embodiments, the NFN and VP64 domains are either directly linked to each other, or they are linked via a linker, or they are separated by a DNA-binding domain. In some embodiments, the NFN and VP64 domains are separated by a DNA-binding domain. 2. Transcriptional Repressor Domains [0444] In some aspects, the transcriptional repressor domain induces, catalyzes, or leads to repressed and/or reduced transcription of a second gene when ectopically recruited to the second sf-6059407 22474-20028.40 gene or DNA regulatory element thereof. In some embodiments, the transcriptional repressor domain induces, catalyzes, or leads to transcription repression, transcription co-repression, histone modification, histone acetylation, histone deacetylation, nucleosome remodeling, chromatin remodeling, heterochromatin formation, proteolysis, ubiquitination, deubiquitination, phosphorylation, dephosphorylation, splicing, DNA methylation, DNA demethylation, histone methylation, histone demethylation, or DNA base oxidation. In some embodiments, the transcriptional repressor domain induces, catalyzes, or leads to transcription repression or transcription co-repression. In some embodiments, the transcriptional repressor domain induces transcription repression. In some embodiments, the transcriptional repressor domain has one of the aforementioned activities itself (i.e., acts directly). In some embodiments, the transcriptional repressor domain recruits and/or interacts with a protein or polypeptide domain that has one of the aforementioned activities (i.e., acts indirectly). [0445] Gene expression of endogenous mammalian genes, such as human genes, can be achieved by targeting a fusion protein comprising a DNA-binding domain, such as a dCas9, and an effector domain, such as a transcriptional repressor domain, to mammalian genes or regulatory DNA elements thereof (e.g., a promoter or enhancer) via one or more gRNAs. Any of a variety of effector domains for transcriptional repression (e.g., transcriptional repressor domains) are known and can be used in accord with the provided embodiments. Transcriptional repressor domains, as well as repression of target genes using Cas fusion proteins with the transcriptional repressor domains, are described, for example, in WO 2014/197748, WO 2017/180915, WO 2021/226077, WO 2013/176772, WO 2014/152432, WO 2014/093661, Adli, M. Nat. Commun.9, 1911 (2018), and Gilbert, L. A. et al. Cell 154(2):442-451 (2013). [0446] In some embodiments, the transcriptional repressor domain comprises a KRAB domain, ERF repressor domain, MXI1 domain, SID4X domain, MAD-SID domain, a DNMT family protein domain (e.g., DNMT3A or DNMT3B), a fusion of one or more DNMT family proteins or domains thereof (e.g., DNMT3A/L, which comprises a fusion of DNMT3A and DNMT3L domains), LSD1, EZH2, a SunTag domain, a partially or fully functional fragment or domain of any of the foregoing, or a combination of any of the foregoing. For example, the second fusion protein may be dCas9-KRAB or dCas9-KRAB-DNMT3A/3L. In some embodiments, the second fusion protein may be dCas9-KRAB, such as dSpCas9-KRAB (e.g., SEQ ID NO:73). In some embodiments, the second fusion protein may be dCas9-KRAB- DNMT3A/3L (e.g., SEQ ID NO:75). sf-6059407 22474-20028.40 [0447] In some embodiments, the transcriptional repressor domain comprises a transcriptional repressor domain described in WO 2021/226077. [0448] In some embodiments, the transcriptional repressor domain comprises a KRAB domain or a variant thereof. The KRAB-containing zinc finger proteins make up the largest family of transcriptional repressors in mammals. The Krüppel associated box (KRAB) domain is a transcriptional repressor domain present in many zinc finger protein-based transcription factors. The KRAB domain comprises charged amino acids and can be divided into sub-domains A and B. The KRAB domain recruits corepressors KAP1 (KRAB-associated protein-1), epigenetic readers such as heterochromatin protein 1 (HP1), and other chromatin modulators to induce transcriptional repression through heterochromatin formation. KRAB-mediated gene repression is associated with loss of histone H3-acetylation and an increase in H3 lysine 9 trimethylation (H3K9me3) at the repressed gene promoters. KRAB domains, including in dCas fusion proteins, have been described, for example, in WO 2017/180915, WO 2014/197748, US 2019/0127713, WO 2013/176772, Urrutia R. et al. Genome Biol.4, 231 (2003), Groner A. C. et al. pLoS Genet.6, e1000869 (2010). In some embodiments, the transcriptional repressor domain comprises at least one KRAB domain or a variant thereof. In some embodiments, an exemplary KRAB domain is set forth in SEQ ID NO:70. In some embodiments, the transcriptional repressor domain comprises the sequence set forth in SEQ ID NO:70, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:70. In some embodiments, an exemplary KRAB domain is set forth in SEQ ID NO:235. In some embodiments, the transcriptional repressor domain comprises the sequence set forth in SEQ ID NO:235, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:235. [0449] In some embodiments, the transcriptional repressor domain comprises at least one ERF repressor domain or a variant thereof. ERF (ETS2 repressor factor) is a strong transcriptional repressor that comprises a conserved ets-DNA-binding domain and represses transcription via a distinct domain at the carboxyl-terminus of the protein. ERF repressor domains, including in dCas fusion proteins, have been described, for example, in WO2017180915, WO2014197748, WO2013176772, Mavrothalassitis, G., Ghysdael, J. Proteins of the ETS family with transcriptional repressor activity. Oncogene 19, 6524–6532 (2000). In some embodiments, the transcriptional repressor domain comprises at least one ERF repressor sf-6059407 22474-20028.40 domain or a variant thereof. An exemplary ERF repressor domain is set forth in SEQ ID NO:128. In some embodiments, the transcriptional repressor domain comprises the sequence set forth in SEQ ID NO:128, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. [0450] In some embodiments, the transcriptional repressor domain comprises at least one MXI1 domain or a variant thereof. The MXI1 domain functions by antagonizing the myc transcriptional activity by competing for binding to myc-associated factor x (MAX). MXI1 domains, including in dCas fusion proteins, have been described, for example, in WO2017180915, WO2014197748, US20190127713. In some embodiments, the transcriptional repressor domain comprises at least one MXI1 domain or a variant thereof. An exemplary MXI1 domain is set forth in SEQ ID NO:129. In some embodiments, the transcriptional repressor domain comprises the sequence set forth in SEQ ID NO:129, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. [0451] In some embodiments, the transcriptional repressor domain comprises at least one SID4X domain or a variant thereof. The mSin3 interacting domain (SID) is present on different transcription repressor proteins. It interacts with the paired amphipathic alpha-helix 2 (PAH2) domain of mSin3, a transcriptional repressor domain that is attached to transcription repressor proteins such as the mSin3 A corepressor. A dCas9 molecule can be fused to four concatenated mSin3 interaction domains (SID4X). SID domains, including in dCas fusion proteins, have been described, for example, in WO2017180915, WO2014197748, WO2014093655. In some embodiments, the transcriptional repressor domain comprises at least one SID domain or a variant thereof. An exemplary SID domain is set forth in SEQ ID NO:236. In some embodiments, the transcriptional repressor domain comprises the sequence set forth in SEQ ID NO:236, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. [0452] In some embodiments, the transcriptional repressor domain comprises at least one MAD domain or a variant thereof. The MAD family proteins, Mad1, Mxi1, Mad3, and Mad4, belong to the basic helix-loop-helix-zipper class and contain a conserved N terminal region (termed Sin3 interaction domain (SID)) necessary for repressional activity. MAD-SID domains, including in dCas fusion proteins, have been described, for example, in WO2017180915, WO2014197748, WO2013176772. In some embodiments, the transcriptional repressor domain sf-6059407 22474-20028.40 comprises at least one MAD-SID domain or a variant thereof. An exemplary MAD-SID domain is set forth in SEQ ID NO:237. In some embodiments, the transcriptional repressor domain comprises the sequence set forth in SEQ ID NO:237, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. [0453] In some embodiments, the transcriptional repressor domain comprises at least one DNMT3 domain or a variant thereof. In some embodiments, the at least one DNMT3 domain, or a variant thereof, is from a DNMT3 or is a portion or a functionally active variant thereof with DNA methyltransferase activity. The DNMT3A and DNMT3B are two DNA methyltransferases that catalyze de novo methylation, which depending on the site may be associated with transcriptional repression. DNMTs, such as DNMT3s, mediate transfer of a methyl group from the universal methyl donor, S-adenosyl-L-methionine (SAM), to the 5-position of cytosine residues. In some aspects, these DNMT3 DNA methyltransferases induce de novo methylation of a cytosine base to methylated 5-methylcytosine. DNMT3, including in dCas fusion proteins, have been described, for example, in US20190127713, Liu, X. S. et al. Cell 167, 233–247.e17 (2016), Lei, Y. et al. Nat. Commun.8, 16026 (2017). DNMT3 proteins, such as DNMT3A and DNMT3B, contain an N-terminal part that is naturally involved in regulatory activity and targeting, and a C-terminal catalytic domain termed the mTase C5-type domain. In some embodiments, the transcriptional repressor domain includes a catalytically active portion of a DNMT3A or a DNMT3B that contains a catalytically active C-terminal domain. In particular, isolated catalytic domains of DNMT3a and DNMT3b are catalytically active (see e.g. Gowher and Jeltsch (2002) J. Biol. Chem., 277:20409). [0454] In some embodiments, the transcriptional repressor domain comprises at least one DNMT3 domain or a variant thereof. In some embodiments, the DNMT3 domain may be an effector domain of DNMT3A or DNMT3B that is catalytically active. In some embodiments, the transcriptional repressor domain may be the full-length of DNMT3A or DNMT3B or a catalytically active portion thereof. In some embodiments, the transcriptional repressor domain is a catalytically active portion that is less than the full-length sequence of DNMT3A or DNMT3B. In some embodiments, a catalytically active portion is a contiguous sequence of amino acids that confers DNA methyltransferase activity, such as by mediating methylation of a cytosine base to methylated 5-methylcytosine. In some embodiments, the contiguous sequence of amino acids is a contiguous C-terminal portion of a DNMT3 protein, such as DNMT3A or sf-6059407 22474-20028.40 DNMT3B, that is from 280 amino acids to 330 amino acids in length. In some embodiments, the contiguous portion is 280 amino acids, 290 amino acids, 300 amino acids, 310 amino acids, 320 amino acids, or 330 amino acids in length, or is a length of any value between any of the foregoing. In some embodiments, a catalytically active portion of a DNMT, such as a DNMT3, includes a SAM-dependent mTase C5-type domain. In some embodiments, the DNMT3 domain, such as a domain of DNMT3A or DNMT3B, is of human origin. In some embodiments, the DNMT3 domain, such as a domain of DNMT3A or DNMT3B, is of non-human origin, such as of mouse origin. [0455] An exemplary DNMT3A domain is set forth in SEQ ID NO:131 or 238. An exemplary DNMT3B domain is set forth in SEQ ID NO:239. In some embodiments, the transcriptional repressor domain comprises the sequence set forth in SEQ ID NO: 131, SEQ ID NO:238, or SEQ ID NO:239, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. [0456] In some embodiments, the DNMT3A domain is set forth in SEQ ID NO:131, or is a catalytically active portion thereof, or is an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:131 or the catalytically active portion thereof that exhibits DNA methyltransferase activity. In some embodiments, the DNMT3A domain is set forth in SEQ ID NO:131. [0457] In some embodiments, the DNMT3A domain is set forth in SEQ ID NO:238, or is a catalytically active portion thereof, or is an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:238 or the catalytically active portion thereof that exhibits DNA methyltransferase activity. In some embodiments, the DNMT3A domain is set forth in SEQ ID NO:238. [0458] In some embodiments, the transcriptional repressor domain is from DNMT3B or a catalytically active portion or variant thereof that exhibits DNA methyltransferase activity. An exemplary DNMT3B domain is set forth in SEQ ID NO:239, or is a catalytically active portion thereof, or is an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:239 or the catalytically active portion thereof that exhibits DNA methyltransferase activity. In some embodiments, the catalytically active portion is a contiguous portion of amino acids of SEQ ID NO:239 that includes the SAM- dependent mTase C5-type domain (e.g., corresponding to amino acids 575-853 of SEQ ID sf-6059407 22474-20028.40 NO:239). In some embodiments, the contiguous sequence of amino acids of SEQ ID NO:239 includes at least 250 amino acids, 275 amino acids, 300 amino acids, or 325 amino acids, or any value between any of the foregoing. In some embodiments, the contiguous sequence of amino acids is a contiguous portion of SEQ ID NO:239 that includes amino acids 575-853 and is from 280 amino acids to 330 amino acids in length. In some embodiments, the contiguous portion is 280 amino acids, 290 amino acids, 300 amino acids, 310 amino acids, 320 amino acids, or 330 amino acids in length, or is a length of any value between any of the foregoing. [0459] Any of a variety of assays are known to assess or monitor methyltransferase (mTase) activity. In some embodiments, exemplary assays to assess DNA methyltransferase activity include radio DNA mTase assays, colorimetric DNA mTase activity assays, fluorescent DNA mTase activity assays, chemiluminescent/bioluminescent DNA mTase activity assays, electrochemical DNA mTase activity assays, and electrogenerated chemiluminescence (ECL) DNA mTase activity assays. Exemplary assays are described in Poh et al. Theranostics, 2016, 6:369-391; Li et al., Methods Appl. Fluoresc., 2017, 5:012002; Deng et al., Anal Chem., 2014, 86:2117-23; and Ma et al. J Mater Chem B., 2020, 8:3488-3501. [0460] In some embodiments, the transcriptional repressor domain includes at least one DNMT3L domain or a variant thereof. The DNMT3L domain or a variant thereof may be a DNMT3L or a portion of DNMT3L, or a variant of DNMT3L or the portion thereof. DNMT3L (DNA (cytosine-5)-methyltransferase 3-like) is a catalytically inactive regulatory factor of DNA methyltransferases that can either promote or inhibit DNA methylation depending on the context. DNMT3L is essential for the function of DNMT3A and DNMT3B; DNMT3L interacts with DNMT3A and DNMT3B and significantly enhances their catalytic activity. For instance, DNMT3L interacts with the catalytic domain of DNMT3A to form a heterodimer, demonstrating that DNMT3L has dual functions of binding an unmethylated histone tail and activating DNA methyltransferase. In some embodiments, reference to a portion or variant of a DNMT3L for purposes herein refers to a sufficient C-terminal sequence portion of DNMT3L that interacts with the catalytic domain of DNMT3A or DNMT3B and is able to stimulate or promote DNA methyltransferase activity of DNMT3A or DNMT3B (see e.g. Jia et al. Nature, 2007, 449:248- 251; Gowher et al. J. Biol. Chem., 2005, 280: 13341-13348). In some embodiments, the DNMT3L or portion thereof is of animal origin. In some embodiments, the domain from DNMT3L is of murine origin. In some embodiments, the domain from DNMT3L is of human origin. sf-6059407 22474-20028.40 [0461] In some embodiments, the DNMT3L domain is a DNMT3L, or a C-terminal portion or variant thereof, that interacts with the catalytic domain of DNMT3A to form a heterodimer to provide for a more active DNA methyltransferase. In some embodiments, the effector domain is a fusion domain of a DNMT3A domain and the DNMT3L domain (DNMT3A/3L). [0462] In some embodiments, the DNMT3L domain is a DNMT3L, or a C-terminal portion or variant thereof, that interacts with the catalytic domain of DNMT3B to form a heterodimer to provide for a more active DNA methyltransferase. In some embodiments, the effector domain is a fusion domain of a DNMT3B domain and the DNMT3L domain (DNMT3B/3L). [0463] In some embodiments, the DNMT3L domain is a C-terminal portion of DNMT3L composed of a contiguous C-terminal portion of the full-length DNMT3L that does not include the N-terminal cysteine-rich ATRX-Dnmt3-Dnmt3L (ADD) domain (e.g. corresponding to residues 41-73 of SEQ ID NO:133, or 75-207 of the sequence set forth in SEQ ID NO:240). In some embodiments, the DNMT3L domain is a contiguous C-terminal portion of DNMT3L that is less than 220 amino acids in length, such as between 100 and 215 amino acids, such as at or about 100, 110, 120, 130, 140, 150, 160, 170, 180, 190, 200, 210 or 215 amino acids in length, or a length between a value of any of the foregoing. In some embodiments, the DNMT3L domain is a contiguous C-terminal portion of DNMT3L that is 205, 206, 207, 208, 209, 210, 211, 212, 213, 214 or 215 amino acids in length. [0464] An exemplary DNMT3L domain is set forth in SEQ ID NO:240, or is a portion thereof, or is an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:240 or the portion thereof. In some embodiments, the DNMT3L domain is a contiguous C-terminal portion of the full-length DNMT3L set forth in SEQ ID NO:240 that does not include the N-terminal cysteine-rich ATRX-Dnmt3-Dnmt3L (ADD) domain (corresponding to residues 75-207 of the sequence set forth in SEQ ID NO:240). In some embodiments, the DNMT3L domain is a contiguous C- terminal portion of the full-length DNMT3L set forth in SEQ ID NO:240 that is less than 220 amino acids in length, such as between 100 and 215 amino acids, such as at or about 100, 110, 120, 130, 140, 150, 160, 170, 180, 190, 200, 210 or 215 amino acids in length, or a length between a value of any of the foregoing. In some embodiments, the DNMT3L domain is a contiguous C-terminal portion of the full-length DNMT3L set forth in SEQ ID NO:240 that is 205, 206, 207, 208, 209, 210, 211, 212, 213, 214 or 215 amino acids in length. sf-6059407 22474-20028.40 [0465] In some embodiments, the DNMT3L domain is set forth in SEQ ID NO:241, or is a portion thereof, or is an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:241. In some embodiments, the DNMT3L domain is set forth in SEQ ID NO:241. In some embodiments, the DNMT3L domain does not contain an N-terminal methionine, such as set forth in SEQ ID NO:241. [0466] In some embodiments, the DNMT3L domain is a human or humanized DNMT3L. Corresponding sequences of human are highly homologous to the Dnmt3L derived from mouse and have a sequence identity of at least 90% with the murine sequence. It is within the level of a skilled artisan to humanize a non-human sequence of a DNMT3L domain, such as a domain of a murine DNMT3L. In some embodiments, the effector domain includes a DNMT3L domain that is a humanized variant of the murine DMT3L set forth in SEQ ID NO:240 or a portion thereof that is able to interact with DNMT3A or DNMT3A. In some embodiments, the effector domain includes a DNMT3L domain that is a humanized variant of the murine C-terminal portion of DNMT3L set forth in SEQ ID NO:241. [0467] An exemplary DNMT3L domain of human origin is set forth in SEQ ID NO:133, or is a portion thereof, or is an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:133 or the portion thereof. In some embodiments, the DNMT3L domain is a contiguous C-terminal portion of the full-length DNMT3L set forth in SEQ ID NO:133 that does not include the N-terminal cysteine-rich ATRX-Dnmt3-Dnmt3L (ADD) domain (corresponding to residues 41-73 of the sequence set forth in SEQ ID NO:133). In some embodiments, the DNMT3L domain is a contiguous C- terminal portion of the full-length DNMT3L set forth in SEQ ID NO:133 that is less than 220 amino acids in length, such as between 100 and 215 amino acids, such as at or about 100, 110, 120, 130, 140, 150, 160, 170, 180, 190, 200, 210 or 215 amino acids in length, or a length between a value of any of the foregoing. In some embodiments, the DNMT3L domain is a contiguous C-terminal portion of the full-length DNMT3L set forth in SEQ ID NO:133 that is 205, 206, 207, 208, 209, 210, 211, 212, 213, 214 or 215 amino acids in length. [0468] An exemplary DNMT3L domain is set forth in SEQ ID NO:133. In some embodiments, a DNMT3L domain comprises the sequence set forth in SEQ ID NO:133, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:133. sf-6059407 22474-20028.40 [0469] In some embodiments, the DNMT3L domain comprises the sequence set forth in SEQ ID NO:242, or is a portion thereof, or is an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:242. In some embodiments, the DNMT3L domain is set forth in SEQ ID NO:242. In some embodiments, the DNMT3L domain contains an N-terminal methionine. [0470] In some embodiments, the transcriptional repressor domain comprises a fusion of DNMT3A and DNMT3L (DNMT3A/L). The fusion protein contains DNMT3A and DNMT3L domains that can be any as described above. In some embodiments, the fusion protein contains the DNMT3A domain set forth in SEQ ID NO:131 and the DNMT3L domain set forth in SEQ ID NO:240, arranged in any order. In some embodiments, the fusion protein contains the DNMT3A domain set forth in SEQ ID NO:131 and the DNMT3L domain set forth in SEQ ID NO:241, arranged in any order. In some embodiments, the fusion protein contains the DNMT3A domain set forth in SEQ ID NO:131 and the DNMT3L domain set forth in SEQ ID NO:242, arranged in any order. In some embodiments, the fusion protein contains the DNMT3A domain set forth in SEQ ID NO:238 and the DNMT3L domain set forth in SEQ ID NO:240, arranged in any order. In some embodiments, the fusion protein contains the DNMT3A domain set forth in SEQ ID NO:238 and the DNMT3L domain set forth in SEQ ID NO:241, arranged in any order. In some embodiments, the fusion protein contains the DNMT3A domain set forth in SEQ ID NO:238 and the DNMT3L domain set forth in SEQ ID NO:242, arranged in any order. In some embodiments, the DNMT3A and DNMT3L domains present in a provided fusion protein are separated from each other in the fusion protein by an intervening sequence, such as the DNA-binding domain, another transcriptional repressor domain, or a linker. In some embodiments, the domains are either directly linked to each other, or they are linked via a linker, such as a peptide linker. In some embodiments, the DNMT3A and DNMT3L domains are connected as a fusion domain via a linker that connects the DNMT3A domain and the DNMT3L domain. Exemplary linkers are described herein. In some embodiments, the linker is the linker set forth in SEQ ID NO:243. [0471] An exemplary DNMT3A/L fusion domain is set forth in SEQ ID NO:135. In some embodiments, the transcriptional repressor domain comprises the sequence set forth in SEQ ID NO:135, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:135. sf-6059407 22474-20028.40 [0472] An exemplary DNMT3A/L fusion domain is set forth in SEQ ID NO:137. In some embodiments, the transcriptional repressor domain comprises the sequence set forth in SEQ ID NO:137, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:137. [0473] In some embodiments, the transcriptional repressor domain may comprise a LSD1 domain. LSD1 (also known as Lysine-specific histone demethylase 1A) is a histone demethylase that can demethylate lysine residues of histone H3, thereby acting as a coactivator or a corepressor, depending on the context. LSD1, including in dCas fusion proteins, has been described, for example, in WO 2013/176772, WO 2014/152432, and Kearns, N. A. et al. Nat. Methods.12(5):401–403 (2015). An exemplary LSD1 polypeptide is set forth in SEQ ID NO:244. In some embodiments, the transcriptional repressor domain comprises the sequence set forth in SEQ ID NO:244, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. [0474] In some embodiments, the transcriptional repressor domain may comprise an EZH2 domain. EZH2 (also known as Histone-lysine N-methyltransferase EZH2) is a Catalytic subunit of the PRC2/EED-EZH2 complex, which methylates 'Lys-9' (H3K9me) and 'Lys-27' (H3K27me) of histone H3, in some aspects leading to transcriptional repression of the affected target gene. EZH2, including in dCas fusion proteins, has been described, for example, in O’Geen, H. et al., Epigenetics Chromatin.12(1):26 (2019). An exemplary EZH2 polypeptide is set forth in SEQ ID NO:245. In some embodiments, the transcriptional repressor domain comprises the sequence set forth in SEQ ID NO:245, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. [0475] In some embodiments, the transcriptional repressor domain may comprise a SunTag domain. SunTag is a repeating peptide array, which can recruit multiple copies of an antibody- fusion protein that binds the repeating peptide. The antibody-fusion protein may comprise an additional transcriptional repressor domain, such as KRAB, to reduce transcription of the target gene. SunTag, including in dCas fusion proteins for gene modulation have been described, for example, in WO 2016/011070 and Tanenbaum, M. et al. Cell.159(3):635–646 (2014). An exemplary SunTag effector domain includes a repeating GCN4 peptide having the amino acid sequence LLPKNYHLENEVARLKKLVGER (SEQ ID NO:246) separated by linkers having the amino acid sequence GGSGG (SEQ ID NO:247). In some embodiments, the transcriptional sf-6059407 22474-20028.40 repressor domain comprises at least one copy of the sequence set forth in SEQ ID NO:246, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. In some embodiments, the SunTag transcriptional repressor domain recruits an antibody-fusion protein that comprises a transcriptional repressor effector domain (e.g., KRAB) and binds the GCN4 peptide, thereby repressing transcription at the target site and acting as a transcriptional repressor domain. E. Fusion Proteins [0476] In some aspects, the DNA-targeting systems provided herein include first and/or second fusion proteins. In some embodiments, the first and/or second fusion protein comprises: (a) a DNA-binding domain capable of being targeted to a target site for one or more genes, and (b) at least one transcriptional effector domain, e.g., a transcriptional activator or repressor domain. In some embodiments, at least one, optionally each, of the first DNA-targeting modules comprises a first fusion protein comprising (i) a first DNA-binding domain for targeting to a target site of one of the one or more first genes and (ii) at least one transcriptional activator domain. In some embodiments, at least one, optionally each, of the second DNA-targeting modules comprises a second fusion protein comprising (i) a second DNA-binding domain for targeting to a target site of one of the one or more second genes and (ii) at least one transcriptional repressor domain. [0477] In some embodiments, the DNA-binding domain and effector domain of the first and/or second fusion protein are heterologous, i.e., the domains are from different species, or at least one of the domains is not found in nature. In some aspects, the first and/or second fusion protein is an engineered fusion protein, i.e., the fusion protein is not found in nature. [0478] In some embodiments, the at least one effector domain of the first and/or second fusion protein is fused to the N-terminus, the C-terminus, or both the N-terminus and the C- terminus, of the DNA-binding domain or a component thereof. The at least one effector domain may be fused to the DNA-binding domain directly, or via any intervening amino acid sequence, such as a linker sequence or a nuclear localization sequence (NLS). [0479] In some embodiments, the first fusion protein comprises, from N- to C-terminal order: a transcriptional activator effector domain and a first DNA-binding domain. In some embodiments, the first fusion protein comprises, from N- to C-terminal order: a first DNA- binding domain and a transcriptional activator effector domain. sf-6059407 22474-20028.40 [0480] In some embodiments, the second fusion protein comprises, from N- to C-terminal order: a transcriptional repressor effector domain and a second DNA-binding domain. In some embodiments, the second fusion protein comprises, from N- to C-terminal order: a second DNA- binding domain and a transcriptional repressor domain. [0481] In some embodiments, the at least one effector domain of the first and/or second fusion protein includes more than one effector domain. In some embodiments, the first and/or second fusion protein includes 2, 3 or 4 effector domains, or more than 4 effector domains. In some embodiments, at least two of the effector domains of the first and/or second fusion protein are different. In some embodiments, each of the effector domains of the first and/or second fusion protein are different. In some embodiments, the at least one effector domain includes two effector domains in which the two effector domains are different. In some embodiments, the effector domains and the DNA-binding domain can be arranged in any order. [0482] In some embodiments, the at least one effector domain of the first and/or second fusion protein includes two different effector domains. The two different effector domains and the DNA-binding domain can be arranged in any order. In some embodiments, each of the effector domains are N-terminal to the DNA-binding domain in which a first effector domain is fused to the N-terminus of the second effector domain and the second effector domain is fused to the N-terminus of the DNA-binding domain. In some embodiments, the first and/or second fusion protein comprises from N- to C-terminal order: a first effector domain, a second effector domain, and the DNA binding domain. In some embodiments, each of the effector domains are C-terminal to the DNA-binding domain in which a first effector domain is fused to the C- terminus of the DNA-binding domain and the second effector domain is fused to the C-terminus of the first effector domain. In some embodiments, the first and/or second fusion protein comprises from N- to C-terminal order: a DNA-binding domain, a first effector domain, and a second effector domain. In some embodiments, the DNA-binding domain is between the effector domains, in which one effector domain is fused to the N-terminus of the DNA-binding domain and the other effector domain is fused to the C-terminus of the DNA-binding domain. In some embodiments, the first and/or second fusion protein comprises from N- to C-terminal order: a first effector domain, a DNA-binding domain, and a second effector domain. In some embodiments, one or more of the components may be fused to each other directly, or via any intervening amino acid sequence, such as via a linker sequence or a nuclear localization sequence (NLS). sf-6059407 22474-20028.40 [0483] In some embodiments, the first and/or second fusion protein comprises one or more linkers. In some embodiments, the linker is a peptide linker. In some embodiments, the one or more linkers connect the DNA-binding domain or a component thereof to the at least one effector domain. A linker may be included anywhere in the polypeptide sequence of the first and/or second fusion protein, for example, between the effector domain and the DNA-binding domain or a component thereof. A linker may be of any length and designed to promote or restrict the mobility of components in the first and/or second fusion protein. A linker may comprise any amino acid sequence of about 2 to about 100, about 5 to about 80, about 10 to about 60, or about 20 to about 50 amino acids. A linker may comprise an amino acid sequence of at least about 2, 3, 4, 5, 10, 15, 20, 25, or 30 amino acids. A linker may comprise an amino acid sequence of less than about 100, 90, 80, 70, 60, 50, or 40 amino acids. A skilled artisan can readily choose an appropriate linker for the connection of two domains. In some embodiments, the linker is a flexible linker. Flexible linkers are generally composed of small, non-polar or polar residues such as glycine, serine, or threonine. A linker may include sequential or tandem repeats of an amino acid sequence that is 2 to 20 amino acids in length. Linkers may be rich in amino acids glycine (G), serine (S), and/or alanine (A). Linkers may include, for example, a GS linker. An exemplary GS linker is represented by the sequence GGGGS (SEQ ID NO:259). A linker may comprise repeats of a sequence, for example repeats of the sequence GGGGS (SEQ ID NO:259). The number of times a linker sequence is repeated can be adjusted to optimize the linker length and achieve appropriate separation of the functional domains. For example, in some embodiments, the linker is the (GGGGS)
n linker, whereby n is an integer of 1 to 10 (SEQ ID NO:442). Other examples of linkers may include, for example, GGGGG (SEQ ID NO:260), GGAGG (SEQ ID NO:261), GGGGSSS (SEQ ID NO:262), and GGGGAAA (SEQ ID NO:263). [0484] In some embodiments, artificial linker sequences can be used. In some embodiments, the linker is EASGSGRASPGIPGSTR (SEQ ID NO:264). In some embodiments, the linker is GIHGVPAA (SEQ ID NO:265). In some embodiments, the linker is SSGNSNANSRGPSFSSGLVPLSLRGSH (SEQ ID NO:243). In some embodiments, the linker is KRPAATKKAGQAKKKKASDAKSLTAWS (SEQ ID NO:266). [0485] In some embodiments, inclusion of a linker in the first and/or second fusion protein leads to enhanced modulation (such as repression or activation) of the target gene. sf-6059407 22474-20028.40 [0486] In some embodiments, the linker is an XTEN linker. In some aspects, an XTEN linker is a recombinant polypeptide (e.g., an unstructured recombinant peptide) lacking hydrophobic amino acid residues. Exemplary XTEN linkers are described in, for example, Schellenberger et al., Nature Biotechnology 27, 1186-1190 (2009) or WO 2021/247570. In some embodiments, inclusion of a linker in the first and/or second fusion protein leads to enhanced repression or activation of the target gene. In some embodiments, a linker comprises the sequence set forth in SEQ ID NO:267, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to SEQ ID NO:267. In some aspects, the linker comprises the sequence set forth in SEQ ID NO:233, or a contiguous portion of SEQ ID NO:267 of at least 5, 10, 15, 20, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70 or 75 amino acids. In some aspects, the linker consists of the sequence set forth in SEQ ID NO:267, or a contiguous portion of SEQ ID NO:267 of at least 5, 10, 15, 20, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70 or 75 amino acids. In some embodiments, the linker comprises the sequence set forth in SEQ ID NO:267. In some embodiments, the linker consists of the sequence set forth in SEQ ID NO:267. In some embodiments, a linker comprises the sequence set forth in SEQ ID NO:268, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. In some aspects, the linker comprises the sequence set forth in SEQ ID NO:268, or a contiguous portion of SEQ ID NO:268 of at least 5, 10, or 15 amino acids. In some aspects, the linker consists of the sequence set forth in SEQ ID NO:268, or a contiguous portion of SEQ ID NO:268 of at least 5, 10, or 15 amino acids. In some embodiments, the linker comprises the sequence set forth in SEQ ID NO:268. In some embodiments, the linker consists of the sequence set forth in SEQ ID NO:268. Appropriate linkers may be selected or designed based on rational criteria known in the art, for example as described in Chen et al. Adv. Drug Deliv. Rev.65(10):1357-1369 (2013). In some embodiments, a linker comprises a linker described in WO 2021/247570. [0487] In some embodiments, the linker comprises the sequence set forth in any one of SEQ ID NOS:243, 361, or 362, or a sequence having at least 90% identity thereto. [0488] In some embodiments, the first and/or second fusion protein comprises one or more nuclear localization signals (NLS). In some embodiments, the first and/or second fusion protein comprises one or more nuclear localization sequences (NLSs), such as about or more than about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more NLSs. When more than one NLS is present, each may be selected independently of the others, such that a single NLS may be present in more than one sf-6059407 22474-20028.40 copy and/or in combination with one or more other NLSs present in one or more copies. Non- limiting examples of NLSs include an NLS sequence derived from the NLS of the SV40 virus large T-antigen, having the amino acid sequence PKKKRKV (SEQ ID NO:269); the NLS from nucleoplasmin, e.g., the nucleoplasmin bipartite NLS with the sequence KRPAATKKAGQAKKKK (SEQ ID NO:270); the c-myc NLS having the amino acid sequence PAAKRVKLD (SEQ ID NO:271) or RQRRNELKRSP (SEQ ID NO:272); the hRNPA1 M9 NLS having the sequence NQSSNFGPMKGGNFGGRSSGPYGGGGQYFAKPRNQGGY (SEQ ID NO:273); the sequence RMRIZFKNKGKDTAELRRRRVEVSVELRKAKKDEQILKRRNV (SEQ ID NO:274) of the IBB domain from importin-alpha; the sequences VSRKRPRP (SEQ ID NO:275) and PPKKARED (SEQ ID NO:276) of the myoma T protein; the sequence PQPKKKPL (SEQ ID NO:277) of human p53; the sequence SALIKKKKKMAP (SEQ ID NO:278) of mouse c-abl IV; the sequences DRLRR (SEQ ID NO:279) and PKQKKRK (SEQ ID NO:280) of the influenza virus NS1; the sequence RKLKKKIKKL (SEQ ID NO:281) of the Hepatitis virus delta antigen; the sequence REKKKFLKRR (SEQ ID NO:282) of the mouse Mx1 protein; the sequence KRKGDEVDGVDEVAKKKSKK (SEQ ID NO:283) of the human poly(ADP-ribose) polymerase; and the sequence RKCLQAGMNLEARKTKK (SEQ ID NO:284) of the steroid hormone receptors (human) glucocorticoid. The NLS may comprise a portion of any of the foregoing. In general, the one or more NLSs are of sufficient strength to drive accumulation of the first and/or second fusion protein in a detectable amount in the nucleus of a eukaryotic cell. In general, strength of nuclear localization activity may derive from the number of NLSs in the first and/or second fusion protein, the particular NLS(s) used, or a combination of these factors. Detection of accumulation in the nucleus may be performed by any suitable technique. For example, a detectable marker may be fused to the first and/or second fusion protein, such that location within a cell may be visualized, such as in combination with a means for detecting the location of the nucleus (e.g., a stain specific for the nucleus such as DAPI). Cell nuclei may also be isolated from cells, the contents of which may then be analyzed by any suitable process for detecting protein, such as immunohistochemistry, Western blot, or enzyme activity assay. Accumulation in the nucleus may also be determined indirectly, such as by an assay for the effect of the first and/or second fusion protein (e.g., an assay for altered gene expression activity in a cell transformed with the DNA-targeting system comprising the first and/or second fusion protein), as compared to a control condition (e.g., an untransformed cell). sf-6059407 22474-20028.40 [0489] In some embodiments, the NLS is a nucleoplasmin NLS or an SV40 NLS. In some embodiments, the NLS comprises the sequence set forth in SEQ ID NO:269 or 270, or a sequence having at least 90% identity thereto. [0490] In some embodiments, the NLS is linked to the N-terminus or the C-terminus of the DNA-binding domain via a linker. In some embodiments, the NLS is linked to the N-terminus or the C-terminus of an effector domain via a linker. The linker may be any linker as described above. In some embodiments, the linker is GIHGVPAA (SEQ ID NO:265). In some embodiments, the NLS and linker has the sequence PKKKRKVGIHGVPAA (SEQ ID NO:285). [0491] In some configurations, the N- or C-terminus of the first and/or second fusion protein can be linked to a moiety for detection and/or purification. In some aspects, the moiety is or includes a Flag tag DYKDDDDK (SEQ ID NO:286), a 3xFlag tag MDYKDHDGDYKDHDIDYKDDDDK (SEQ ID NO:287), an HA tag YPYDVPDYA (SEQ ID NO:288) or a His tag, such as HHHHHH (SEQ ID NO:289). 1. Split Fusion Proteins [0492] In some embodiments, the first and/or second fusion protein is a split protein, i.e., comprises two or more separate polypeptide domains that interact or self-assemble to form a functional fusion protein. In some aspects, the split fusion protein comprises a dCas9 and an effector domain. In some aspects, the first and/or second fusion protein comprises a split dCas9- effector domain fusion protein. [0493] In some embodiments, the split fusion protein is assembled from separate polypeptide domains comprising trans-splicing inteins. Inteins are internal protein elements that self-excise from their host protein and catalyze ligation of flanking sequences with a peptide bond. In some embodiments, the split fusion protein is assembled from a first polypeptide comprising an N-terminal intein and a second polypeptide comprising a C-terminal intein. In an exemplary embodiment, the N terminal intein is the N terminal Npu Intein set forth in SEQ ID NO:290. In some embodiments, the C terminal intein is the C terminal Npu intein set forth in SEQ ID NO:291. [0494] In some embodiments, the split fusion protein comprises a split dCas9-effector domain fusion protein assembled from two polypeptides. In an exemplary embodiment, the first polypeptide comprises an effector domain catalytic domain and an N-terminal fragment of dSpCas9, followed by an N terminal Npu Intein (effector domain-dSpCas9-573N), and the second polypeptide comprises a C terminal Npu Intein, followed by a C-terminal fragment of sf-6059407 22474-20028.40 dSpCas9 (dSpCas9-573C). The N- and C-terminal fragments of the fusion protein are split at position 573Glu of the SpCas9 molecule, with reference to SEQ ID NO:126 (corresponding to residue 572Glu of the dSpCas9 molecule set forth in SEQ ID NO:127). In some aspects, the N- terminal Npu Intein (SEQ ID NO:290) and C-terminal Npu Intein (set forth in SEQ ID NO:291) may self-excise and ligate the two fragments, thereby forming the full-length dSpCas9-effector domain fusion protein when expressed in a cell. [0495] In some embodiments, the polypeptides of a split protein may interact non-covalently to form a complex that recapitulates the activity of the non-split protein. For example, two domains of a Cas enzyme expressed as separate polypeptides may be recruited by a gRNA to form a ternary complex that recapitulates the activity of the full-length Cas enzyme in complex with the gRNA, for example as described in Wright et al. PNAS 112(10):2984-2989 (2015). In some embodiments, assembly of the split protein is inducible (e.g. light inducible, chemically inducible, small-molecule inducible). [0496] In some aspects, the two polypeptides of a split fusion protein may be delivered and/or expressed from separate vectors, such as any of the vectors described herein. In some embodiments, the two polypeptides of a split fusion protein may be delivered to a cell and/or expressed from two separate AAV vectors, i.e. using a split AAV-based approach, for example as described in WO 2017/197238. [0497] Approaches for the rationale design of split proteins and their delivery, including Cas proteins and fusions thereof, are described, for example, in WO 2016/114972, WO 2017/197238, Zetsche. et al. Nat. Biotechnol.33(2):139-42 (2015), Wright et al. PNAS 112(10):2984-2989 (2015), Truong. et al. Nucleic Acids Res.43, 6450–6458 (2015), and Fine et al. Sci. Rep.5, 10777 (2015). 2. Fusion Proteins for Transcriptional Activation [0498] In some embodiments, the first fusion protein is a dCas-VP64 fusion protein, such as dSpCas9-2xVP64, which is a fusion of dSpCas9 fused to two copies of VP64. In some embodiments, the first fusion protein is dSpCas9-2xVP64. In some embodiments, the first fusion protein comprises the sequence set forth in SEQ ID NO:77, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the first fusion protein comprises the sequence set forth in SEQ ID NO:77. In some embodiments, the first fusion protein is encoded by the nucleotide sequence set forth in SEQ ID NO:76. sf-6059407 22474-20028.40 [0499] In some embodiments, the first fusion protein is a dCas-VP64 fusion protein, such as dSaCas9-2xVP64, which is a fusion of dSaCas9 fused to two copies of VP64. In some embodiments, the first fusion protein is dSaCas9-2xVP64. In some embodiments, the first fusion protein comprises the sequence set forth in SEQ ID NO:386, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the first fusion protein comprises the sequence set forth in SEQ ID NO:386. In some embodiments, the first fusion protein is encoded by the nucleotide sequence set forth in SEQ ID NO:385. [0500] In some embodiments, the first fusion protein is a dCas-NFN-VP64 fusion protein, such as dSaCas9-NFN-VP64, which is a fusion of dSaCas9 fused to one copy of VP64 and one NFN domain, as described in Section I.D.1. In some embodiments, the first fusion protein is dSaCas9-NFN-VP64. In some embodiments, the first fusion protein comprises the sequence set forth in SEQ ID NO:529, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the first fusion protein comprises the sequence set forth in SEQ ID NO:529. In some embodiments, the first fusion protein is encoded by the nucleotide sequence set forth in SEQ ID NO:530. [0501] In some embodiments, the first fusion protein is a dCas-NFN fusion protein, such as dSaCas9-NFN, which is a fusion of dSaCas9 fused to one NFN domain, as described in Section I.D.1. In some embodiments, the first fusion protein is dSaCas9-NFN. In some embodiments, the first fusion protein comprises the sequence set forth in SEQ ID NO:541, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the first fusion protein comprises the sequence set forth in SEQ ID NO:541. In some embodiments, the first fusion protein is encoded by the nucleotide sequence set forth in SEQ ID NO:540. [0502] In some embodiments, the first fusion protein is a ZFP-NFN fusion protein, such as an IL-2 targeting ZFP, e.g., any IL-2 targeting ZFP described herein in Section I.C.1, fused to a NFN domain. Exemplary IL-2 targeting ZFP-NFN fusion proteins include IL2_R4_A-NFN, IL2-R4_B-NFN, and IL2-R5_A-NFN fusion proteins. In some embodiments, the first fusion protein is IL2_R4_A-NFN. In some embodiments, the first fusion protein comprises the sequence set forth in SEQ ID NO:514, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the first fusion protein comprises the sequence set forth in SEQ ID NO:514. In sf-6059407 22474-20028.40 some embodiments, the first fusion protein is encoded by the nucleotide sequence set forth in SEQ ID NO:521. In some embodiments, the first fusion protein is IL2_R4_B-NFN. In some embodiments, the first fusion protein comprises the sequence set forth in SEQ ID NO:515, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the first fusion protein comprises the sequence set forth in SEQ ID NO:515. In some embodiments, the first fusion protein is encoded by the nucleotide sequence set forth in SEQ ID NO:522. In some embodiments, the first fusion protein is IL2_R5_A-NFN. In some embodiments, the first fusion protein comprises the sequence set forth in SEQ ID NO:516, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the first fusion protein comprises the sequence set forth in SEQ ID NO:516. In some embodiments, the first fusion protein is encoded by the nucleotide sequence set forth in SEQ ID NO:523. 3. Fusion Proteins for Transcriptional Repression [0503] In some embodiments, the second fusion protein is a dCas-KRAB fusion protein, such as dSpCas9-KRAB. In some embodiments, the second fusion protein is dSpCas9-KRAB. In some embodiments, the second fusion protein comprises the sequence set forth in SEQ ID NO:70, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the second fusion protein comprises the sequence set forth in SEQ ID NO:70. In some embodiments, the second fusion protein is encoded by the nucleotide sequence set forth in SEQ ID NO:71. [0504] In some embodiments, the second fusion protein is a dCas-KRAB-DNMT3A/3L fusion protein, such as dSpCas9-KRAB-DNMT3A/3L. In some embodiments, the second fusion protein is dSpCas9-KRAB-DNMT3A/3L. In some embodiments, the second fusion protein comprises the sequence set forth in SEQ ID NO:75, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the second fusion protein comprises the sequence set forth in SEQ ID NO:75. In some embodiments, the second fusion protein is encoded by the nucleotide sequence set forth in SEQ ID NO:74. [0505] In some embodiments, the second fusion protein is a dCas-KRAB-DNMT3A/3L fusion protein comprising an XTEN linker. In some embodiments, the second fusion protein comprises the sequence set forth in SEQ ID NO:140 or 141, or an amino acid sequence that has sf-6059407 22474-20028.40 at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the second fusion protein comprises the sequence set forth in SEQ ID NO:140. In some embodiments, the second fusion protein comprises the sequence set forth in SEQ ID NO:141. [0506] In some embodiments, the second fusion protein comprises from N-terminus to C- terminus: (i) a second DNA-binding domain, and (ii) a KRAB domain or EZH2 domain. In some embodiments, the second fusion protein comprises from N-terminus to C-terminus: (i) a DNMT3A domain, (ii) a DNMT3L domain, (iii) a second DNA-binding domain, and (iv) a KRAB domain or EZH2 domain. In some embodiments, the second fusion protein comprises from N-terminus to C-terminus: (i) a DNMT3A domain, (ii) a DNMT3L domain, (iii) a KRAB domain or EZH2 domain, and (iv) a second DNA-binding domain. In some embodiments, the second fusion protein comprises from N-terminus to C-terminus: (i) a DNMT3B domain, (ii) a DNMT3L domain, (iii) a second DNA-binding domain, and (iv) a KRAB domain or EZH2 domain. In some embodiments, the second fusion protein comprises: a second DNA-binding domain, a DNMT3B domain, and a DNMT3L domain. In some embodiments, the second fusion protein comprises: a second DNA-binding domain, a DNMT3B domain, a DNMT3L domain, and a KRAB domain or EZH2 domain. In some embodiments, the second DNA-binding domain is a dCas protein, such as dSpCas9. [0507] In some embodiments, the DNMT3A domain comprises the sequence set forth in SEQ ID NO:131 or 238, or a sequence having at least 90% identity thereto. In some embodiments, the DNMT3A domain is encoded by the nucleotide sequence set forth in SEQ ID NO: 130. In some embodiments, the DNMT3L domain comprises the sequence set forth in any one of SEQ ID NOS: 133 and 240-242, or a sequence having at least 90% identity thereto. In some embodiments, the DNMT3L domain is encoded by the nucleotide sequence set forth in SEQ ID NO: 132. In some embodiments, the DNMT3B domain comprises the sequence set forth in SEQ ID NO:239 or 360, or a sequence having at least 90% identity thereto. In some embodiments, the second fusion protein comprises a DNMT3A-DNMT3L fusion domain. In some embodiments, the DNMT3A-DNMT3L fusion domain comprises the sequence set forth in SEQ ID NO:135 or 137, or a sequence having at least 90% identity thereto. In some embodiments, the second fusion protein comprises a DNMT3B-DNMT3L fusion domain. In some embodiments, the DNMT3B-DNMT3L fusion domain comprises the sequence set forth in SEQ ID NO:363, or a sequence having at least 90% identity thereto. In some embodiments, the sf-6059407 22474-20028.40 KRAB domain is selected from a KRAB domain from KOX1, a KRAB domain from ZIM3, and a KRAB domain from ZNF324. In some embodiments, the KRAB domain comprises the sequence set forth in any one of SEQ ID NOS:70, 235, and 355-358, or a sequence having at least 90% identity thereto. In some embodiments, the EZH2 domain comprises the sequence set forth in SEQ ID NO:359, or a sequence having at least 90% identity thereto. [0508] In some embodiments, the second fusion protein comprises the sequence set forth in any one of SEQ ID NOS:332-351, a portion thereof, or a sequence having at least 90% sequence identity to any of the foregoing. In some embodiments, the second fusion protein comprises the sequence set forth in any one of SEQ ID NOS:365-384, or a sequence having at least 90% sequence identity to any of the foregoing. In some embodiments, the second fusion protein comprises the sequence set forth in any one of SEQ ID NOS:365-384. [0509] In some embodiments, the second fusion protein is a ZFP-KRAB-DNMT3A/L fusion protein, such as a MED12- or TGFBR2-targeting ZFP, e.g., any MED12 or TGFBR2 targeting ZFP described herein in Section C.1, fused to a KRAB domain and a DNMT3A/L domain. Exemplary MED12 targeting ZFP-KRAB-DNMT3A/L fusion proteins include MED12_A- KRAB-DNMT3A/L, MED12_B-KRAB-DNMT3A/L, and MED12_C-KRAB-DNMT3A/L fusion proteins. Exemplary TGFBR2 targeting ZFP-KRAB-DNMT3A/L fusion proteins include TGFBR2_A-KRAB-DNMT3A/L fusion proteins. [0510] In some embodiments, the second fusion protein is MED12_A-KRAB-DNMT3A/L. In some embodiments, the second fusion protein comprises the sequence set forth in SEQ ID NO:517, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the first fusion protein comprises the sequence set forth in SEQ ID NO:517. In some embodiments, the first fusion protein is encoded by the nucleotide sequence set forth in SEQ ID NO:524. In some embodiments, the second fusion protein is MED12_B-KRAB-DNMT3A/L. In some embodiments, the second fusion protein comprises the sequence set forth in SEQ ID NO:518, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the first fusion protein comprises the sequence set forth in SEQ ID NO:518. In some embodiments, the first fusion protein is encoded by the nucleotide sequence set forth in SEQ ID NO:525. In some embodiments, the second fusion protein is MED12_C-KRAB-DNMT3A/L. In some embodiments, the second fusion protein comprises the sequence set forth in SEQ ID NO:519, or an amino acid sequence that has sf-6059407 22474-20028.40 at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the first fusion protein comprises the sequence set forth in SEQ ID NO:519. In some embodiments, the first fusion protein is encoded by the nucleotide sequence set forth in SEQ ID NO:526. [0511] In some embodiments, the second fusion protein is TGFBR2_A-KRAB-DNMT3A/L. In some embodiments, the second fusion protein comprises the sequence set forth in SEQ ID NO:520, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. In some embodiments, the first fusion protein comprises the sequence set forth in SEQ ID NO:520. In some embodiments, the first fusion protein is encoded by the nucleotide sequence set forth in SEQ ID NO:527. II. POLYNUCLEOTIDES, VECTORS, AND RELATED METHODS FOR DELIVERY [0512] In some aspects, provided are polynucleotides encoding any of the DNA-targeting systems described herein in Section I or a portion or a component of any of the foregoing. In some aspects, the polynucleotides can encode any of the components of the DNA-targeting systems, and/or any nucleic acid or proteinaceous molecule necessary to carry out aspects of the methods of the disclosure. In particular embodiments, provided are polynucleotides encoding any of the first and/or second fusion proteins described herein, for example in Section I.E. Also provided herein are polynucleotides encoding any of the first and/or second gRNAs described herein, for example in Section I.B.2. [0513] In some embodiments, provided are polynucleotides comprising the first and/or second gRNAs described herein. In some embodiments, the first and/or second gRNA is transcribed from a genetic construct (i.e. vector or plasmid) in the target cell. In some embodiments, the first and/or second gRNA is produced by in vitro transcription and delivered to the target cell. In some embodiments, the first and/or second gRNA comprises one or more modified nucleotides for increased stability. In some embodiments, the first and/or second gRNA is delivered to the target cell pre-complexed as a RNP with the first and/or second fusion protein. [0514] In some embodiments, a provided polynucleotide encodes a first and/or second fusion protein as described herein that includes (a) a DNA-binding domain capable of being targeted to a target site of a target gene as described; and (b) at least one effector domain capable of modulating (such as increasing or decreasing) transcription of the first and/or second gene. In sf-6059407 22474-20028.40 some embodiments, the first and/or second fusion protein includes a fusion protein of a Cas protein and at least one effector domain capable of modulating (such as increasing or decreasing) transcription of a first and/or second gene. In particular examples, the Cas is a dCas, such as dCas9. In some embodiments, the dCas9 is a dSpCas9, such as the polynucleotide encoding a dSpCas9 set forth in SEQ ID NO:127. Examples of such domains and fusion proteins include any as described in Section I. [0515] In some embodiments, the polynucleotide is RNA or DNA. In some embodiments, the polynucleotide, such as a polynucleotide encoding a provided fusion protein, is mRNA. The mRNA can be 5′ capped and/or 3′ polyadenylated. In another embodiment, a polynucleotide provided herein, such as a polynucleotide encoding a provided first and/or second fusion protein, is DNA. The DNA can be present in a vector. A. Exemplary Polynucleotides [0516] In some embodiments, the at least one effector domain capable of modulation transcription of the first and/or second gene is described herein, such as any effector domain described in Section I.D. In some embodiments, the polynucleotide encodes an effector domain that is a DNMT3A domain. An exemplary polynucleotide encoding a DNMT3A domain is set forth in SEQ ID NO: 130. In some embodiments, the polynucleotide comprises a sequence set forth in SEQ ID NO: 130, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide comprises a sequence set forth in SEQ ID NO: 130. [0517] In some embodiments, the polynucleotide encodes an effector domain that is a DNMT3L domain. An exemplary polynucleotide encoding a DNMT3L domain is set forth in SEQ ID NO: 132. In some embodiments, the polynucleotide comprises a sequence set forth in SEQ ID NO: 132, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide comprises a sequence set forth in SEQ ID NO: 132. [0518] In some embodiments, the polynucleotide encodes an effector domain that is a DNMT3A/L domain. Exemplary polynucleotides encoding a DNMT3A/L domain are set forth in SEQ ID NO: 134 and SEQ ID NO: 136. In some embodiments, the polynucleotide comprises a sequence set forth in SEQ ID NO: 134, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide comprises a sequence set forth in SEQ ID NO: 134. In some sf-6059407 22474-20028.40 embodiments, the polynucleotide comprises a sequence set forth in SEQ ID NO: 136, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide comprises a sequence set forth in SEQ ID NO: 136. [0519] In some embodiments, the polynucleotide encodes a dCas-VP64 fusion protein, such as dSpCas9-2xVP64. In some embodiments, the polynucleotide comprises the sequence set forth in SEQ ID NO:76, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide is set forth in SEQ ID NO:76. In some embodiments, the polynucleotide encodes an amino acid sequence comprising SEQ ID NO:77, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO:77. [0520] In some embodiments, the polynucleotide encodes a dCas-NFN-VP64 fusion protein, such as dSpCas9-NFN-VP64. In some embodiments, the polynucleotide comprises the sequence set forth in SEQ ID NO:530, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide is set forth in SEQ ID NO:530. In some embodiments, the polynucleotide encodes an amino acid sequence comprising SEQ ID NO:529, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO:529. [0521] In some embodiments, the polynucleotide encodes a dCas-NFN fusion protein, such as dSpCas9-NFN. In some embodiments, the polynucleotide comprises the sequence set forth in SEQ ID NO:540, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide is set forth in SEQ ID NO:540. In some embodiments, the polynucleotide encodes an amino acid sequence comprising SEQ ID NO:541, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO:541. sf-6059407 22474-20028.40 [0522] In some embodiments, the polynucleotide encodes a dCas-KRAB fusion protein, such as dSpCas9-KRAB. In some embodiments, the polynucleotide encodes an amino acid sequence comprising SEQ ID NO:138, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO:138. [0523] In some embodiments, the polynucleotide encodes a dCas-KRAB-DNMT3A/3L fusion protein, such as dSpCas9-KRAB-DNMT3A/3L. In some embodiments, the polynucleotide encodes an amino acid sequence comprising SEQ ID NO:139, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO:139. [0524] In some embodiments, the polynucleotide encodes a dCas-KRAB-DNMT3A/3L fusion protein, such as dSpCas9-KRAB-DNMT3A/3L. In some embodiments, the polynucleotide encodes an amino acid sequence comprising SEQ ID NO:140, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO:140. [0525] In some embodiments, the polynucleotide encodes a dCas-KRAB-DNMT3A/3L fusion protein, such as dSpCas9-KRAB-DNMT3A/3L. In some embodiments, the polynucleotide encodes an amino acid sequence comprising SEQ ID NO:141, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO:141. [0526] In some embodiments, the polynucleotide encodes a ZFP-NFN fusion protein, such as an IL-2-targeting ZFP-NFN. In some embodiments, the polynucleotide comprises any one of SEQ ID NOs:521-523 and 545, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide comprises SEQ ID NO: 521, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide comprises SEQ ID NO: 521. In some embodiments, the polynucleotide comprises SEQ ID NO: 522, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence sf-6059407 22474-20028.40 identity thereto. some embodiments, the polynucleotide comprises SEQ ID NO: 522. In some embodiments, the polynucleotide comprises SEQ ID NO: 523, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide comprises SEQ ID NO: 523. In some embodiments, the polynucleotide comprises SEQ ID NO: 545, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide comprises SEQ ID NO: 545. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NOs:514-516, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO: 514, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO: 514. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO: 515, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO: 515. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO: 516, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO: 516. [0527] In some embodiments, the polynucleotide encodes an ZFP-KRAB-DNMT3A/3L fusion protein, such as a MED12 or TGFBR2-targeting ZFP-KRAB-DNMT3A/3L. In some embodiments, the polynucleotide comprises any one of SEQ ID NOs:524-527 and 546, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide comprises SEQ ID NO: 524, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide comprises SEQ ID NO: 524. In some embodiments, the polynucleotide comprises SEQ ID NO: 525, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some sf-6059407 22474-20028.40 embodiments, the polynucleotide comprises SEQ ID NO: 525. In some embodiments, the polynucleotide comprises SEQ ID NO: 526, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide comprises SEQ ID NO: 526. In some embodiments, the polynucleotide comprises SEQ ID NO: 527, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide comprises SEQ ID NO: 527. In some embodiments, the polynucleotide comprises SEQ ID NO: 546, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide comprises SEQ ID NO: 546. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in any one of SEQ ID NOs:517-520, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO: 517, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO: 517. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO: 518, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO: 518. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO: 519, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO: 519. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO: 520, or a sequence having at least about 80%, 85%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity thereto. In some embodiments, the polynucleotide encodes the amino acid sequence set forth in SEQ ID NO: 520. B. Multicistronic Expressing Polynucleotides [0528] In some embodiments, a polynucleotide provided herein can comprise nucleic acids encoding any two or more of the fusion proteins (such as ZFP fusion proteins or dCas fusion proteins described in Section I.E) described herein. In some embodiments, a polynucleotide sf-6059407 22474-20028.40 provided herein can comprise nucleic acids encoding any two or more of the dCas fusion proteins described herein including any containing two or more dCas fusion proteins described in Section I.B.1. In some embodiments, such polynucleotides can be packaged in a vector in combination with a gRNA (e.g., gRNAs described in Section I.B.2) for use in provided systems. In some embodiments, a polynucleotide provided herein can comprise nucleic acids encoding any two or more of the zinc finger fusion proteins described herein including any containing two or more ZFPs described in Section I.C. [0529] In some embodiments, a provided polynucleotide comprises a first nucleic acid sequence encoding at least one activator DNA-targeting module comprising a fusion protein protein comprising (i) a first zinc finger protein (ZFP) for targeting to a target site of one of the one or more activation genes, such as any described in Section I.A.2, and (ii) at least one transcriptional activator domain; and a second nucleic acid sequence encoding at least one repressor DNA-targeting module comprising a fusion protein comprising (i) a second zinc finger protein (ZFP) for targeting to a target site of one of the one or more repression genes, such as any described in Section I.A.3, and (ii) at least one transcriptional repressor domain. In provided embodiments, the first and second nucleic acid sequences of the polynucleotide can be in any order. In some embodiments, the polynucleotide comprises a nucleic acid sequence encoding at least one activator DNA-targeting module comprises a fusion protein comprising a first ZFP that targets the target site set forth in any one of SEQ ID NOs: 451-453; and a nucleic acid sequence encoding at least one repressor DNA-targeting module comprising a fusion protein comprising a second ZFP that targets the target site set forth in any one of SEQ ID NOs:454-457. In some embodiments, the polynucleotide comprises a nucleic acid sequence encoding at least one activator DNA-targeting module comprising a fusion protein comprising a first ZFP that comprises the sequence set forth in any of SEQ ID NOs: 458-460; and a nucleic acid sequence encoding at least one repressor DNA-targeting module comprising a fusion protein comprising a second ZFP comprises the sequence set forth in any one of SEQ ID NOs: 461-464. In some embodiments, the polynucleotide comprises a nucleic acid encoding at least one activator DNA- targeting module comprising a fusion protein comprising at least one transcriptional activator domain comprising a sequence set forth in SEQ ID NO: 549; and a nucleic acid encoding at least one repressor DNA-targeting module comprising a fusion protein comprising at least one transcriptional repressor domain comprising sequences set forth in SEQ ID NO: 70 and SEQ ID NO: 135. In some embodiments, the polynucleotide comprises a nucleic acid sequence encoding sf-6059407 22474-20028.40 at least one activator DNA-targeting module comprising a fusion protein comprising any one of the sequences set forth in SEQ ID NOs: 514-516; and a nucleic acid sequence encoding at least one repressor DNA-targeting module comprising a fusion protein comprising any one of the sequences set forth in SEQ ID NOs: 517-520. [0530] In some embodiments, the polynucleotide comprises a nucleic acid sequence encoding at least one activator DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 514 and a nucleic acid sequence encoding at least one repressor DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 517. In some embodiments, the polynucleotide comprises a nucleic acid sequence encoding at least one activator DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 514 and a nucleic acid sequence encoding at least one repressor DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 518. In some embodiments, the polynucleotide comprises a nucleic acid sequence encoding at least one activator DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 514 and a nucleic acid sequence encoding at least one repressor DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 519. In some embodiments, the polynucleotide comprises a nucleic acid sequence encoding at least one activator DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 514 and a nucleic acid sequence at least one repressor DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 520. In some embodiments, the polynucleotide comprises a nucleic acid sequence encoding at least one activator DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 515 and a nucleic acid sequence encoding at least one repressor DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 517. In some embodiments, the polynucleotide comprises a nucleic acid sequence encoding at least one activator DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 515 and a nucleic acid sequence encoding at least one repressor DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 518. In some embodiments, the polynucleotide comprises a nucleic acid sequence encoding at least one activator DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 515 and a nucleic acid sequence encoding at least one repressor DNA-targeting module comprising a fusion protein comprising the sequence sf-6059407 22474-20028.40 set forth in SEQ ID NO: 519. In some embodiments, the polynucleotide comprises a nucleic acid sequence encoding at least one activator DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 515 and a nucleic acid sequence encoding at least one repressor DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 520. In some embodiments, the polynucleotide comprises a nucleic acid sequence encoding at least one activator DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 516 and a nucleic acid sequence encoding at least one repressor DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 517. In some embodiments, the polynucleotide comprises a nucleic acid sequence encoding at least one activator DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 516 and a nucleic acid sequence encoding at least one repressor DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 518. In some embodiments, the polynucleotide comprises a nucleic acid encoding at least one activator DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 516 and a nucleic acid encoding at least one repressor DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 519. In some embodiments, the polynucleotide comprises a nucleic acid sequence encoding at least one activator DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 516 and a nucleic acid sequence encoding at least one repressor DNA-targeting module comprising a fusion protein comprising the sequence set forth in SEQ ID NO: 520. [0531] In some embodiments, the polynucleotide comprises a cleavable linker sequence located between the nucleic acid sequence encoding the at least one activator DNA-targeting module and the nucleic acid sequence encoding the at least one repressor DNA-targeting module. In some embodiments, the cleavable linker sequence encodes a self-cleaving peptide. In some embodiments, the self-cleaving peptide is a 2A peptide sequence (e.g., such as any 2A or 2A-like sequence described in US 11,279,949). An exemplary 2A peptide sequence is a P2A peptide sequence, which is set forth in SEQ ID NO: 352. [0532] In some embodiments, the polynucleotide comprises an IRES located between the nucleic acid sequence encoding the at least one activator DNA-targeting module and the nucleic acid sequence encoding the at least one repressor DNA-targeting module. sf-6059407 22474-20028.40 [0533] In some embodiments, the polynucleotide comprises a single promoter that is operably linked to the first and second nucleic acid sequence to control their expression. IN some embodiments, the promoter is any promoter described herein. In some embodiments, the promoter is a constitutive promoter, such as an SV40 promoter, E1-Fα promoter or a CMV promoter. An exemplary promoter is a CMV promoter whose sequence is set forth in SEQ ID NO: 546. [0534] In some embodiments, the polynucleotide comprises from N-terminus to C-terminus, a sequence comprising: (a) a promoter sequence, such as described above; (b) a nucleic acid encoding the at least one activator DNA-targeting module, (c) the cleavable linker sequence or IRES, and (d) a nucleic acid encoding the at least one repressor DNA-targeting module. In some embodiments, the polynucleotide comprises, from N-terminus to C-terminus, a sequence comprising: (a) a promoter sequence; (b) a nucleic acid sequence coding the at least one activator DNA-targeting module comprising a fusion protein as set forth in SEQ ID NO: 516, (c) a nucleic acid sequence encoding the self-cleaving peptide as set forth in SEQ ID NO: 352, and (d) a nucleic acid sequence encoding the at least one repressor DNA-targeting module comprising a fusion protein as sest forth in SEQ ID NO: 517. An exemplary polynucleotide comprises the sequenceset forth in SEQ ID NO: 543. In some embodiments, the polynucleotide set forth in SEQ ID NO:543 is operably linked to the promoter sequence. [0535] In some embodiments, the polynucleotide comprises, from N-terminus to C- terminus, a sequence comprising: (a) a promoter sequence, such as any described herein; (b) a nucleic acid encoding the at least one repressor DNA-targeting module, (b) the self-cleaving peptide, (c) a cleavable linker sequence or IRES and (d) a nucleic acid sequence encoding the at least one activator DNA-targeting module. In some embodiments, the polynucleotide comprises, from N-terminus to C-terminus, a sequence comprising: (a) a nucleic acid encoding the at least one repressor DNA-targeting module comprising a fusion protein as sest forth in SEQ ID NO: 517, (b) a nucleic acid sequence encoding the self-cleaving peptide as set forth in SEQ ID NO: 352, and (c) a nucleic acid sequence encoding the at least one activator DNA-targeting module comprising a fusion protein as set forth in SEQ ID NO: 516. An exemplary polynucleotide comprises the sequenceset forth in SEQ ID NO: 542. In some embodiments, the polynucleotide set forth in SEQ ID NO:542 is operably linked to the promoter sequence. sf-6059407 22474-20028.40 C. Vectors and Delivery Methods [0536] Also provided herein is a vector that contains any of the provided polynucleotides. In some embodiments, the vector comprises a genetic construct, such as a plasmid or an expression vector. [0537] In some embodiments, a vector provided herein can comprise nucleic acids encoding any of the DNA-binding domains described herein, such as Cas proteins described in Section I.B.1 or ZFPs described in Section I.C, fusion proteins (such as ZFP fusion proteins or dCas fusion proteins described in Section I.E), gRNAs (such as gRNAs described in Section I.B.2), and DNA-targeting systems (such as DNA-targeting systems described in Section I) provided herein, or a portion or component thereof, or a combination of any of the foregoing. [0538] In some embodiments, the expression vector comprising the sequence encoding the first and/or second fusion protein of a DNA-targeting system provided herein can further comprise a polynucleotide sequence encoding at least one first and/or second gRNA. The sequence encoding the first and/or second gRNA can be operably linked to at least one transcriptional control sequence for expression of the first and/or second gRNA in the cell. For example, DNA encoding the first and/or second gRNA can be operably linked to a promoter sequence that is recognized by RNA polymerase III (Pol III). Examples of suitable Pol III promoters include, but are not limited to, mammalian U6, U3, H1, and 7SL RNA promoters. [0539] In some embodiments, provided is a vector containing a polynucleotide that encodes a first and/or second fusion protein comprising a DNA-binding domain comprising a dCas and at least one effector domain capable of modulating (e.g. increasing or decreasing) transcription of a first and/or second gene, and a polynucleotide(s) encoding at least one first and/or second gRNA. In some embodiments, the dCas is a dCas9, such as dSpCas9. In some embodiments, the polynucleotide encodes a first and/or second fusion protein that includes a dSpCas9 set forth in SEQ ID NO:127. In some embodiments, the polynucleotide encoding at least one first and/or second gRNA encodes a first and/or second gRNA as described in Section I.B.2. For example, the polynucleotide can encode a first and/or second gRNA comprising a spacer sequence selected from any one of SEQ ID NOS:35-40, 44-67, 91-101, 113-123, 212-223, 296-299, 303- 305, and 309-311, or a contiguous portion thereof of at least 14 nt. In another example, the polynucleotide can encode a first and/or second gRNA comprising a spacer sequence selected from any one of SEQ ID NOS:41-43, 79, 157-169, 171, 178-183, and 192-199, or a contiguous portion thereof of at least 14 nt. sf-6059407 22474-20028.40 [0540] In some embodiments, the polynucleotide encodes the first and/or second fusion protein and the at least one first and/or second gRNA. [0541] In some embodiments, the polynucleotide as provided herein can be codon optimized for efficient translation into protein in the eukaryotic cell or animal of interest. For example, codons can be optimized for expression in humans, mice, rats, hamsters, cows, pigs, cats, dogs, fish, amphibians, plants, yeast, insects, and so forth. Programs for codon optimization are available as freeware. Commercial codon optimization programs are also available. [0542] In some embodiments, a polynucleotide described herein can comprise one or more transcription and/or translation control elements. Depending on the host/vector system utilized, any of a number of suitable transcription and translation control elements, including constitutive and inducible promoters, transcription enhancer elements, transcription terminators, etc. can be used in the expression vector. [0543] Non-limiting examples of suitable eukaryotic promoters (i.e., promoters functional in a eukaryotic cell) include those from cytomegalovirus (CMV) immediate early, herpes simplex virus (HSV) thymidine kinase, early and late SV40, long terminal repeats (LTRs) from retrovirus, human elongation factor-1 promoter (EF1), a hybrid construct comprising the cytomegalovirus (CMV) enhancer fused to the chicken beta-actin promoter (CAG), murine stem cell virus promoter (MSCV), phosphoglycerate kinase-1 locus promoter (PGK), and mouse metallothionein-I. [0544] For expressing small RNAs, including guide RNAs used in connection with the DNA-targeting systems, various promoters such as RNA polymerase III promoters, including for example U6 and H1, can be advantageous. Descriptions of and parameters for enhancing the use of such promoters are known in the art, and additional information and approaches are regularly being described; see, e.g., Ma, H. et al., Molecular Therapy—Nucleic Acids 3, e161 (2014) doi:10.1038/mtna.2014.12. [0545] The expression vector can also contain a ribosome binding site for translation initiation and a transcription terminator. The expression vector can also comprise appropriate sequences for amplifying expression. The expression vector can also include nucleotide sequences encoding non-native tags (e.g., histidine tag, hemagglutinin tag, green fluorescent protein, etc.) that are fused to the site-directed polypeptide, thus resulting in a fusion protein. [0546] A promoter can be an inducible promoter (e.g., a heat shock promoter, tetracycline- regulated promoter, steroid-regulated promoter, metal-regulated promoter, estrogen receptor- sf-6059407 22474-20028.40 regulated promoter, etc.). The promoter can be a constitutive promoter (e.g., CMV promoter, UBC promoter). In some cases, the promoter can be a spatially restricted and/or temporally restricted promoter (e.g., a tissue specific promoter, a cell type specific promoter (e.g. a T cell specific promoter), etc.). [0547] Expression vectors contemplated include, but are not limited to, viral vectors based on vaccinia virus, poliovirus, adenovirus, adeno-associated virus, SV40, herpes simplex virus, human immunodeficiency virus, retrovirus (e.g., Murine Leukemia Virus, spleen necrosis virus, and vectors derived from retroviruses such as Rous Sarcoma Virus, Harvey Sarcoma Virus, avian leukosis virus, a lentivirus, human immunodeficiency virus, myeloproliferative sarcoma virus, and mammary tumor virus) and other recombinant vectors. Other vectors contemplated for eukaryotic target cells include, but are not limited to, the vectors pXT1, pSG5, pSVK3, pBPV, pMSG, and pSVLSV40 (Pharmacia). Other vectors can be used so long as they are compatible with the host cell. [0548] In some embodiments, the vector is a viral vector, such as an adeno-associated virus (AAV) vector, a retroviral vector, a lentiviral vector, or a gammaretroviral vector. In some embodiments In some embodiments, the viral vector is an adeno-associated virus (AAV) vector. In some embodiments, the AAV vector is selected from among an AAV1, AAV2, AAV3, AAV4, AAV5, AAV6, AAV7, AAV8, or AAV9 vector. In some embodiments, the vector is a lentiviral vector. In some embodiments, the vector is a non-viral vector, for example a lipid nanoparticle, a liposome, an exosome, or a cell penetrating peptide. In some embodiments, the vector comprises one vector, or two or more vectors. [0549] In some embodiments, a vector described herein is or comprises a lipid nanoparticle (LNP). Among provided embodiments is a lipid nanoparticle that contains any of the provided polynucleotides for delivery of an epigenetic-modifying DNA-targeting system. In some embodiments, the LNP contains a polynucleotide that encodes a first and/or second fusion protein as provided herein that includes (a) a DNA-binding domain capable of being targeted to a target site for one or more first and/or second genes; and (b) at least one effector domain. In some embodiments, the DNA-binding domain is a Cas (e.g. dCas) and the LNP further includes a gRNA. In some embodiments, the polynucleotide encoding the first and/or second fusion protein is an mRNA and the first and/or second gRNA is provided as an RNA. [0550] In some embodiments, any of the epigenetic-modifying DNA-targeting systems, gRNAs, Cas-gRNA combinations, polynucleotides, fusion proteins, or components thereof sf-6059407 22474-20028.40 described herein, are incorporated in lipid nanoparticles (LNPs), such as for delivery. In some embodiments, the lipid nanoparticle is a vector for delivery. In some embodiments, the nanoparticle may comprise at least one lipid. The lipid may be selected from, but is not limited to, dLin-DMA, dLin-K-DMA, 98N12- 5, C12-200, dLin-MC3-DMA, dLin-KC2-DMA, DODMA, PLGA, PEG, PEG-DMG and PEGylated lipids. In another aspect, the lipid may be a cationic lipid such as, but not limited to, dLin-DMA, dLin-D-DMA, dLin-MC 3 -DMA, dLin- KC2-DMA and DODMA. Typically, the LNPs are composed of two or more lipids, such as 3, 4 or 5 lipids. In some embodiments, at least lipid is either ionoizable cationic or cationic. [0551] Lipid nanoparticles can be used for the delivery of encapsulated or associated (e.g., complexed) therapeutic agents, including nucleic acids and proteins, such as those encoding and/or comprising CRISPR/Cas systems. See, e.g., US Patent No.10,723,692, US Patent No. 10,941,395, and WO 2015/035136. [0552] In some embodiments, the provided methods involve use of a lipid nanoparticle (LNP) comprising mRNA, such as mRNA encoding a protein component of any of the provided DNA-targeting systems, for example any of the fusion proteins provided herein. In some embodiments, the mRNA can be produced using methods known in the art such as in vitro transcription. In some embodiments of the method, the mRNA comprises a 5' cap. In some embodiments, the 5’ cap is an altered nucleotide on the 5’ end of primary transcripts such as messenger RNA. In some aspects, the 5’ caps of the mRNA improves one or more of RNA stability and processing, mRNA metabolism, the processing and maturation of an RNA transcript in the nucleus, transport of mRNA from the nucleus to the cytoplasm, mRNA stability, and efficient translation of mRNA to protein. In some embodiments, a 5’ cap can be a naturally- occurring 5’ cap or one that differs from a naturally-occurring cap of an mRNA. A 5’ cap may be any 5' cap known to a skilled artisan. In certain embodiments, the 5' cap is selected from the group consisting of an Anti-Reverse Cap Analog (ARCA) cap, a 7-methyl-guanosine (7mG) cap, a CleanCap® analog, a vaccinia cap, and analogs thereof. For instance, the 5’ cap may include, without limitation, an anti-reverse cap analogs (ARCA) (US7074596), 7-methyl- guanosine, CleanCap® analogs, such as Cap 1 analogs (Trilink; San Diego, CA), or enzymatically capped using, for example, a vaccinia capping enzyme or the like. In some embodiments, the mRNA may be polyadenylated. The mRNA may contain various 5’ and 3’ untranslated sequence elements to enhance expression of the encoded protein and/or stability of the mRNA itself. Such elements can include, for example, posttranslational regulatory elements sf-6059407 22474-20028.40 such as a woodchuck hepatitis virus post-transcriptional regulatory element (WPRE). In some embodiments, the mRNA comprises at least one nucleoside modification. The mRNA may contain modifications of naturally-occurring nucleosides to nucleoside analogs. Any nucleoside analogs known in the art are envisioned. Such nucleoside analogs can include, for example, those described in US 8,278,036. In certain embodiments of the method, the nucleoside modification is selected from the group consisting of a modification from uridine to pseudouridine and uridine to Nl- methyl pseudouridine. In particular embodiments of the method the nucleoside modification is from uridine to pseudouridine. [0553] In some embodiments, LNPs useful for in the present methods comprise a cationic lipid selected from dLin-DMA ( 1 ,2-dilinoleyloxy-3 -dimethylaminopropane) , dLin-MC3 -DM A (dilinoleylmethyl-4-dimethylaminobutyrate), dLin-KC2-DMA (2,2-dilinoleyl-4-(2- dimethylaminoethyl)-[l,3]-dioxolane), DODMA (1,2- dioleyloxy-N,N-dimethyl-3- aminopropane), SS-OP (Bis[2-(4-{2-[4-(cis-9 octadecenoyloxy)phenylacetoxy]ethyl}piperidinyl)ethyl] disulfide), and derivatives thereof. dLin-MC3-DMA and derivatives thereof are described, for example, in WO 2010/144740. DODMA and derivatives thereof are described, for example, in US 7,745,651 and Mok et al. (1999), Biochimica et Biophysica Acta, 1419(2): 137-150. dLin-DMA and derivatives thereof are described, for example, in US 7,799,565. dLin-KC2-DMA and derivatives thereof are described, for example, in US 9,139,554. SS-OP (NOF America Corporation, White Plains, NY) is described, for example, at https://www.nofamerica.com/store/index.php?dispatch=products.view&product_id=962. Additional and non-limiting examples of cationic lipids include methylpyridiyl-dialkyl acid (MPDACA), palmitoyl-oleoyl-nor-arginine (PONA), guanidino-dialkyl acid (GUADACA), l,2- di-0-octadecenyl-3-trimethylammonium propane (DOTMA), 1,2- dioleoyl-3- trimethylammonium-propane (DOTAP), Bis{2-[N-methyl-N-(a-D- tocopherolhemisuccinatepropyl)amino]ethyl} disulfide (SS-33/3AP05), Bis{2-[4-(a-D- tocopherolhemisuccinateethyl)piperidyl] ethyl} disulfide (SS33/4PE15), Bis{2-[4-(cis-9- octadecenoateethyl)-l-piperidinyl] ethyl} disulfide (SS18/4PE16), and Bis{2-[4-(cis,cis-9,12- octadecadienoateethyl)-l-piperidinyl] ethyl} disulfide (SS18/4PE13). In further embodiments, the lipid nanoparticles also comprise one or more non-cationic lipids and a lipid conjugate. [0554] In some embodiments, the molar concentration of the cationic lipid is from about 20% to about 80%, from about 30% to about 70%, from about 40% to about 60%, from about sf-6059407 22474-20028.40 45% to about 55%, or about 20%, about 25%, about 30%, about 35%, about 40%, about 45%, about 50%, about 55%, about 60%, about 65%, about 70%, about 75%, or about 80% of the total lipid molar concentration, wherein the total lipid molar concentration is the sum of the cationic lipid, the non-cationic lipid, and the lipid conjugate molar concentrations. In certain embodiments, the lipid nanoparticles comprise a molar ratio of cationic lipid to any of the polynucleotides of from about 1 to about 20, from about 2 to about 16, from about 4 to about 12, from about 6 to about 10, or about 1, about 2, about 3, about 4, about 5, about 6, about 7, about 8, about 9, about 10, about 11, about 12, about 13, about 14, about 15, about 16, about 17, about 18, about 19, or about 20. [0555] In some embodiments, the lipid nanoparticles can comprise at least one non-cationic lipid. In particular embodiments, the molar concentration of the non-cationic lipids is from about 20% to about 80%, from about 30% to about 70%, from about 40% to about 70%, from about 40% to about 60%, from about 46% to about 50%, or about 20%, about 25%, about 30%, about 35%, about 40%, about 45%, about 48.5%, about 50%, about 55%, about 60%, about 65%, about 70%, about 75%, or about 80% of the total lipid molar concentration. Non-cationic lipids include, in some embodiments, phospholipids and steroids. [0556] In some embodiments, phospholipids useful for the lipid nanoparticles described herein include, but are not limited to, l,2-Distearoyl-sn-glycero-3-phosphocholine (DSPC), l,2- Didecanoyl-sn-glycero-3- phosphocholine (DDPC), l,2-Dierucoyl-sn-glycero-3- phosphate(Sodium Salt) (DEPA-NA), l,2-Dierucoyl-sn-glycero-3-phosphocholine (DEPC), l,2- Dierucoyl-sn-glycero-3- phosphoethanolamine (DEPE), l,2-Dierucoyl-sn-glycero-3[Phospho- rac-(l-glycerol)(Sodium Salt) (DEPG-NA), l,2-Dilinoleoyl-sn-glycero-3-phosphocholine (DLOPC), 1,2-Dilauroyl-sn- glycero-3-phosphate(Sodium Salt) (DLPA-NA), l,2-Dilauroyl-sn- glycero-3-phosphocholine (DLPC), l,2-Dilauroyl-sn-glycero-3-phosphoethanolamine (DLPE), 1,2-Dilauroyl-sn- glycero-3[Phospho-rac-(l-glycerol...)(Sodium Salt) (DLPG-NA), 1,2- Dilauroyl-sn-glycero- 3[Phospho-rac-(l-glycerol)(Ammonium Salt) (DLPG-NH4), l,2- Dilauroyl-sn-glycero-3- phosphoserine(Sodium Salt) (DLPS-NA), l,2-Dimyristoyl-sn-glycero-3- phosphate(SodiumSalt) (DMPA-NA), l,2-Dimyristoyl-sn-glycero-3-phosphocholine (DMPC), 1,2-Dimyristoyl- sn-glycero-3-phosphoethanolamine (DMPE), l,2-Dimyristoyl-sn-glycero- 3[Phospho-rac-(l- glycerol)(Sodium Salt) (DMPG-NA), l,2-Dimyristoyl-sn-glycero-3[Phospho- rac-(l- glycerol)(Ammonium Salt) (DMPG-NH4), l,2-Dimyristoyl-sn-glycero-3[Phospho-rac-(l- glycerol)(Sodium/ Ammonium Salt) (DMPG-NH4/NA), l,2-Dimyristoyl-sn-glycero-3- sf-6059407 22474-20028.40 phosphoserine(Sodium Salt) (DMPS-NA), l,2-Dioleoyl-sn-glycero-3-phosphate(Sodium Salt) (DOPA-NA), l,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-Dioleoyl-sn- glycero-3- phosphoethanolamine (DOPE), l,2-Dioleoyl-sn-glycero-3[Phospho-rac-(l- glycerol)(Sodium Salt) (DOPG-NA), l,2-Dioleoyl-sn-glycero-3-phosphoserine(Sodium Salt) (DOPS-NA), l,2- Dipalmitoyl-sn-glycero-3-phosphate(Sodium Salt) (DPPA-NA), 1,2- Dipalmitoyl-sn-glycero-3- phosphocholine (DPPC), 1 ,2-Dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE), 1 ,2- Dipalmitoyl-sn-glycero- 3[Phospho-rac-(l-glycerol)(Sodium Salt) (DPPG-NA), 1,2-Dipalmitoyl- sn-glycero- 3[Phospho-rac-(l-glycerol)(Ammonium Salt) (DPPG-NH4), l,2-Dipalmitoyl-sn- glycero-3- phosphoserine(Sodium Salt) (DPPS-NA), l,2-Distearoyl-sn-glycero-3- phosphate(Sodium Salt) (DSPA-NA), l,2-Distearoyl-sn-glycero-3-phosphoethanolamine (DSPE), 1,2- Distearoyl-sn-glycero-3[Phospho-rac-(l-glycerol)(Sodium Salt) (DSPG-NA), 1,2- Distearoyl- sn-glycero-3[Phospho-rac-(l-glycerol)(Ammonium Salt) (DSPG-NH4), 1,2- Distearoyl-sn- glycero-3-phosphoserine(Sodium Salt) (DSPS-NA), Egg-PC (EPC), Hydrogenated Egg PC (HEPC), Hydrogenated Soy PC (HSPC), l-Myristoyl-sn-glycero-3- phosphocholine (LY S OPCM YRIS TIC ) , l-Palmitoyl-sn-glycero-3-phosphocholine (LYSOPCPALMITIC), 1- Stearoyl-sn-glycero-3-phosphocholine (LYSOPC STEARIC), l- Myristoyl-2-palmitoyl-sn- glycero3-phosphocholine (MPPC), l-Myristoyl-2-stearoyl-sn-glycero- 3-phosphocholine (MSPC), l-Palmitoyl-2-myristoyl-sn-glycero-3-phosphocholine (PMPC), l- Palmitoyl-2- oleoyl-sn-glycero-3-phosphocholine (POPC), l-Palmitoyl-2-oleoyl-sn-glycero-3- phosphoethanolamine (POPE), l-Palmitoyl-2-oleoyl-sn-glycero-3[Phospho-rac-(l- glycerol)] (Sodium Salt) (POPG-NA), l-Palmitoyl-2-stearoyl-sn-glycero-3-phosphocholine (PS PC), l- Stearoyl-2-myristoyl-sn-glycero-3-phosphocholine (SMPC), l-Stearoyl-2-oleoyl- sn-glycero-3- phosphocholine (SOPC), and l-Stearoyl-2-palmitoyl-sn-glycero-3- phosphocholine (SPPC). In particular embodiments, the phospholipid is DSPC. In particular embodiments, the phospholipid is DOPE. In particular embodiments, the phospholipid is DOPC. [0557] In some embodiments, the non-cationic lipids comprised by the lipid nanoparticles include one or more steroids. Steroids useful for the lipid nanoparticles described herein include, but are not limited to, cholestanes such as cholesterol, cholanes such as cholic acid, pregnanes such as progesterone, androstanes such as testosterone, and estranes such as estradiol. Further steroids include, but are not limited to, cholesterol (ovine), cholesterol sulfate, desmosterol-d6, cholesterol-d7, lathosterol-d7, desmosterol, stigmasterol, lanosterol, dehydrocholesterol, dihydrolanosterol, zymosterol, lathosterol, zymosterol-d5, 14-demethyl-lanosterol, 14-demethyl- sf-6059407 22474-20028.40 lanosterol-d6, 8(9)- dehydrocholesterol, 8(14)-dehydrocholesterol, diosgenin, DHEA sulfate, DHEA, lanosterol- d6, dihydrolanosterol-d7, campesterol-d6, sitosterol, lanosterol-95, Dihydro FF-MAS-d6, zymostenol-d7, zymostenol, sitostanol, campestanol, campesterol, 7- dehydrodesmosterol, pregnenolone, sitosterol-d7, Dihydro T-MAS, Delta 5-avenasterol, Brassicasterol, Dihydro FF-MAS, 24-methylene cholesterol, cholic acid derivatives, cholesteryl esters, and glycosylated sterols. In particular embodiments, the lipid nanoparticles comprise cholesterol. [0558] In some embodiments, the lipid nanoparticles comprise a lipid conjugate. Such lipid conjugates include, but are not limited to, ceramide PEG derivatives such as C8 PEG2000 ceramide, C16 PEG2000 ceramide, C8 PEG5000 ceramide, C16 PEG5000 ceramide, C8 PEG750 ceramide, and C16 PEG750 ceramide, phosphoethanolamine PEG derivatives such as 16:0 PEG5000PE, 14:0 PEG5000 PE, 18:0 PEG5000 PE, 18:1 PEG5000 PE, 16:0 PEG3000 PE, 14:0 PEG3000 PE, 18:0 PEG3000 PE, 18:1 PEG3000 PE, 16:0 PEG2000 PE, 14:0 PEG2000 PE, 18:0 PEG2000 PE, 18:1 PEG2000 PE 16:0 PEG1000 PE, 14:0 PEG1000 PE, 18:0 PEG1000 PE, 18:1 PEG 1000 PE, 16:0 PEG750 PE, 14:0 PEG750 PE, 18:0 PEG750 PE, 18:1 PEG750 PE, 16:0 PEG550 PE, 14:0 PEG550 PE, 18:0 PEG550 PE, 18:1 PEG550 PE, 16:0 PEG350 PE, 14:0 PEG350 PE, 18:0 PEG350 PE, and 18:1 PEG350, sterol PEG derivatives such as Chol- PEG600, and glycerol PEG derivatives such as DMG-PEG5000, DSG-PEG5000, DPG- PEG5000, DMG-PEG3000, DSG-PEG3000, DPG-PEG3000, DMG-PEG2000, DSG- PEG2000, DPG-PEG2000, DMG-PEG1000, DSG-PEG1000, DPG-PEG1000, DMG- PEG750, DSG- PEG750, DPG-PEG750, DMG-PEG550, DSG-PEG550, DPG-PEG550, DMG-PEG350, DSG- PEG350, and DPG-PEG350. In some embodiments, the lipid conjugate is a DMG-PEG. In some particular embodiments, the lipid conjugate is DMG- PEG2000. In some particular embodiments, the lipid conjugate is DMG-PEG5000. [0559] It is within the level of a skilled artisan to select the cationic lipids, non-cationic lipids and/or lipid conjugates which comprise the lipid nanoparticle, as well as the relative molar ratio of such lipids to each other, such as based upon the characteristics of the selected lipid(s), the nature of the delivery to the intended target cells, and the characteristics of the nucleic acids and/or proteins to be delivered. Additional considerations include, for example, the saturation of the alkyl chain, as well as the size, charge, pH, pKa, fusogenicity and toxicity of the selected lipid(s). Thus, the molar ratios of each individual component may be adjusted accordingly. sf-6059407 22474-20028.40 [0560] The lipid nanoparticles for use in the method can be prepared by various techniques which are known to a skilled artisan. Nucleic acid-lipid particles and methods of preparation are disclosed in, for example, U.S. Patent Publication Nos.20040142025 and 20070042031. [0561] In some embodiments, the lipid nanoparticles will have a size within the range of about 25 to about 500 nm. In some embodiments, the lipid nanoparticles have a size from about 50 nm to about 300 nm, or from about 60 nm to about 120 nm. The size of the lipid nanoparticles may be determined by quasi-electric light scattering (QELS) as described in Bloomfield, Ann. Rev. Biophys. Bioeng., 10:421A150 (1981). A variety of methods are known in the art for producing a population of lipid nanoparticles of particular size ranges, for example, sonication or homogenization. One such method is described in U.S. Pat. No.4,737,323. [0562] In some embodiments, the lipid nanoparticles comprise a cell targeting molecule such as, for example, a targeting ligand (e.g., antibodies, scFv proteins, DART molecules, peptides, aptamers, and the like) anchored on the surface of the lipid nanoparticle that selectively binds the lipid nanoparticles to the targeted cell, such as any cell described herein, e.g. a T cell. [0563] In some embodiments, the vector exhibits immune cell or T cell tropism. [0564] In some aspects, provided herein are pluralities of vectors that comprise any of the vectors described herein, and one or more additional vectors comprising one or more additional polynucleotides encoding an additional portion or an additional component of any of the DNA- targeting systems described herein, any of the gRNAs described herein, any of the first and/or second fusion proteins described herein, or a portion or a component of any of the foregoing. [0565] Provided are pluralities of vectors, that include: a first vector comprising any of the polynucleotides described herein; and a second vector comprising any of the polynucleotides described herein. [0566] In some aspects, vectors provided herein may be referred to as delivery vehicles. In some aspects, any of the DNA-targeting systems, components thereof, or polynucleotides disclosed herein can be packaged into or on the surface of delivery vehicles for delivery to cells. Delivery vehicles contemplated include, but are not limited to, nanospheres, liposomes, quantum dots, nanoparticles, polyethylene glycol particles, hydrogels, and micelles. As described in the art, a variety of targeting moieties can be used to enhance the preferential interaction of such vehicles with desired cell types or locations. sf-6059407 22474-20028.40 [0567] Methods of introducing a nucleic acid into a host cell are known in the art, and any known method can be used to introduce a nucleic acid (e.g., an expression construct) into a cell. Suitable methods include, include e.g., viral or bacteriophage infection, transfection, conjugation, protoplast fusion, lipofection, electroporation, calcium phosphate precipitation, polyethyleneimine (PEI)-mediated transfection, DEAE-dextran mediated transfection, liposome- mediated transfection, particle gun technology, calcium phosphate precipitation, direct micro injection, nanoparticle-mediated nucleic acid delivery, and the like. In some embodiments, the composition may be delivered by mRNA delivery and ribonucleoprotein (RNP) complex delivery. Direct delivery of the RNP complex, including the DNA-binding domain complexed with the sgRNA, can eliminate the need for intracellular transcription and translation and can offer a robust platform for host cells with low transcriptional and translational activity. The RNP complexes can be introduced into the host cell by any of the methods known in the art. [0568] In some embodiments, the method of introducing a nucleic acid into a host cell is a method comprising transient delivery. [0569] Nucleic acids or RNPs of the disclosure can be incorporated into a host using virus- like particles (VLP). VLPs contain normal viral vector components, such as envelope and capsids, but lack the viral genome. For instance, nucleic acids expressing the Cas and sgRNA can be fused to the viral vector components such as gag and introduced into producer cells. The resulting virus-like particles containing the sgRNA-expressing vectors can infect the host cell for efficient editing. [0570] Introduction of the complexes, polypeptides, and nucleic acids of the disclosure can occur by protein transduction domains (PTDs). PTDs, including the human immunodeficiency virus-1 TAT, herpes simplex virus-1 VP22, Drsophila Antennapedia Antp, and the poluarginines, are peptide sequences that can cross the cell membrane, enter a host cell, and deliver the complexes, polypeptides, and nucleic acids into the cell. [0571] Introduction of the complexes, polypeptides, and nucleic acids of the disclosure into cells can occur by viral or bacteriophage infection, transfection, conjugation, protoplast fusion, lipofection, electroporation, nucleofection, calcium phosphate precipitation, polyethyleneimine (PEI)-mediated transfection, DEAE-dextran mediated transfection, liposome-mediated transfection, particle gun technology, calcium phosphate precipitation, direct micro-injection, nanoparticle-mediated nucleic acid delivery, and the like, for example as described in WO sf-6059407 22474-20028.40 2017/193107, WO 2016/123578, WO 2014/152432, WO 2014/093661, WO 2014/093655, or WO 2021/226555. [0572] Various methods for the introduction of polynucleotides are well known and may be used with the provided methods and compositions. Exemplary methods include those for transfer of polynucleotides encoding the DNA targeting systems provided herein, including via viral, e.g., retroviral or lentiviral, transduction, transposons, and electroporation. [0573] In some embodiments, polynucleotides can be cloned into a suitable vector, such as an expression vector or vectors. The expression vector can be any suitable recombinant expression vector, and can be used to transform or transfect any suitable cell. Suitable vectors include those designed for propagation and expansion or for expression or both, such as plasmids and viruses. [0574] In some embodiments, the vector can a vector of the pUC series (Fermentas Life Sciences), the pBluescript series (Stratagene, LaJolla, Calif.), the pET series (Novagen, Madison, Wis.), the pGEX series (Pharmacia Biotech, Uppsala, Sweden), or the pEX series (Clontech, Palo Alto, Calif.). In some embodiments, animal expression vectors include pEUK- Cl, pMAM and pMAMneo (Clontech). In some embodiments, a viral vector is used, such as a lentiviral or retroviral vector. In some embodiments, the recombinant expression vectors can be prepared using standard recombinant DNA techniques. In some embodiments, vectors can contain regulatory sequences, such as transcription and translation initiation and termination codons, which are specific to the type of host into which the vector is to be introduced, as appropriate and taking into consideration whether the vector is DNA- or RNA- based. In some embodiments, the vector can contain a nonnative promoter operably linked to the nucleotide sequence encoding the recombinant receptor. In some embodiments, the promoter can be a non- viral promoter or a viral promoter, such as a cytomegalovirus (CMV) promoter, an SV40 promoter, an RSV promoter, and a promoter found in the long-terminal repeat of the murine stem cell virus. Other promoters known to a skilled artisan also are contemplated. [0575] In some embodiments, recombinant nucleic acids are transferred into cells using recombinant infectious virus particles, such as, e.g., vectors derived from simian virus 40 (SV40), adenoviruses, or adeno-associated virus (AAV). In some embodiments, recombinant nucleic acids are transferred into cells (e.g. T cells) using recombinant lentiviral vectors or retroviral vectors, such as gamma-retroviral vectors (see, e.g., Koste et al. (2014) Gene Therapy 2014 Apr 3. doi: 10.1038/gt.2014.25; Carlens et al. (2000) Exp Hematol 28(10): 1137-46; sf-6059407 22474-20028.40 Alonso-Camino et al. (2013) Mol Ther Nucl Acids 2, e93; Park et al., Trends Biotechnol.2011 November 29(11): 550-557. [0576] In some embodiments, the retroviral vector has a long terminal repeat sequence (LTR), e.g., a retroviral vector derived from the Moloney murine leukemia virus (MoMLV), myeloproliferative sarcoma virus (MPSV), murine embryonic stem cell virus (MESV), murine stem cell virus (MSCV), spleen focus forming virus (SFFV), or adeno-associated virus (AAV). Most retroviral vectors are derived from murine retroviruses. In some embodiments, the retroviruses include those derived from any avian or mammalian cell source. The retroviruses typically are amphotropic, meaning that they are capable of infecting host cells of several species, including humans. In one embodiment, the gene to be expressed replaces the retroviral gag, pol and/or env sequences. A number of illustrative retroviral systems have been described (e.g., U.S. Pat. Nos.5,219,740; 6,207,453; 5,219,740; Miller and Rosman (1989) BioTechniques 7:980-990; Miller, A. D. (1990) Human Gene Therapy 1:5-14; Scarpa et al. (1991) Virology 180:849-852; Burns et al. (1993) Proc. Natl. Acad. Sci. USA 90:8033-8037; and Boris-Lawrie and Temin (1993) Cur. Opin. Genet. Develop.3: 102-109. [0577] In some embodiments, the vector is a lentiviral vector. In some embodiments, the lentiviral vector is an integrase-deficient lentiviral vector. In some embodiments, the lentiviral vector is a recombinant lentiviral vector. In some embodiments, the lentivirus is selected or engineered for a desired tropism (e.g. for T cell or immune cell tropism). Methods of lentiviral production, transduction, and engineering are known, for example as described in Kasaraneni, N. et al. Sci. Rep.8(1):10990 (2018), Ghaleh, H.E.G. et al. Biomed. Pharmacother.128:110276 (2020), and Milone, M.C. et al. Leukemia.32(7):1529-1541 (2018). Additional methods for lentiviral transduction are described, for example in Wang et al. (2012) J. Immunother.35(9): 689-701; Cooper et al. (2003) Blood.101: 1637- 1644; Verhoeyen et al. (2009) Methods Mol Biol.506: 97-114; and Cavalieri et al. (2003) Blood.102(2): 497-505. [0578] In some embodiments, recombinant nucleic acids are transferred into cells (e.g. T cells) via electroporation {see, e.g., Chicaybam et al, (2013) PLoS ONE 8(3): e60298 and Van Tedeloo et al. (2000) Gene Therapy 7(16): 1431-1437). In some embodiments, recombinant nucleic acids are transferred into cells via transposition (see, e.g., Manuri et al. (2010) Hum Gene Ther 21(4): 427-437; Sharma et al. (2013) Molec Ther Nucl Acids 2, e74; and Huang et al. (2009) Methods Mol Biol 506: 115-126). Other methods of introducing and expressing genetic material into immune cells include calcium phosphate transfection (e.g., as described in Current sf-6059407 22474-20028.40 Protocols in Molecular Biology, John Wiley & Sons, New York. N.Y.), protoplast fusion, cationic liposome-mediated transfection; tungsten particle-facilitated microparticle bombardment (Johnston, Nature, 346: 776-777 (1990)); and strontium phosphate DNA co- precipitation (Brash et al., Mol. Cell Biol., 7: 2031-2034 (1987)). III. PHARMACEUTICAL COMPOSITIONS AND FORMULATIONS OF DNA- TARGETING SYSTEMS OR ENCODING POLYNUCLEOTIDES OR VECTORS [0579] In some aspects, provided herein are compositions, such as pharmaceutical compositions and formulations for administration, that include any of the DNA-targeting systems described herein, for example in Section I, or any of the polynucleotides or vectors encoding the same, for example as described in Section II. In some aspects, the pharmaceutical composition contains one or more DNA-targeting systems provided herein or a component thereof. In some aspects, the pharmaceutical composition comprises one or more vectors, e.g., viral vectors that contain polynucleotides that encode one or more components of the DNA- targeting systems provided herein. Such compositions can be used in accord with the provided methods, and/or with the provided articles of manufacture or compositions, such as in the prevention or treatment of diseases, conditions, and disorders, or in detection, diagnostic, and prognostic methods. [0580] The term “pharmaceutical formulation” refers to a preparation which is in such form as to permit the biological activity of an active ingredient contained therein to be effective, and which contains no additional components which are unacceptably toxic to a subject or a cell to which the formulation would be administered. [0581] In some embodiments, the pharmaceutical composition may further comprise a pharmaceutically acceptable excipient. The pharmaceutically acceptable excipient may be functional molecules as vehicles, adjuvants, carriers, or diluents. [0582] A “pharmaceutically acceptable carrier” refers to an ingredient in a pharmaceutical formulation, other than an active ingredient, which is nontoxic to a subject. A pharmaceutically acceptable carrier includes, but is not limited to, a buffer, excipient, stabilizer, or preservative. [0583] In some aspects, the choice of carrier is determined in part by the particular agent and/or by the method of administration. Accordingly, there are a variety of suitable formulations. For example, the pharmaceutical composition can contain preservatives. Suitable preservatives may include, for example, methylparaben, propylparaben, sodium benzoate, and benzalkonium chloride. In some aspects, a mixture of two or more preservatives is used. The sf-6059407 22474-20028.40 preservative or mixtures thereof are typically present in an amount of about 0.0001% to about 2% by weight of the total composition. Carriers are described, e.g., by Remington’s Pharmaceutical Science
s 16th edition, Osol, A. Ed. (1980). Pharmaceutically acceptable carriers are generally nontoxic to recipients at the dosages and concentrations employed, and include, but are not limited to: buffers such as phosphate, citrate, and other organic acids; antioxidants including ascorbic acid and methionine; preservatives (such as octadecyldimethylbenzyl ammonium chloride; hexamethonium chloride; benzalkonium chloride; benzethonium chloride; phenol, butyl or benzyl alcohol; alkyl parabens such as methyl or propyl paraben; catechol; resorcinol; cyclohexanol; 3-pentanol; and m-cresol); low molecular weight (less than about 10 residues) polypeptides; proteins, such as serum albumin, gelatin, or immunoglobulins; hydrophilic polymers such as polyvinylpyrrolidone; amino acids such as glycine, glutamine, asparagine, histidine, arginine, or lysine; monosaccharides, disaccharides, and other carbohydrates including glucose, mannose, or dextrins; chelating agents such as EDTA; sugars such as sucrose, mannitol, trehalose or sorbitol; salt-forming counter-ions such as sodium; metal complexes (e.g. Zn-protein complexes); and/or non-ionic surfactants such as polyethylene glycol (PEG). [0584] In some embodiments, the pharmaceutically acceptable excipient may be a transfection facilitating agent, which may include surface active agents, such as immune- stimulating complexes (ISCOMS), Freunds incomplete adjuvant, LPS analog including monophosphoryl lipid A, muramyl peptides, quinone analogs, vesicles such as squalene and squalene, hyaluronic acid, lipids, liposomes, calcium ions, viral proteins, polyanions, polycations, or nanoparticles, or other known transfection facilitating agents. [0585] In some embodiments, the transfection facilitating agent is a polyanion, polycation, including poly-L-glutamate (LGS), or lipid. In some embodiments, the transfection facilitating agent is poly-L-glutamate. In some embodiments, the transfection facilitating agent may also include surface active agents such as immune-stimulating complexes (ISCOMS), Freunds incomplete adjuvant, LPS analog including monophosphoryl lipid A, muramyl peptides, quinone analogs and vesicles such as squalene and squalene, and hyaluronic acid may also be used administered in conjunction with the genetic construct. In some embodiments, the DNA vector encoding the DNA-targeting system may also include a transfection facilitating agent such as lipids, liposomes, including lecithin liposomes or other liposomes known in the art, as a DNA- liposome mixture (see for example WO9324640), calcium ions, viral proteins, polyanions, sf-6059407 22474-20028.40 polycations, or nanoparticles, or other known transfection facilitating agents. In some embodiments, the transfection facilitating agent is a polyanion, polycation, including poly-L- glutamate (LGS), or lipid. [0586] Compositions in some embodiments are provided as sterile liquid preparations, e.g., isotonic aqueous solutions, suspensions, emulsions, dispersions, or viscous compositions, which may in some aspects be buffered to a selected pH. Liquid preparations are normally easier to prepare than gels, other viscous compositions, and solid compositions. Additionally, liquid compositions are somewhat more convenient to administer, especially by injection. Viscous compositions, on the other hand, can be formulated within the appropriate viscosity range to provide longer contact periods with specific tissues. Liquid or viscous compositions can comprise carriers, which can be a solvent or dispersing medium containing, for example, water, saline, phosphate buffered saline, polyol (for example, glycerol, propylene glycol, liquid polyethylene glycol) and suitable mixtures thereof. [0587] Sterile injectable solutions can be prepared by incorporating the agent in a solvent, such as in admixture with a suitable carrier, diluent, or excipient such as sterile water, physiological saline, glucose, dextrose, or the like. The formulations to be used for in vivo or ex vivo administration or use are generally sterile. Sterility may be readily accomplished, e.g., by filtration through sterile filtration membranes. [0588] The pharmaceutical composition in some embodiments contains components in amounts effective to treat or prevent the disease or condition, such as a therapeutically effective or prophylactically effective amount. Therapeutic or prophylactic efficacy in some embodiments is monitored by periodic assessment of treated subjects. For repeated administrations over several days or longer, depending on the condition, the treatment is repeated until a desired suppression of disease symptoms occurs. However, other dosage regimens may be useful and can be determined. The desired dosage can be delivered by a single bolus administration of the composition, by multiple bolus administrations of the composition, or by continuous infusion administration of the composition. [0589] In some embodiments, the composition can be administered to a subject by any suitable means, for example, by bolus infusion or by injection, e.g., by intravenous or subcutaneous injection. In some embodiments, a given dose is administered by a single bolus administration of the composition. In some embodiments, the composition is administered by multiple bolus administrations of the composition, for example, over a period of no more than 3 sf-6059407 22474-20028.40 days, or by continuous infusion administration of the composition. In some embodiments, the composition is administered parenterally, for example by intravenous, intramuscular, subcutaneous, or intraperitoneal administration. In some embodiments, the composition is administered to a subject using peripheral systemic delivery by intravenous, intraperitoneal, or subcutaneous injection. [0590] In some embodiments, the composition is contacted with our introduced into cells (e.g. primary T cells) from a subject ex vivo, and the cells are subsequently administered to the same subject or to a different subject. [0591] For the prevention or treatment of disease, the appropriate dosage may depend on the type of disease to be treated, the type of agent or agents, the type of cells or recombinant receptors, the severity and course of the disease, whether the agent or cells are administered for preventive or therapeutic purposes, previous therapy, the subject’s clinical history and response to the agent or the cells, and the discretion of the attending physician. The compositions are in some embodiments suitably administered to the subject at one time or over a series of treatments. IV. METHODS OF EPIGENETICALLY MODIFYING LYMPHOID CELLS AND RELATED MODIFIED CELLS AND COMPOSITIONS THEREOF [0592] Provided herein are modified lymphoid cells (e.g. T cells) that have one or more phenotypic and/or epigenetic modifications (also referred to as changes or alterations) in their epigenome. In some embodiments, the epigenetic change is a change relative to a comparable unmodified lymphoid cell. Reference to a comparable unmodified cell is understood to refer to the same or similar cell but that has not been introduced with a provided epigenome-modifying DNA-targeting system (such as any described in Section I) or that or that does not contain the same epigenetic changes (e.g. methylation or histone modification) of the target gene or regulatory region thereof. [0593] In some embodiments, the phenotypic and/or epigenetic modification includes increased transcription of one or more first genes (such as any described in Section I.A.2), or a phenotype or epigenetic modification resulting from the increased transcription. In some embodiments, the epigenetic modification includes decreased transcription of one or more second genes (such as any described in Section I.A.3), or a phenotype or epigenetic modification resulting from the decreased transcription. [0594] In some embodiments, the lymphoid cells that are modified by the provided DNA- sf-6059407 22474-20028.40 binding systems can include T cells, NK cells, or NKT cells. Such cells can include cells that have been enriched or isolated from a primary population of cells from a subject, or can include any cells that have been differentiated from stem cells into such lymphoid cells and/or have been differentiated from progenitor cells, such as common lymphoid progenitors (CLPs). In some embodiments, the lymphoid cells are differentiated from stem cells, such as hematopoietic stem or progenitor cells, or progenitor cells. In some embodiments, the lymphoid cells are trans- differentiated from a non-pluripotent cell of non-hematopoietic lineage. In particular embodiments, the cells are modified T cells that have been modified by the provided DNA- binding systems. [0595] Provided herein are modified T cells (e.g. CD4+ T cell or CD8+ T cell) that have one or more modifications (also referred to as changes or alterations) in their phenotype and/or epigenome. In some embodiments, the modification increases or promotes a phenotype in the T cell, such as any phenotype described herein, for example in Section I. In some embodiments, the modified cell is a modified T cell that has increased T cell effector function upon T cell stimulation. In some embodiments, the epigenetic change is a change relative to a comparable unmodified T cell. Reference to a comparable unmodified T cells is understood to refer to the same or similar T cell but that has not been introduced with a provided epigenome-modifying DNA-targeting system or that or that does not contain the same epigenetic changes (e.g. methylation or histone modification) of the target gene or regulatory region thereof. [0596] In some embodiments, the epigenetic change comprises a change in at least one of: DNA accessibility, histone methylation, acetylation, phosphorylation, ubiquitylation, sumoylation, ribosylation, citrullination, and DNA methylation. In some embodiments, the epigenetic change is an altered DNA methylation of a target site in a target gene or a regulatory element thereof as described herein. In some embodiments, the epigenetic change is a histone modification of a target site in a target gene or a regulatory element thereof as described herein. [0597] Provided herein are methods of phenotypically and/or epigenetically modifying a lymphoid cell or a population of lymphoid cells. The methods provided herein include use of one or more DNA-targeting system provided herein (e.g. as described in Section I), or polynucleotide or vector for delivery of same (e.g. as described in Section II) to the lymphoid cell or compositions of any of the foregoing. In some embodiments, the DNA-targeting system (or polynucleotides or vectors for delivery of same to the lymphoid cell or compositions of any of the foregoing) is contacted with a lymphoid cell or a population of lymphoid cells. In some sf-6059407 22474-20028.40 embodiments, the contacting introduces the epigenome-modifying DNA-targeting system (or polynucleotides or vectors for delivery of same to the lymphoid cell or compositions of any of the foregoing) into the lymphoid cell, such as where it is able to translocate or localize to the nucleus of the lymphoid cell. In some embodiments, the methods increase the expression of one or more of the described target genes in lymphoid cells (e.g. T cells) in the population of cells. Also provided herein is a population of lymphoid cells containing a plurality of any of the provided modified lymphoid cells. [0598] Provided herein are methods of phenotypically and/or epigenetically modifying a T cell or a population of T cells. In some embodiments, such methods promote a phenotype, such as any described herein, for example by altering the differentiation fate of the T cell. In some embodiments, such methods increase or enrich the phenotype among a population of T cells. Also provided herein are methods of promoting a phenotype in a T cell or a population of T cells. The methods provided herein include use of one or more DNA-targeting system provided herein, or polynucleotide or vector for delivery of same to the T cell or compositions of any of the foregoing. In some embodiments, the DNA-targeting system (or polynucleotides or vectors for delivery of same to the T cell or compositions of any of the foregoing) is contacted with a T cell or a population of T cells. In some embodiments, the contacting introduces the DNA- targeting system (or polynucleotides or vectors for delivery of same to the T cell or compositions of any of the foregoing) into the T cell, such as where it is able to translocate or localize to the nucleus of the T cell. In some embodiments, the methods promote a phenotype in the T cell or one or more T cells in the population. In some embodiments, the methods increase the percentage of T cells with the phenotype in the population of T cells. [0599] In some embodiments, the epigenome-modifying DNA-targeting system (or polynucleotides or vectors for delivery of same to the T cell or compositions of any of the foregoing) can be introduced into a T cell or a population of T cells. In some embodiments, the epigenome-modifying DNA-targeting system (or polynucleotides or vectors for delivery of same to the T cell or compositions of any of the foregoing) can be cultured with a T cell or a population of T cells under conditions in which the epigenome-modifying DNA-targeting system (or polynucleotides or vectors for delivery of same to the T cell or compositions of any of the foregoing) will be introduced into or delivered to the T cell or one or more T cells in the population. [0600] In some embodiments, the methods can be carried out in vitro. In other sf-6059407 22474-20028.40 embodiments, the methods can be carried out ex vivo on T cells or a population containing T cells isolated from a subject. In other embodiments the epigenome-modifying DNA-targeting system (or polynucleotides or vectors for delivery of same to the T cell or compositions of any of the foregoing) can be administered to a subject, and then T cells can be isolated from the subject, such as for subsequent engineering. In still other embodiments the epigenome- modifying DNA-targeting system (or polynucleotides or vectors for delivery of same to the T cell or compositions of any of the foregoing) can be administered to a subject, and the T cells modified in vivo in the subject. [0601] In any of the provided methods, the T cells can be T cells for use as a T cell based immunotherapy, such as for ACT. In certain embodiments, the population of lymphocytes is derived from peripheral blood mononuclear cells (PBMCs) isolated from the circulation of a subject. In certain embodiments, the population of lymphocytes is derived from lymphocytes isolated from a tumor (tumor infiltrating lymphocytes) of an individual. In certain embodiments, the population of lymphocytes comprises T lymphocytes (T cells). These cell populations can be heterogeneous comprised of a variety of lymphocytes, or they can be further subject to isolation/purification using density centrifugation (e.g., Percoll), fluorescently activated cell sorting (FACS), leukapheresis, or antibody based selection methods (positive or negative). T cells can be generally marked by expression of CD3, and further subdivided into cytotoxic (CD8+) or helper (CD4+) populations. When isolated/purified the cell population can comprise CD3+ cells at least 80%, 90%, or 95% pure. In certain embodiments, the population comprises CD3+, CD4+ T cells at least 80%, 90%, or 95% pure. In certain embodiments, the population comprises CD3+, CD8+ T cells at least 80%, 90%, or 95% pure. [0602] In some embodiments, an isolated or purified cell population containing T cells can be further stimulated and, in some cases, expanded using standard methods, such as, incubation with anti-CD3 or CD28 antibody and/or co-culture with cytokines such as IL-2, IL-7 and/or IL- 15. For instance, a population of isolated cells containing T cells can be further expanded using standard methods such as incubation with anti-CD3 or CD28 antibody and/or co-culture with cytokines such as IL-2, IL-7 and/or IL-15. [0603] After the cells have been expanded the cells can comprise greater than 60%, 70%, 80%, 90%, or 95% CD3+ cells, CD3+CD4+ cells, or CD3+CD8+ cells. In certain embodiments, an aliquot of the cells can be tested for efficacy after expansion. [0604] There are numerous methods available for isolating or expanding T cells or T-cell sf-6059407 22474-20028.40 populations taken from an individual. Certain non-limiting methods of expanding and/or isolating T-cell populations are disclosed in U.S. Pat. Nos.5,827,642; 6,316,257; 6,399,054; 7,745,140; 8,383,099; US 2003/0134341; US 2004/0241162; all of which are incorporated by reference herein in their entireties. [0605] In some embodiments, the cells, such as T cells, may be further engineered with a recombinant antigen receptor, such as a chimeric antigen receptor (CAR) or an engineered TCR. In some embodiments, the cells may be stimulated (e.g. with anti-CD3 or CD28 antibody and/or IL-2, IL-7 and/or IL-15 cytokines) prior to engineering the cells, such as T cells, with the recombinant receptor. In some embodiments, the cells may be further expanded after engineering the cells, such as T cells, with the recombinant receptor. [0606] In some embodiments, the cells, such as T cells, are engineered with a CAR. In some embodiments, the CAR is a chimeric receptor that contains an extracellular antigen targeting domain (e.g., an antibody Fab or single chain variable fragment) fused to a transmembrane domain, and an intracellular signaling domain that induces activation of the cell, such as T cell, upon interaction of the CD3 zeta signaling domain and a costimulatory signaling domain. Non-limiting examples of a costimulatory signaling domain include a CD28 intracellular domain or a 4-1BB intracellular domain. In some embodiments, the extracellular targeting domain is specific for a tumor associated antigen (TAA). Non-limiting examples of TAAs include, for example, CD19, glioma-associated antigen, carcinoembryonic antigen (CEA), β-human chorionic gonadotropin, alphafetoprotein (AFP), lectin-reactive AFP, thyroglobulin, RAGE-1, MN-CA IX, human telomerase reverse transcriptase, RU1, RU2 (AS), intestinal carboxyl esterase, mut hsp70-2, M-CSF, prostase, prostate-specific antigen (PSA), PAP, NY-ESO-1, LAGE-1a, p53, prostein, PSMA, Her2/neu, survivin and telomerase, prostate- carcinoma tumor antigen-1 (PCTA-1), MAGE, ELF2M, neutrophil elastase, ephrinB2, CD22, insulin growth factor (IGF)-I, IGF-II, IGF-I receptor and mesothelin, MART-1, Lewis Y antigen (LeY), tyrosinase and GP 100, prostatic acid phosphatase (PAP) prostate-specific antigen (PSA), ROR1, MUC16, CD171 (LICAM), B-cell maturation antigen (BCMA), WT1, HER- 2/Neu/ErbB-2, CD19, CD20, or CD37. Current FDA approved CAR T cell therapies include CAR-T cell therapies for targeting CD19, such as for treating lymphoma, including axicabtagene ciloleucel (Yescarta™), tisagenlecleucel (Kymriah™) and lisocabtagene maraleucel (Breyanzi
®). Current FDA approved CAR T cell therapies also include CAR-T cell therapies for targeting BCMA, such as for treating multiple myeloma, including idecabtagene sf-6059407 22474-20028.40 vicleucel (Abecma
®) and ciltacabtagene autoleucel (Carvkti™). CAR constructs and methods of their use are described in, by way of non-limiting example, US20130287748A1; US 2014/0234348A1; or US 2014/0050708, all of which are incorporated by reference herein in their entirety. [0607] In some embodiments, the T cells are engineered with a TCR. In some embodiments, the TCR is specific for a TAA. In particular embodiments, the TCR is a recombinant TCR that is introduced into the T cell and is heterologous to the T cell. The TCR can be specific for a TAA, such as, glioma-associated antigen, carcinoembryonic antigen (CEA), β-human chorionic gonadotropin, alphafetoprotein (AFP), lectin-reactive AFP, thyroglobulin, RAGE-1, MN-CA IX, human telomerase reverse transcriptase, RU1, RU2 (AS), intestinal carboxyl esterase, mut hsp70-2, M-CSF, prostase, prostate-specific antigen (PSA), PAP, NY- ESO-1, LAGE-1a, p53, prostein, PSMA, Her2/neu, survivin and telomerase, prostate-carcinoma tumor antigen-1 (PCTA-1), MAGE, ELF2M, neutrophil elastase, ephrinB2, CD22, insulin growth factor (IGF)-I, IGF-II, IGF-I receptor and mesothelin, MART-1, Lewis Y antigen (LeY), tyrosinase and GP 100, prostatic acid phosphatase (PAP) prostate-specific antigen (PSA), ROR1, MUC16, CD171 (LICAM), B-cell maturation antigen (BCMA), WT1, HER- 2/Neu/ErbB-2, CD19, CD20, or CD37. In some embodiments, the TAA is cancer-testis (CT) antigen. In some embodiments, the TAA is neoantigen or oncoviral antigen. Exemplary target antigens of a TCR include, but are not limited to, a MAGE (e.g. MAGE-A1, MAGE-A2, MAGE-A3, MAGE-A4, MAGE-A6, MAGE-A10, and MAGE-A12, such as MAGE-A4, MAGE-A10, MAGE-A3/MAGE-A6), glycoprotein (gp100), melanoma antigen recognized by T cells (MART-1), Wilms tumor 1 (WT1), PRAME, NY-ESO-1, mesothelin, α-fetoprotein (AFP) or Human papillomavirus (HPV) E6 protein and HPV E7 protein. TCRs and method of their use in ACT are known and described in, by way of non-limiting example, Tsimberidou, AM., et al. J Hematol Oncol 14, 102 (2021). [0608] In some embodiments, the recombinant antigen receptor, such as a CAR or TCR, can be engineered into the cell, such as T cell, by viral transduction of a nucleic acid encoding ther recombinant antigen receptor into a primary T-cell population, using for example a retroviral, adenoviral, or AAV-vector; or transfection via a lipid-based reagent or electroporation. In some embodiments, the methods described herein involve engineering a population of lymphoid cells, such as a T-cell population, with the recombinant antigen receptor (e.g. CAR or TCR) before contacting the population with the epigenome-modifying DNA-targeting system (or sf-6059407 22474-20028.40 polynucleotides or vectors for delivery of same to the T cell or compositions of any of the foregoing). In certain embodiments, the methods involve engineering a population of lymphoid cells, such as a T-cell population, with the recombinant antigen receptor (e.g. CAR or TCR) after contacting the cells with the epigenome-modifying DNA-targeting system (or polynucleotides or vectors for delivery of same to the T cell or compositions of any of the foregoing). In some embodiments, when the engineered lymphocytes, such as T cells (e.g. CAR- T cells or eTCR-T cells), are generated from a primary lymphocyte population the cells are often autologous to the patient being treated. In some embodiments, a process for engineering T cells with a recombinant receptor (e.g. CAR or TCR) includes isolating the T cells from a subject, stimulating the T cells in culture using a conventional method such as CD3/CD28 antibodies prior to transduction with a viral vector encoding the recombinant antigen receptor (e.g. CAR or TCR) and, if necessary, expanding the cells to generate sufficient cells for subsequent administration to the subject. In some embodiments, contacting the T cells with the epigenome- modifying DNA-targeting system (or polynucleotides or vectors for delivery of same to the T cell or compositions of any of the foregoing) can be prior to or during any step of stimulating, transducing or expanding the T cells. [0609] In certain embodiments, an isolated or purified cell population containing T cells is incubated with peptide antigen and, in some cases also irradiated feeder cells or other agents, to expand one or more T cells of a certain antigen specificity. In certain embodiments, the peptide antigen comprises a tumor associated antigen. In some embodiments, such an isolated or purified cell population includes tumor infiltrating lymphocytes (TILs) such as for TIL therapy. In some embodiments, the population can be stimulated or activated by a specific tumor- associated antigen either before or after contact with epigenome-modifying DNA-targeting system (or polynucleotides or vectors for delivery of same to the T cell or compositions of any of the foregoing). A tumor associated antigen (TAA) is one that is exclusively expressed or highly expressed by a neoplastic cell compared to a normal cell of the same origin. Known tumor-associated antigens include, for example, glioma-associated antigen, carcinoembryonic antigen (CEA), β-human chorionic gonadotropin, alphafetoprotein (AFP), lectin-reactive AFP, thyroglobulin, RAGE-1, MN-CA IX, human telomerase reverse transcriptase, RU1, RU2 (AS), intestinal carboxyl esterase, mut hsp70-2, M-CSF, prostase, prostate-specific antigen (PSA), PAP, NY-ESO-1, LAGE-1a, p53, prostein, PSMA, Her2/neu, survivin and telomerase, prostate- carcinoma tumor antigen-1 (PCTA-1), MAGE, ELF2M, neutrophil elastase, ephrinB2, CD22, sf-6059407 22474-20028.40 insulin growth factor (IGF)-I, IGF-II, IGF-I receptor and mesothelin, MART-1, Lewis Y antigen (LeY), tyrosinase and GP 100, prostatic acid phosphatase (PAP) prostate-specific antigen (PSA), ROR1, MUC16, CD171 (LICAM), B-cell maturation antigen (BCMA), WT1, HER- 2/Neu/ErbB-2, CD19, CD20, CD37, or patient specific idiotype. In certain embodiments, greater than 50%, 60%, 70%, 80%, 90%, or 95% of the T-cell population can be specific for a tumor associated antigen (as defined by tetramer staining for example). In certain embodiments, the T- cell population may not be stimulated with TAA, but may possess specificity for the TAA, as indicated for example, by tetramer staining. [0610] In some embodiments, the population of cells, such as T cells, may be autologous to a subject to be treated. For instance, the population of lymphoid cells, such as T-cell populations, to be contacted with an epigenome-modifying DNA-targeting system (or polynucleotides or vectors for delivery of same to the T cell or compositions of any of the foregoing) can be derived from an individual that will ultimately be treated with the cell-based immunotherapeutic (e.g., an autologous population). In certain embodiments, when an autologous cell population is used the cell population has been contacted in vitro with the epigenome-modifying DNA-targeting system (or polynucleotides or vectors for delivery of same to the cells, such as T cell, or compositions of any of the foregoing). In certain embodiments, when an autologous cell population is used a subject to be treated has been administered the epigenome-modifying DNA-targeting system (or polynucleotides or vectors for delivery of same to the cell, such as T cell, or compositions of any of the foregoing) on one or more occasions prior to isolation of the cell population. [0611] In other embodiments, the population of lymphoid cells, such as population of T cells, may be for allogeneic therapy. In such an example, the population of lymphoid cells, such as T-cell population, to be contacted with an epigenome-modifying DNA-targeting system (or polynucleotides or vectors for delivery of same to the cells, such as T cell, or compositions of any of the foregoing) can be derived from a different individual (e.g., a heterologous population) than is to be treated. In certain embodiments, when a heterologous cell population is used it is from an HLA matched individual (e.g., syngeneic) or an HLA mismatched individual (e.g., allogeneic). In certain embodiments, when a heterologous cell population is used it is from an HLA mismatched donor. In certain embodiments, when a heterologous cell population is used it is a T cell line that can be established from an autologous or heterologous source. [0612] T cell populations can also be derived from hematopoietic stem cells (HSCs) or sf-6059407 22474-20028.40 induced pluripotent stem cells (iPSCs) using methods known in the art. In certain embodiments, T-cell populations are derived/differentiated from iPSCs. The source of the iPSCs can be either autologous or heterologous. In certain embodiments, T-cell populations are derived/differentiated from (HSCs) cells. The source of the HSCs can be either autologous or heterologous. [0613] In some embodiments, the modified T cell comprises an epigenetic or phenotypic modification resulting from being contacted by any of the DNA-targeting systems described herein (for example in Section I), including any including any gRNA described herein (for example in Section I.B.2). [0614] In some embodiments, the modified T cell is derived from a cell from a subject, such as a primary T cell, a T cell progenitor, a pluripotent stem cell, or an induced pluripotent stem cell. In some embodiments, the modified T cell is derived from a primary T cell. [0615] In some embodiments, the modified T cell is derived from a subject. In some embodiments, the subject has or is suspected of having cancer. [0616] In some aspects, provided herein are methods for modulating (e.g. increasing or decreasing transcription of) the expression of a gene in a cell (e.g. a T cell), such as a target gene as described in Section I.A. In some embodiments, the method includes: introducing into the cell any of the DNA-targeting systems described herein, or a polynucleotide or vector containing or encoding the same. [0617] In some embodiments, the expression of the one or more first genes, such as one or more target genes described in Section I.A.2, is increased in comparison to a comparable unmodified cell (e.g. T cell) not subjected to the method, i.e. not contacted or introduced with the DNA-targeting system described herein. In some embodiments, the expression of the one or more first genes is increased by at least about 1.2-fold, 1.25-fold, 1.3-fold, 1.4-fold, 1.5-fold, 1.6-fold, 1.7-fold, 1.75-fold, 1.8-fold, 1.9-fold, 2-fold, 2.5-fold, 3-fold, 4-fold, 5-fold, 10-fold, 20-fold, 30-fold, 40-fold, 50-fold, 100-fod, 200-fold, 300-fold, 400-fold, 500-fold, 1000-fold or greater. In some embodiments, the expression is stably increased or transiently increased. In some embodiments, the increased expression of the one or more first genes promotes increased T cell effector function in a T cell. [0618] In some embodiments, the expression of the one or more second genes, such as one or more target genes described in Section I.A.3, is decreased in comparison to a comparable unmodified cell (e.g. T cell) not subjected to the method, i.e. not contacted or introduced with sf-6059407 22474-20028.40 the DNA-targeting system described herein. In some embodiments, the expression of the one or more second genes is decreased by at least about 1.2-fold, 1.25-fold, 1.3-fold, 1.4-fold, 1.5-fold, 1.6-fold, 1.7-fold, 1.75-fold, 1.8-fold, 1.9-fold, 2-fold, 2.5-fold, 3-fold, 4-fold, 5-fold, 10-fold, 20-fold, 30-fold, 40-fold, 50-fold, 100-fod, 200-fold, 300-fold, 400-fold, 500-fold, 1000-fold or greater. In some embodiments, the expression is stably decreased or transiently decreased. In some embodiments, the decreased expression of the one or more second genes promotes increased T cell effector function in a T cell. [0619] In some embodiments, the one or more modifications in the epigenome of the modified lymphoid cells, such as a T cell, NK cell or NK T cell, or any cells that have been differentiated from stem cells into such lymphoid cells and/or have been differentiated from progenitor cells, such as common lymphoid progenitors (CLPs), is by targeting one or more genes as described herein with a provided epigenome-modifying DNA-targeting system to change the epigenome of the cell. In some embodiments, the one or more modifications in the epigenome of the modified T cell is by targeting target genes as described herein with a provided epigenome-modifying DNA-targeting system to change the epigenome of the T cell. In some embodiments, the modified cell, such as modified T cell, includes an epigenetic change in a first gene selected from the list consisting of: BATF, CD28, EOMES, IL-2, IL2RB, IRF4, LAT, LCP2, TBX21, and VAV1. In some embodiments, the modified cell, such as modified T cell, includes an epigenetic change in a second gene selected from the list consisting of: CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and RASA2. [0620] In some embodiments, the modified cell, such as modified T cell has increased expression of one or more first genes selected from the list consisting of: BATF, CD28, EOMES, IL-2, IL2RB, IRF4, LAT, LCP2, TBX21, and VAV1. In some embodiments, the expression of the first gene in the modified cell is increased 1.5-fold or more compared to the expression of the same first gene in a comparable unmodified T cell, such as increased by at or about or greater than 2-fold, 3-fold, 4-fold, 5-fold, 6-fold, 7-fold, 8-fold, 9-fold, 10-fold or more. [0621] In some embodiments, the modified T cell exhibits increased expression of one or more first genes to promote increased T cell effector function upon T cell stimulation, in comparison to a comparable unmodified T cell, such as a T cell not subjected to the method, i.e. not contacted or introduced with the DNA-targeting system described herein. In some embodiments, the modified T cell has increased expression of one more first genes selected from sf-6059407 22474-20028.40 the list consisting of: BATF, CD28, EOMES, IL-2, IL2RB, IRF4, LAT, LCP2, TBX21, and VAV1. In some embodiments, the expression of the first gene in the modified T cell is increased 1.5-fold or more compared to the expression of the same first gene in a comparable unmodified T cell, such as increased by at or about or greater than 2-fold, 3-fold, 4-fold, 5-fold, 6-fold, 7-fold, 8-fold, 9-fold, 10-fold or more. [0622] In some embodiments, the modified cell, such as modified T cell has decreased expression of one or more second genes selected from the list consisting of: CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and RASA2. In some embodiments, the expression of the second gene in the modified cell is decreased 1.5-fold or more compared to the expression of the same second gene in a comparable unmodified T cell, such as decreased by at or about or greater than 2-fold, 3-fold, 4-fold, 5-fold, 6-fold, 7-fold, 8-fold, 9-fold, 10-fold or more. [0623] In some embodiments, the modified T cell exhibits decreased expression of one or more second genes to promote increased T cell effector function upon T cell stimulation, in comparison to a comparable unmodified T cell, such as a T cell not subjected to the method, i.e. not contacted or introduced with the DNA-targeting system described herein. In some embodiments, the modified T cell has decreased expression of one more second genes selected from the list consisting of: CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and RASA2. In some embodiments, the expression of the second gene in the modified T cell is decreased 1.5-fold or more compared to the expression of the same second gene in a comparable unmodified T cell, such as decreased by at or about or greater than 2-fold, 3-fold, 4-fold, 5-fold, 6-fold, 7-fold, 8-fold, 9-fold, 10-fold or more. [0624] Also provided herein is a population of T cells containing a plurality of any of the provided modified T cells. In some embodiments, the population of T cells is enriched for cells that have a phenotype. In some embodiments, the phenotype is increased T cell effector function upon T cell stimulation. In some embodiments, the population of T cells contains greater than at or about 40%, greater than at or about 50%, greater than at or about 60%, greater than at or about 70%, greater than at or about 80% or greater than at or about 90% of cells that have the phenotype. In some embodiments, the population of T cells has an increased percentage of cells of the phenotype compared to a comparable population of unmodified cell (e.g. T cell) not subjected to the method, i.e. not contacted or introduced with the DNA-targeting system sf-6059407 22474-20028.40 described herein. In some embodiments, the population of T cells has an increased percentage of cells having the phenotype compared to the percentage of cells having the phenotype prior to contacting the population of T cells with the DNA-targeting system described herein. In some embodiments, the increased percentage is by at or about or greater than 1.5-fold, 2-food, 3-fold, 4-fold, 5-fold or more. In some aspects, the phenotype comprises expression of IL-2, IFN- gamma, and/or TNF-alpha. [0625] In some embodiments, the population of cells, such as population of T cells, contains at least at or about 40%, at least at or about 50%, at least at or about 60%, at least at or about 70%, at least at or about 80% or at least at or about 90% of cells that have an epigenetic change (e.g. methylation or histone modification) at or near a target site in a target gene. In some embodiments, the population of cells, such as population of T cells, has an increased percentage of cells (e.g. T cells) that have an epigenetic change at or near a target site in a target gene compared to a comparable population of unmodified cell (e.g. T cell) not subjected to the method, i.e. not contacted or introduced with the DNA-targeting system described herein. In some embodiments, the epigenetic change is a change, such as on average in cells in the population, of at least one of: DNA accessibility, histone methylation, acetylation, phosphorylation, ubiquitylation, sumoylation, ribosylation, citrullination, and DNA methylation, compared to a comparable population of unmodified cell (e.g. T cell) not subjected to the method, i.e. not contacted or introduced with the DNA-targeting system described herein. In some embodiments the population of cells is a population of T cells. In some embodiments, the population of T cells contains at least at or about 40%, at least at or about 50%, at least at or about 60%, at least at or about 70%, at least at or about 80% or at least at or about 90% of cells that have an epigenetic change (e.g. methylation or histone modification) at or near a target site in a target gene and exhibits increased T cell effector function upon stimulation. [0626] In some embodiments, provided herein is a population of cells that contains at least at or about 40%, at least at or about 50%, at least at or about 60%, at least at or about 70%, at least at or about 80% or at least at or about 90% of cells that have an epigenetic change (e.g. methylation or histone modification) at a target gene and that are IL-2+, or that are IL-2- and IFN-gamma+. In some embodiments, the population of cells is a population of T cells. [0627] In some embodiments, the modified T cell or a composition containing a plurality of modified T cells is capable of a stronger and/or more persistent immune response (e.g. an anti- tumor immune response in vivo), in comparison to a comparable unmodified T cell or sf-6059407 22474-20028.40 composition of unmodified T cells. In some embodiments, a subject having received administration of a composition of T cells containing provided modified T cells as a T cell therapy, e.g. CAR-T cell, is monitored for the presence, absence or level of T cells of the therapy in the subject, such as in a biological sample of the subject, e.g. in the blood of the subject. In some embodiments, the provided methods result in T cells of the adoptive T cell therapy with increased T cell effector function, including increased cytokine production, proliferation, killing of target cells, and/or persistence. In some embodiments, the T cell effector function of the adoptively transferred T cells, such as CAR-expressing T cells, in the subject is greater as compared to that which would be achieved by alternative methods, such as those involving administration of a T cell therapy but without having been treated or contacted with a provided DNA-targeting system. In some embodiments, the T cell effector function, such as any described herein, is increased at least or about at least 1.5-fold, 2-fold, 3-fold, 4-fold, 5-fold, 6- fold, 7-fold, 8-fold, 9-fold, 10-fold, 20-fold, 30-fold, 50-fold, 60-fold, 70-fold, 80-fold, 90-fold, 100-fold or more. [0628] In some embodiments, the degree or extent of persistence of administered cells can be detected or quantified after administration to a subject. For example, in some aspects, quantitative PCR (qPCR) is used to assess the quantity of cells expressing the recombinant receptor (e.g., CAR or recombinant TCR) or other surrogate marker expressed by T cells of the therapy in the blood or serum or organ or tissue (e.g., disease site) of the subject. In some aspects, persistence is quantified as copies of DNA or plasmid encoding the recombinant receptor (e.g., CAR or recombinant TCR) or surrogate marker per microgram of DNA or per microliter of the sample, e.g., of blood or serum, or per total number of peripheral blood mononuclear cells (PBMCs) or white blood cells or T cells per microliter of the sample. In some embodiments, flow cytometric assays using antibodies specific for the recombinant receptor or surrogate marker also can be performed to detect the adoptively transferred cells. Cell-based assays may also be used to detect the number or percentage of functional cells, such as cells capable of binding to and/or neutralizing and/or inducing responses, e.g., cytotoxic responses, against cells of the disease or condition or expressing the antigen recognized by the receptor. In any of such embodiments, the extent or level of expression of any marker (e.g. surrogate marker, CAR, recombinant TCR) known to be expressed by the adoptively transferred T cells but not endogenous T cells can be used to distinguish the administered cells from endogenous cells in a subject. sf-6059407 22474-20028.40 [0629] In some embodiments, the modified T cell or a composition containing a plurality of modified T cells, such a produced by any of the provided methods, exhibits a reduction in features associated with T cell exhaustion in comparison to a comparable unmodified T cell or composition of unmodified T cells. In some embodiments, the T cells, such as a composition containing a modified T cell or a composition of modified T cell provided herein, exhibits reduced exhaustion following long-term stimulation with antigen, either in vitro or in vivo. For example, an assay for assessing long-term stimulation with antigen may include a serial restimulation assay (see e.g. Jensen et al. Immunol. Rev.2014; 257:127-144; Win et al. Journal of Immunotherapy, 2020; 43:107-120). In some embodiments, the percentage of T cells that exhibit an exhausted phenotype is reduced 2-fold, 3-fold, 4-fold, 5-fold, 6-fold, 7-fold, 8-fold, 9- fold, 10-fold or more. [0630] Various assays are known and can be used to assess or determine if the T cells exhibit features of exhaustion or a reduction in features of exhaustion in comparison to a comparable unmodified T cell or composition of unmodified T cells. In some cases, exhaustion can be assessed by monitoring loss of T cell function, such as reduced or decreased antigen- specific or antigen receptor-driven activity, such as a reduced or decreased ability to produce cytokines or to drive cytolytic activity against target antigen. In some cases, exhaustion also can be assessed by monitoring expression of surface markers on T cells (e.g. CD4 and/or CD4 T cells) that are associated with an exhaustion phenotype. In some embodiments, the exhaustion marker is any one or more of PD-1, CTLA-4, TIM-3, LAG-3, BTLA, 2B4, CD160, CD39, VISTA, and TIGIT Among exhaustion markers are inhibitory receptors such as PD-1, CTLA- 4, LAG-3 and TIM-3. In certain embodiments, the biological activity of the cells is measured by assaying expression and/or secretion of one or more cytokines, such as CD107a, IFNγ, IL-2, GM-CSF and TNFα, and/or by assessing cytolytic activity. In some embodiments, assays for the activity, phenotypes, proliferation and/or function of the T cells include, but are not limited to, ELISPOT, ELISA, cellular proliferation, cytotoxic lymphocyte (CTL) assay, binding to the T cell epitope, antigen or ligand, or intracellular cytokine staining, proliferation assays, lymphokine secretion assays, direct cytotoxicity assays, and limiting dilution assays. In some embodiments, proliferative responses of the T cells can be measured, e.g. by incorporation of
3H-thymidine, BrdU (5-Bromo-2’-Deoxyuridine) or 2’-deoxy-5-ethynyluridine (EdU) into their DNA or dye dilution assays, using dyes such as carboxyfluorescein diacetate succinimidyl ester (CFSE), CellTrace Violet, or membrane dye PKH26. sf-6059407 22474-20028.40 [0631] Also provided herein are compositions containing a modified lymphoid cell or a plurality of or population of modified lymphoid cells provided herein, such as modified T cells, NK cell, NKT cell, or such cells that are modified and have been differentiated from stem cells into such lymphoid cells and/or have been differentiated from progenitor cells, such as common lymphoid progenitors (CLPs). Also provided herein are compositions containing a modified T cell or a plurality of or population of modified T cells provided herein. In some embodiments, the composition is a pharmaceutical composition and further contains a pharmaceutically acceptable carrier. Such compositions can be used in accord with the provided methods, and/or with the provided articles of manufacture or compositions, such as in the prevention or treatment of diseases, conditions, and disorders, or in detection, diagnostic, and prognostic methods. [0632] Among the compositions are pharmaceutical compositions and formulations for administration, such as for adoptive cell therapy. In some embodiments, the engineered cells are formulated with a pharmaceutically acceptable carrier. [0633] A pharmaceutically acceptable carrier can include all solvents, dispersion media, coatings, antibacterial and antifungal agents, isotonic and absorption delaying agents, and the like, compatible with pharmaceutical administration (Gennaro, 2000, Remington: The science and practice of pharmacy, Lippincott, Williams & Wilkins, Philadelphia, PA). Examples of such carriers or diluents include, but are not limited to, water, saline, Ringer's solutions, dextrose solution, and 5% human serum albumin. Liposomes and non-aqueous vehicles such as fixed oils may also be used. Supplementary active compounds can also be incorporated into the compositions. The pharmaceutical carrier should be one that is suitable for T cells, such as a saline solution, a dextrose solution or a solution comprising human serum albumin. [0634] In some embodiments, the pharmaceutically acceptable carrier or vehicle for such compositions is any non-toxic aqueous solution in which the cells, such as T cells, can be maintained, or remain viable, for a time sufficient to allow administration of live cells, such as live T cells. For example, the pharmaceutically acceptable carrier or vehicle can be a saline solution or buffered saline solution. The pharmaceutically acceptable carrier or vehicle can also include various bio materials that may increase the efficiency of the cells, such as T cells. Cell vehicles and carriers can, for example, include polysaccharides such as methylcellulose (M. C. Tate, D. A. Shear, S. W. Hoffman, D. G. Stein, M. C. LaPlaca, Biomaterials 22, 1113, 2001, which is incorporated herein by reference in its entirety), chitosan (Suh J K F, Matthew H W T. Biomaterials, 21, 2589, 2000; Lahiji A, Sohrabi A, Hungerford D S, et al., J Biomed Mater sf-6059407 22474-20028.40 Res, 51, 586, 2000, each of which is incorporated herein by reference in its entirety), N- isopropylacrylamide copolymer P(NIPAM-co-AA) (Y. H. Bae, B. Vernon, C. K. Han, S. W. Kim, J. Control. Release 53, 249, 1998; H. Gappa, M. Baudys, J. J. Koh, S. W. Kim, Y. H. Bae, Tissue Eng.7, 35, 2001, each of which is incorporated herein by reference in its entirety), as well as Poly(oxyethylene)/poly(D,L-lactic acid-co-glycolic acid) (B. Jeong, K. M. Lee, A. Gutowska, Y. H. An, Biomacromolecules 3, 865, 2002, which is incorporated herein by reference in its entirety), P(PF-co-EG) (Suggs L J, Mikos A G. Cell Trans, 8, 345, 1999, which is incorporated herein by reference in its entirety), PEO/PEG (Mann B K, Gobin A S, Tsai A T, Schmedlen R H, West J L., Biomaterials, 22, 3045, 2001; Bryant S J, Anseth K S. Biomaterials, 22, 619, 2001, each of which is incorporated herein by reference in its entirety), PVA (Chih-Ta Lee, Po-Han Kung and Yu-Der Lee, Carbohydrate Polymers, 61, 348, 2005, which is incorporated herein by reference in its entirety), collagen (Lee C R, Grodzinsky A J, Spector M., Biomaterials 22, 3145, 2001, which is incorporated herein by reference in its entirety), alginate (Bouhadir K H, Lee K Y, Alsberg E, Damm K L, Anderson K W, Mooney D J. Biotech Prog 17, 945, 2001; Smidsrd O, Skjak-Braek G., Trends Biotech, 8, 71, 1990, each of which is incorporated herein by reference in its entirety). [0635] In some embodiments, the cells, such as T cells, can be present in the composition in an effective amount. In some embodiments, the composition contains an effective amount of T cells, such as containing modified T cells produced by the provided methods. In some embodiments, the composition of T cells are enriched in T cells with increased T cell effector function. An effective amount of cells can vary depending on the patient, as well as the type, severity and extent of disease. Thus, a physician can determine what an effective amount is after considering the health of the subject, the extent and severity of disease, and other variables. [0636] In some embodiments, the composition, including pharmaceutical composition, is sterile. In some embodiments, isolation, enrichment, or culturing of the cells is carried out in a closed or sterile environment, for example and for instance in a sterile culture bag, to minimize error, user handling and/or contamination. In some embodiments, sterility may be readily accomplished, e.g., by filtration through sterile filtration membranes. In some embodiments, culturing is carried out using a gas permeable culture vessel. In some embodiments, culturing is carried out using a bioreactor. [0637] Also provided herein are compositions that are suitable for cryopreserving the provided lymphoid cells, such as modified cells including such lymphoid cells produced by any sf-6059407 22474-20028.40 of the provided methods. In some embodiments, the lymphoid cells are cryopreserved in a serum-free cryopreservation medium. [0638] Also provided herein are compositions that are suitable for cryopreserving the provided T cells, such as modified T cells including T cells produced by any of the provided methods. In some embodiments, the T cells are cryopreserved in a serum-free cryopreservation medium. [0639] In some embodiments, the composition comprises a cryoprotectant. In some embodiments, the cryoprotectant is or comprises DMSO and/or s glycerol. In some embodiments, the cryopreservation medium is between at or about 5% and at or about 10% DMSO (v/v). In some embodiments, the cryopreservation medium is at or about 5% DMSO (v/v). In some embodiments, the cryopreservation medium is at or about 6% DMSO (v/v). In some embodiments, the cryopreservation medium is at or about 7% DMSO (v/v). In some embodiments, the cryopreservation medium is at or about 8% DMSO (v/v). In some embodiments, the cryopreservation medium is at or about 9% DMSO (v/v). In some embodiments, the cryopreservation medium is at or about 10% DMSO (v/v). In some embodiments, the cryopreservation medium contains a commercially available cryopreservation solution (CryoStor™ CS10). CryoStor™ CS10 is a cryopreservation medium containing 10% dimethyl sulfoxide (DMSO).In some embodiments, compositions formulated for cryopreservation can be stored at low temperatures, such as ultra low temperatures, for example, storage with temperature ranges from -40 ºC to -150 ºC, such as or about 80 ºC ± 6.0 º C. [0640] In some embodiments, the cryopreserved cells, such as T cells, are prepared for administration by thawing. In some cases, the cells, such as T cells, can be administered to a subject immediately after thawing. In such an embodiment, the composition is ready-to-use without any further processing. In other cases, the cells, such as T cells, are further processed after thawing, such as by resuspension with a pharmaceutically acceptable carrier, incubation with an activating or stimulating agent, or are activated washed and resuspended in a pharmaceutically acceptable buffer prior to administration to a subject. V. METHODS OF TREATMENT [0641] Provided herein are methods of treatment, e.g., including administering any of the compositions described herein, such as DNA-targeting systems (e.g. as described in Section I), polynucleotides and vectors (e.g. as described in Section II), and pharmaceutical compositions and formulations (e.g. as described in Section III). In some aspects, also provided are methods sf-6059407 22474-20028.40 of administering any of the compositions described herein to a subject, such as a subject that has a disease or disorder. The compositions, such as pharmaceutical compositions, described herein are useful in a variety of therapeutic, diagnostic and prophylactic indications. For example, the compositions are useful in treating a variety of diseases and disorders in a subject. Such methods and uses include therapeutic methods and uses, for example, involving administration of the compositions, to a subject having a disease, condition, or disorder, such as a tumor or cancer. In some embodiments, the compositions are administered in an effective amount to effect treatment of the disease or disorder. Uses include uses of the compositions in such methods and treatments, and in the preparation of a medicament in order to carry out such therapeutic methods. In some embodiments, the methods are carried out by administering the compositions to the subject having or suspected of having the disease or condition. In some embodiments, the methods thereby treat the disease or condition or disorder in the subject. Also provided are therapeutic methods for administering the cells and compositions to subjects, e.g., patients. [0642] In some embodiments, the compositions include a DNA-targeting system provided herein (e.g. as described in Section I), or a polynucleotide or vector encoding the same (e.g. as described in Section II), in which delivery of the composition to a subject modulates one or more activities or function of lymphoid cells in a subject to thereby treat a disease or condition. For instance, in some embodiments, the subject has been previously treated with an adoptive cell therapy involving administration of a population of lymphoid cells (e.g. T cell, NK or NKT cell therapy, including primary cells or cells differentiated from stem cells or progenitor cells such as common lymphoid cells) for treating a disease or disorder, and administration of a provided DNA-targeting system, or a polynucleotide or vector encoding the same, modulates a phenotype or function of the adoptively transferred cells in the subject for treating the disease or condition. In some embodiments, the cells may include a T cell infiltrating lymphocyte (TIL) therapy. In som embodiments, the cells are engineered with an antigen receptor, such as a chimeric antigen receptor or T cell receptor, targeting an antigen associated with the disease or condition. In some embodiments, administration or use of a composition that includes a DNA-targeting system provided herein, or a polynucleotide or vector encoding the same, increases expression of one or more target genes as described herein in the lymphoid cell. [0643] In some embodiments, the compositions include a DNA-targeting system provided herein, or a polynucleotide or vector encoding the same, in which delivery of the composition to a subject modulates one or more activities or function of T cells in a subject to thereby treat a sf-6059407 22474-20028.40 disease or condition. For instance, in some embodiments, the subject has been previously treated with an adoptive T cell therapy for treating a disease or disorder, such as a TIL therapy or a CAR- or TCR-engineered T cell therapy, and administration of a provided DNA-targeting system, or a polynucleotide or vector encoding the same, modulates a phenotype or function of the adoptively transferred T cells in the subject for treating the disease or condition. In some embodiments, administration or use of a composition that includes a DNA-targeting system provided herein, or a polynucleotide or vector encoding the same, increases expression of one or more first genes that promotes a phenotype comprising increased T cell effector function upon stimulation in a T cell. In some embodiments, administration or use of a composition that includes a DNA-targeting system provided herein, or a polynucleotide or vector encoding the same, decreases expression of one or more second genes that repress a phenotype comprising increased T cell effector function upon stimulation in a T cell. In some embodiments, the percentage of T cells of the adoptive cell therapy in the subject that has the phenotype is increased in the subject compared to prior to the administration of the composition that includes the DNA-targeting system or a polynucleotide or vector encoding the same. [0644] In some aspects, also provided herein are methods of promoting a phenotype in a T cell or in T cells in a subject, according to any description of a phenotype comprising increased T cell effector function provided herein. In some embodiments, the percentage of T cells that have the phenotype is increased in the subject compared to prior to the administration of the composition containing the DNA-targeting system or a polynucleotide or vector encoding the same. In some embodiments, the T cells include T cells of a previously administered adoptive cell therapy, such as CAR-expressing or recombinant TCR-expressing T cells. [0645] In some embodiments, the methods of administering a composition containing the DNA-targeting system or a polynucleotide or vector encoding the same to a subject as provided herein are carried out in vivo (i.e. in a subject). [0646] In some embodiments, methods of contacting a cell (e.g. T cell) with a composition containing the DNA-targeting system or a polynucleotide or vector encoding the same provided herein are carried out ex vivo on a cell from a subject, for example a primary T cell, a T cell progenitor, a pluripotent stem cell, or an induced pluripotent stem cell, such as by methods described in Section IV. In some embodiments, the methods provided herein are carried out ex vivo on a primary T cell. In some embodiments, when the methods are carried out ex vivo, such as by methods described in Section IV, the provided methods of treatment include administering sf-6059407 22474-20028.40 a dose of the modified cells (e.g. T cells) to the subject for treating a disease or disorder. In some embodiments, the modified cells are modified T cells that have been epigenetically modified by the provided methods and enriched in T cells that have the phenotype comprising increased T cell effector function. [0647] In some embodiments, also provided herein are methods include administering to a subject a composition containing an epigenetically modified cells (e.g. epigenetically modified T cells) provided herein. In some embodiments, administration of an effective dose of epigenetically modified cells treats a disease or condition in the subject. In some embodiments, the dose of epigenetically modified cells (e.g. T cells) is for use in adoptive cell therapy. In some embodiments, the epigenetically modified cell is a tumor infiltrating lymphocyte (TIL) therapy. In some embodiments, the epigenetically modified cell is a T cell that has been engineered with a recombinant antigen receptor, such as a chimeric antigen receptor or a T cell receptor (TCR) in which targeting of the antigen by the recombinant receptor (e.g. CAR or TCR)-engineered T cell treats the disease or condition. [0648] In some aspects, provided is a method for treating a disease in a subject, comprising administering to the subject a cellular composition that comprises any of the modified T cells described herein. In some aspects, the modified cell (e.g. T cell) is one that has been obtained from or derived from a cell from a subject and modified by contacting the cells with a provided DNA-targeting system or a polynucleotide or vector encoding the same. In some aspects, the modified cell (e.g. T cell) is obtained from or derived from a cell from a subject, and administered to the same subject (i.e. autologous adoptive cell therapy). In some aspects, the modified cell (e.g. T cell) is obtained from or derived from a cell from a subject, and administered to a different subject (i.e. allogeneic adoptive cell therapy). [0649] In some embodiments, the methods of treatment or uses involve administration to a subject of an effective amount of a composition containing modified cells (e.g. T cells) provided herein. In some embodiments, the effective amount may include a dose of cells (e.g. T cells) of the composition from at or about 10
5 to at about 10
12, or from at or about 10
5 and at or about 10
8, or from at or about 10
6 and at or about 10
12, or from at or about 10
8 and at or about 10
11, or from at or about 10
9 and at or about 10
10 of such. In some embodiments, the provided compositions containing modified cells (e.g. T cells) provided herein can be administered to a subject by any convenient route including parenteral routes such as subcutaneous, intramuscular, intravenous, sf-6059407 22474-20028.40 and/or epidural routes of administration. In particular embodiments, the modified T cells are administered by intravenous infusion to the subject. [0650] In some embodiments, the methods of treatment or uses involve administration to a subject of an effective amount of a composition containing modified cells T cells provided herein, including any such composition that is enriched in T cells having a phenotype comprising increased T cell effector function as produced by the provided methods. In some embodiments, the effective amount may include a dose of T cells of the composition from at or about 10
5 to at about 10
12, or from at or about 10
5 and at or about 10
8, or from at or about 10
6 and at or about 10
12, or from at or about 10
8 and at or about 10
11, or from at or about 10
9 and at or about 10
10 of such. In some embodiments, the provided compositions containing modified T cells provided herein can be administered to a subject by any convenient route including parenteral routes such as subcutaneous, intramuscular, intravenous, and/or epidural routes of administration. In particular embodiments, the modified T cells are administered by intravenous infusion to the subject. [0651] In some embodiments, the provided methods can be used to treat any disease or disorder in which treatment is contemplated by the adoptive cell therapy. For instance, in the case of a CAR or a TCR, the disease or condition to be treated is any disease or condition that is associated with expression of an antigen that is recognized or targeted by the CAR- or TCR-cell therapy. In other embodiments, for the case of a TIL therapy the disease or condition is a tumor, and typically is a tumor present in the subject from which the TIL therapy was derived. Methods for adoptive T cell therapy are known, see e.g. for CAR-T cell therapy: US7446190, US7741465, WO2016109410, WO2012079000, WO2017015427, WO2017040930, WO2017149515, WO201716568; WO2017181119; ; for TCR-T cell therapy: US20160137715, US 20190321478; WO2015184228, WO2017158103; for TIL therapy: US2003194804, US20120244133, US20210220457, US20210189339, U.S. Pat. No.5,126,132, and U.S. Patent No.11,083,752. Any of such methods or other similar methods can be used in connection with the present disclosure. In some embodiments, the provided methods are performed ex vivo during the process of manufacturing or preparing the T cells for adoptive transfer to a subject, such as using methods described in Section IV, and then the modified T cells are administered to the subject for treating a disease or disorder. In other embodiments, the provided methods are performed by administering to the subject a composition containing the DNA-targeting system or a polynucleotide or vector encoding the same in combination with adoptive transfer of a T sf-6059407 22474-20028.40 cell therapy. In such methods, the composition containing the DNA-targeting system or a polynucleotide or vector encoding the same is administered prior to, simultaneously with or after administration of the adoptive T cell therapy. [0652] In some embodiments, the disease, condition, or disorder to be treated is cancer, viral infection, autoimmune disease, or graft-versus-host disease. In some embodiments, the subject to be treated has undergone or is expected to undergo organ transplantation. [0653] In some embodiments, the disease or condition to be treated is a cancer. In some embodiments, the cancer is a hematologic cancer. In some embodiments, the cancer is a B cell malignancy. In some embodiments, the cancer is a myeloma, a lymphoma or a leukemia. In some embodiments, the methods can be used to treat a non-Hodgkin lymphoma (NHL), an acute lymphoblastic leukemia (ALL), a chronic lymphocytic leukemia (CLL), a diffuse large B-cell lymphoma (DLBCL), acute myeloid leukemia (AML), or a myeloma, e.g., a multiple myeloma (MM). [0654] In some embodiments, the cancer is a solid tumor cancer. In some embodiments, the cancer is a bladder, lung, brain, melanoma (e.g. small-cell lung, melanoma), breast, cervical, ovarian, colorectal, pancreatic, endometrial, esophageal, kidney, liver, prostate, skin, thyroid, or uterine cancers. In some embodiments, the cancer is a pancreatic cancer, bladder cancer, colorectal cancer, breast cancer, prostate cancer, renal cancer, hepatocellular cancer, lung cancer, ovarian cancer, cervical cancer, pancreatic cancer, rectal cancer, thyroid cancer, uterine cancer, gastric cancer, esophageal cancer, head and neck cancer, melanoma, neuroendocrine cancers, CNS cancers, brain tumors, bone cancer, or soft tissue sarcoma. [0655] In some aspects, the provided methods can further include administering one or more lymphodepleting therapies, such as prior to or simultaneous with initiation of administration of the adoptive T cell therapy, such as a composition containing modified T cells provided herein. In some embodiments, the lymphodepleting therapy comprises administration of a phosphamide, such as cyclophosphamide. In some embodiments, the lymphodepleting therapy can include administration of fludarabine. [0656] In some aspects, preconditioning subjects with immunodepleting (e.g., lymphodepleting) therapies can improve the effects of adoptive cell therapy (ACT). In some embodiments, the lymphodepleting therapy includes combinations of cyclosporine and fludarabine. sf-6059407 22474-20028.40 [0657] Thus in some embodiments, the provided method further involves administering a lymphodepleting therapy to the subject. In some embodiments, the method involves administering the lymphodepleting therapy to the subject prior to the administration of the dose of cells. In some embodiments, the lymphodepleting therapy contains a chemotherapeutic agent such as fludarabine and/or cyclophosphamide. In some embodiments, the administration of the cells and/or the lymphodepleting therapy is carried out via outpatient delivery. [0658] In some embodiments, the methods include administering a preconditioning agent, such as a lymphodepleting or chemotherapeutic agent, such as cyclophosphamide, fludarabine, or combinations thereof, to a subject prior to the administration of the dose of cells. For example, the subject may be administered a preconditioning agent, such as a lymphodepleting or chemotherapeutic agent, such as cyclophosphamide, fludarabine, or combinations thereof, at least 2 days prior, such as at least 3, 4, 5, 6, or 7 days prior, to the first or subsequent dose. In some embodiments, the subject is administered a preconditioning agent, such as a lymphodepleting or chemotherapeutic agent, such as cyclophosphamide, fludarabine, or combinations thereof, no more than 7 days prior, such as no more than 6, 5, 4, 3, or 2 days prior, to the administration of the dose of cells. In some embodiments, the subject is administered a preconditioning agent, such as a lymphodepleting or chemotherapeutic agent, such as cyclophosphamide, fludarabine, or combinations thereof, no more than 14 days prior, such as no more than 13, 12, 11, 10, 9 or 8 days prior, to the administration of the dose of cells. [0659] In some embodiments, the subject is preconditioned with cyclophosphamide at a dose between or between about 20 mg/kg and 100 mg/kg, such as between or between about 40 mg/kg and 80 mg/kg. In some aspects, the subject is preconditioned with or with about 60 mg/kg of cyclophosphamide. In some embodiments, the fludarabine can be administered in a single dose or can be administered in a plurality of doses, such as given daily, every other day or every three days. In some embodiments, the cyclophosphamide is administered once daily for one or two days. [0660] In some embodiments, where the lymphodepleting agent comprises fludarabine, the subject is administered fludarabine at a dose between or between about 1 mg/m
2 and 100 mg/m
2, such as between or between about 10 mg/m
2 and 75 mg/m
2, 15 mg/m
2 and 50 mg/m
2, 20 mg/m
2 and 30 mg/m
2, or 24 mg/m
2 and 26 mg/m
2. In some instances, the subject is administered 25 mg/m
2 of fludarabine. In some embodiments, the fludarabine can be administered in a single dose or can be administered in a plurality of doses, such as given daily, every other day or every sf-6059407 22474-20028.40 three days. In some embodiments, fludarabine is administered daily, such as for 1-5 days, for example, for 3 to 5 days. [0661] In some embodiments, the lymphodepleting agent comprises a combination of agents, such as a combination of cyclophosphamide and fludarabine. Thus, the combination of agents may include cyclophosphamide at any dose or administration schedule, such as those described above, and fludarabine at any dose or administration schedule, such as those described above. For example, in some aspects, the subject is administered 60 mg/kg (~2 g/m
2) of cyclophosphamide and 3 to 5 doses of 25 mg/m
2 fludarabine prior to the dose of cells. [0662] In some embodiments, prior to the administration of adoptive T cell therapy, such as a composition containing modified T cells described herein, the subject has received a lymphodepleting therapy. In some embodiments, the lymphodepleting therapy includes fludarabine and/or cyclophosphamide. In some embodiments, the lymphodepleting includes the administration of fludarabine at or about 20-40 mg/m
2 body surface area of the subject, optionally at or about 30 mg/m
2, daily, for 2-4 days, and/or cyclophosphamide at or about 200- 400 mg/m
2 body surface area of the subject, optionally at or about 300 mg/m
2, daily, for 2-4 days. [0663] In some embodiments, the lymphodepleting therapy includes fludarabine and cyclophosphamide. In some embodiments, the lymphodepleting therapy includes the administration of fludarabine at or about 30 mg/m
2 body surface area of the subject, daily, and cyclophosphamide at or about 300 mg/m
2 body surface area of the subject, daily, each for 2-4 days, optionally 3 days. [0664] In some embodiments, the administration of the preconditioning agent prior to infusion of the dose of cells improves an outcome of the treatment. For example, in some aspects, preconditioning, such as a lymphodepleting or chemotherapeutic agent, such as cyclophosphamide, fludarabine, or combinations thereof, improves the efficacy of treatment with the dose or increases the persistence of the T cells in the subject. In some embodiments, preconditioning treatment increases disease-free survival, such as the percent of subjects that are alive and exhibit no minimal residual or molecularly detectable disease after a given period of time following the dose of cells. In some embodiments, the time to median disease-free survival is increased. [0665] Once the cells are administered to the subject (e.g., human), the biological activity of the engineered cell populations in some aspects is measured by any of a number of known sf-6059407 22474-20028.40 methods. Parameters to assess include specific binding of an engineered or natural T cell or other immune cell to antigen, in vivo, e.g., by imaging, or ex vivo, e.g., by ELISA or flow cytometry. In certain embodiments, the ability of the T cells to destroy target cells can be measured using any suitable method known in the art, such as cytotoxicity assays described in, for example, Kochenderfer et al., J. Immunotherapy, 32(7): 689-702 (2009) , and Herman et al. J. Immunological Methods, 285(1): 25-40 (2004). In certain embodiments, the biological activity of the cells also can be measured by assaying expression and/or secretion of certain cytokines or other effector molecules, such as IFNγ and TNF. [0666] In some aspects the biological activity is measured by assessing clinical outcome, such as reduction in tumor burden or load. In some aspects, toxic outcomes, persistence and/or expansion of the cells, and/or presence or absence of a host immune response, are assessed. In some embodiments, exemplary parameters for determination include particular clinical outcomes indicative of amelioration or improvement in the disease or condition, e.g., tumor. Such parameters include: duration of disease control, including complete response (CR), partial response (PR) or stable disease (SD) (see, e.g., Response Evaluation Criteria In Solid Tumors (RECIST) guidelines), objective response rate (ORR), progression-free survival (PFS) and overall survival (OS). Specific thresholds for the parameters can be set to determine the efficacy of the method of combination therapy provided herein. VI. KITS AND ARTICLES OF MANUFACTURE [0667] Also provided are articles of manufacture, systems, apparatuses, and kits useful in performing the provided embodiments. In some embodiments, the provided articles of manufacture or kits contain any of the DNA-targeting systems described herein, any of the gRNAs described herein, any of the fusion proteins described herein, any of the polynucleotides described herein, any of the pluralities of polynucleotides described herein, any of the vectors described herein, any of the pluralities of vectors described herein, any of the cells (e.g., modified T cells) described herein, or a portion or a component of any of the foregoing, or any combination thereof. In some embodiments, the articles of manufacture or kits include polypeptides, polynucleotides, nucleic acids, vectors, and/or cells useful in performing the provided methods. [0668] In some embodiments, the articles of manufacture or kits include one or more containers, typically a plurality of containers, packaging material, and a label or package insert sf-6059407 22474-20028.40 on or associated with the container or containers and/or packaging, generally including instructions for use, e.g., instructions for introducing or administering. [0669] Also provided are articles of manufacture, systems, apparatuses, and kits useful in administering the provided compositions, e.g., pharmaceutical compositions, e.g., for use in therapy or treatment. In some embodiments, the articles of manufacture or kits provided herein contain vectors and/or plurality of vectors, such as any vectors and/or plurality of vectors described herein. In some aspects, the articles of manufacture or kits provided herein can be used for administration of the vectors and/or plurality of vectors, and can include instructions for use. [0670] The articles of manufacture and/or kits containing cells or cell compositions for therapy, may include a container and a label or package insert on or associated with the container. Suitable containers include, for example, bottles, vials, syringes, IV solution bags, etc. The containers may be formed from a variety of materials, such as glass or plastic. The container in some embodiments holds a composition which is by itself or combined with another composition effective for treating, preventing, and/or diagnosing the condition. In some embodiments, the container has a sterile access port. Exemplary containers include an intravenous solution bags, vials, including those with stoppers pierceable by a needle for injection, or bottles or vials for orally administered agents. The label or package insert may indicate that the composition is used for treating a disease or condition. The article of manufacture may further include a package insert indicating that the compositions can be used to treat a particular condition. Alternatively, or additionally, the article of manufacture may further include another or the same container comprising a pharmaceutically acceptable buffer. It may further include other materials such as other buffers, diluents, filters, needles, and/or syringes. VII. DEFINITIONS [0671] Unless defined otherwise, all terms of art, notations and other technical and scientific terms or terminology used herein are intended to have the same meaning as is commonly understood by one of ordinary skill in the art to which the claimed subject matter pertains. In some cases, terms with commonly understood meanings are defined herein for clarity and/or for ready reference, and the inclusion of such definitions herein should not necessarily be construed to represent a substantial difference over what is generally understood in the art. sf-6059407 22474-20028.40 [0672] As used herein, the singular forms “a,” “an,” and “the” include plural referents unless the context clearly dictates otherwise. For example, “a” or “an” means “at least one” or “one or more.” It is understood that aspects and variations described herein include “consisting” and/or “consisting essentially of” aspects and variations. [0673] Throughout this disclosure, various aspects of the claimed subject matter are presented in a range format. It should be understood that the description in range format is merely for convenience and brevity and should not be construed as an inflexible limitation on the scope of the claimed subject matter. Accordingly, the description of a range should be considered to have specifically disclosed all the possible sub-ranges as well as individual numerical values within that range. For example, where a range of values is provided, it is understood that each intervening value, between the upper and lower limit of that range and any other stated or intervening value in that stated range is encompassed within the claimed subject matter. The upper and lower limits of these smaller ranges may independently be included in the smaller ranges, and are also encompassed within the claimed subject matter, subject to any specifically excluded limit in the stated range. Where the stated range includes one or both of the limits, ranges excluding either or both of those included limits are also included in the claimed subject matter. This applies regardless of the breadth of the range. [0674] The term “about” as used herein refers to the usual error range for the respective value readily known. Reference to “about” a value or parameter herein includes (and describes) embodiments that are directed to that value or parameter per se. For example, description referring to “about X” includes description of “X”. In some embodiments, “about” may refer to ±25%, ±20%, ±15%, ±10%, ±5%, or ±1%. [0675] As used herein, recitation that nucleotides or amino acid positions “correspond to” nucleotides or amino acid positions in a disclosed sequence, such as set forth in the Sequence listing, refers to nucleotides or amino acid positions identified upon alignment with the disclosed sequence to maximize identity using a standard alignment algorithm, such as the GAP algorithm. By aligning the sequences, corresponding residues can be identified, for example, using conserved and identical amino acid residues as guides. In general, to identify corresponding positions, the sequences of amino acids are aligned so that the highest order match is obtained (see, e.g. : Computational Molecular Biology, Lesk, A.M., ed., Oxford University Press, New York, 1988; Biocomputing: Informatics and Genome Projects, Smith, D.W., ed., Academic Press, New York, 1993; Computer Analysis of Sequence Data, Part I, sf-6059407 22474-20028.40 Griffin, A.M., and Griffin, H.G., eds., Humana Press, New Jersey, 1994; Sequence Analysis in Molecular Biology, von Heinje, G., Academic Press, 1987; and Sequence Analysis Primer, Gribskov, M. and Devereux, J., eds., M Stockton Press, New York, 1991; Carrillo et al. (1988) SIAM J Applied Math 48: 1073). [0676] A “gene,” includes a DNA region encoding a gene product. Thus, the gene typically refers to coding and/or transcribed sequences. The sequence of a gene is typically present at a fixed chromosomal position or locus on a chromosome in the cell. [0677] A “regulatory element” or “DNA regulatory element,” which terms are used interchangeably herein, in reference to a gene refers to DNA regions which regulate the production of a gene product, whether or not such regulatory sequences are adjacent to coding and/or transcribed sequences. Accordingly, a regulatory element includes, but is not necessarily limited to, promoter sequences, terminators, translational regulatory sequences such as ribosome binding sites and internal ribosome entry sites, enhancers, silencers, insulators, boundary elements, replication origins, matrix attachment sites and locus control regions. [0678] As used herein, a “target site” or “target nucleic acid sequence” is a nucleic acid sequence that defines a portion of a nucleic acid to which a binding molecule (e.g. a DNA- binding domain disclosed herein) will bind, provided sufficient conditions for binding exist. [0679] The term “expression” with reference to a gene or “gene expression” refers to the conversion of the information, contained in a gene, into a gene product. A gene product can be the direct transcriptional product of a gene (e.g., mRNA, tRNA, rRNA, antisense RNA, ribozyme, structural RNA or any other type of RNA) or can be a protein produced by translation of an mRNA. For instance, expression includes the transcription and/or translation of a particular nucleotide sequence drive by its promoter. Gene products also include RNAs which are modified, by processes such as capping, polyadenylation, methylation, and editing, and proteins modified by, for example, methylation, acetylation, phosphorylation, ubiquitination, ADP-ribosylation, myristoylation, and glycosylation. Hence, reference to expression or gene expression includes protein (or polypeptide) expression or expression of a transcribable product of or a gene such as mRNA. The protein expression may include intracellular expression or surface expression of a protein. Typically, expression of a gene product, such as mRNA or protein, is at a level that is detectable in the cell. [0680] As used herein, a “detectable” expression level, means a level that is detectable by standard techniques known to a skilled artisan, and include for example, differential display, RT sf-6059407 22474-20028.40 (reverse transcriptase)-coupled polymerase chain reaction (PCR), Northern Blot, and/or RNase protection analyses as well as immunoaffinity-based methods for protein detection, such as flow cytometry, ELISA, or western blot. The degree of expression levels need only be large enough to be visualized or measured via standard characterization techniques. [0681] As used herein, the term “increased expression”, “enhanced expression” or “overexpression” means any form of expression that is additional to the expression in an original or source cell that does not contain the modification for modulating a particular gene expression by a DNA-targeting system, for instance a wild-type expression level (which can be absence of expression or immeasurable expression as well). Reference herein to “increased expression,” “enhanced expression” or “overexpression” is taken to mean an increase in gene expression relative to the level in a cell that does not contain the modification, such as the original source cell prior to contacting with, or engineering to introduce, the Dna-targeting system into the T cell, such as an unmodified cell or a wild-type T cell. The increase in expression can be at least 5%, 10%, 20%, 30%, 40% or 50%, 60%, 70%, 80%, 85%, 90%, or 100% or even more. In some cases, the increase in expression can be at least 2-fold, 5-fold, 10-fold, 20-fold, 30-fold, 40-fold, 50-fold, 60-fold, 70-fold, 80-fold, 90-fold, 100-fold, 200-fold, 300-fold, 400-food, 500- fold, 1000-fold or more. [0682] As used herein, the term “increased transcription” refers to the level of transcription of a gene that is additional to the transcription of the gene in an original or source cell that does not contain the modification for modulating transcription by a DNA-targeting system, for instance a wild-type transcription level of a gene. Reference to increased transcription can refer to an increase in the levels of a transcribable product of a gene such as mRNA. Any of a variety of methods can be used to monitor or quantitate a level of a transcribable product such as mRNA, including but not limited to, real-time quantitative RT (reverse transcriptase)- polymerase chain reaction (qRT-PCR), Northern Blot, microarray analysis, or RNA sequencing (RNA-Seq). The increase in transcription can be at least 5%, 10%, 20%, 30%, 40% or 50%, 60%, 70%, 80%, 85%, 90%, or 100% or even more. In some cases, the increase in transcription can be at least 2-fold, 5-fold, 10-fold, 20-fold, 30-fold, 40-fold, 50-fold, 60-fold, 70-fold, 80- fold, 90-fold, 100-fold, 200-fold or more. [0683] As used herein, an “epigenetic modification” refers to changes in the gene expression that are non-genetic modifications, i.e. not caused by changes in the DNA sequences, but are due to epigenetic changes such as events like DNA methylations or histone modifications. An sf-6059407 22474-20028.40 epigenetic modification may result in a heritable change in gene activity and expression that occur without alteration in DNA sequence. For instance, epigenetic modifications include non- genetic modifications such as chemical modifications to the cytosine residues of DNA (DNA methylation) and histone proteins associated with DNA (histone modifications). [0684] As used herein, the term “modification” or “modified” with reference to a T cell refers to any change or alteration in a cell that impacts gene expression in the cell. In some embodiments, the modification is an epigenetic modification that directly changes the epigenetic state of a gene or regulatory elements thereof to alter (e.g. increase) expression of a gene product. In some embodiments, a modification described herein results in increased expression of a target gene or selected polynucleotide sequence. [0685] As used herein, a “fusion” molecule is a molecule in which two or more subunit molecules are linked, such as covalently. Examples of a fusion molecule include, but are not limited to, fusion proteins (for example, a fusion between a DNA-binding domain such as a ZFP, TALE DNA-binding domain or CRISPR-Cas protein and one or more effector domains, such as a transactivation domain). The fusion molecule also may be part of a system in which a polynucleotide component associates with a polypeptide component to form a functional system (e.g., a CRISPR/Cas system in which a single guide RNA associates with a functional domain to modulate gene expression). Fusion molecules also include fusion nucleic acids, for example, a nucleic acid encoding the fusion protein. Expression of a fusion protein in a cell can result from delivery of the fusion protein to the cell or by delivery of a polynucleotide encoding the fusion protein to a cell, where the polynucleotide is transcribed, and the transcript is translated, to generate the fusion protein. [0686] The term “vector,” as used herein, refers to a nucleic acid molecule capable of propagating another nucleic acid to which it is linked. The term includes the vector as a self- replicating nucleic acid structure as well as the vector incorporated into the genome of a host cell into which it has been introduced. Certain vectors are capable of directing the expression of nucleic acids to which they are operatively linked. Such vectors are referred to herein as “expression vectors.” Among the vectors are viral vectors, such as adenoviral vectors or lentiviral vectors. [0687] The term “expression vector" refers to a vector comprising a recombinant polynucleotide comprising expression control sequences operatively linked to a nucleotide sequence to be expressed. An expression vector comprises sufficient cis-acting elements for sf-6059407 22474-20028.40 expression; other elements for expression can be supplied by the host cell or in an in vitro expression system. Expression vectors include, but are not limited to, cosmids, plasmids (e.g., naked or contained in liposomes) and viruses (e.g., lentiviruses, retroviruses, adenoviruses, and adeno-associated viruses) that incorporate the recombinant polynucleotide. [0688] The term “isolated” means altered or removed from the natural state. For example, a nucleic acid or a peptide naturally present in a living animal is not "isolated," but the same nucleic acid or peptide partially or completely separated from the coexisting materials of its natural state is “isolated.” An isolated nucleic acid or protein can exist in substantially purified form, or can exist in a non-native environment such as, for example, a host cell. [0689] The term "polynucleotide" refers to a chain of nucleotides. Furthermore, nucleic acids are polymers of nucleotides. Thus, nucleic acids and polynucleotides as used herein are interchangeable. One skilled in the art has the general knowledge that nucleic acids are polynucleotides, which can be hydrolyzed into the monomelic "nucleotides." The monomelic nucleotides can be hydrolyzed into nucleosides. As used herein polynucleotides include, but are not limited to, all nucleic acid sequences which are obtained by any means available in the art, including, without limitation, recombinant means, i.e., the cloning of nucleic acid sequences from a recombinant library or a cell genome, using ordinary cloning technology and PCR™, and the like, and by synthetic means. [0690] As used herein, the terms “peptide,” “polypeptide,” and “protein” are used interchangeably, and refer to a compound comprised of amino acid residues covalently linked by peptide bonds. A protein or peptide must contain at least two amino acids, and no limitation is placed on the maximum number of amino acids that can comprise a protein's or peptide's sequence. Polypeptides include any peptide or protein comprising two or more amino acids joined to each other by peptide bonds. As used herein, the term refers to both short chains, which also commonly are referred to in the art as peptides, oligopeptides and oligomers, for example, and to longer chains, which generally are referred to in the art as proteins, of which there are many types. “Polypeptides” include, for example, biologically active fragments, substantially homologous polypeptides, oligopeptides, homodimers, heterodimers, variants of polypeptides, modified polypeptides, derivatives, analogs, fusion proteins, among others. The polypeptides include natural peptides, recombinant peptides, synthetic peptides, or a combination thereof. [0691] As used herein, “percent (%) amino acid sequence identity” and “percent identity” sf-6059407 22474-20028.40 when used with respect to an amino acid sequence (reference polypeptide sequence) is defined as the percentage of amino acid residues in a candidate sequence (e.g., the subject antibody or fragment) that are identical with the amino acid residues in the reference polypeptide sequence, after aligning the sequences and introducing gaps, if necessary, to achieve the maximum percent sequence identity, and not considering any conservative substitutions as part of the sequence identity. Alignment for purposes of determining percent amino acid sequence identity can be achieved in various known ways, in some embodiments, using publicly available computer software such as BLAST, BLAST-2, ALIGN or Megalign (DNASTAR) software. Appropriate parameters for aligning sequences can be determined, including any algorithms needed to achieve maximal alignment over the full length of the sequences being compared. [0692] In some embodiments, “operably linked” may include the association of components, such as a DNA sequence, (e.g. a heterologous nucleic acid) and a regulatory sequence(s), in such a way as to permit gene expression when the appropriate molecules (e.g. transcriptional activator proteins) are bound to the regulatory sequence. Hence, it means that the components described are in a relationship permitting them to function in their intended manner. [0693] An amino acid substitution may include replacement of one amino acid in a polypeptide with another amino acid. The substitution may be a conservative amino acid substitution or a non-conservative amino acid substitution. Amino acid substitutions may be introduced into a binding molecule, e.g., antibody, of interest and the products screened for a desired activity, e.g., retained/improved antigen binding, decreased immunogenicity, or improved ADCC or CDC. [0694] Amino acids generally can be grouped according to the following common side- chain properties: (1) hydrophobic: Norleucine, Met, Ala, Val, Leu, Ile; (2) neutral hydrophilic: Cys, Ser, Thr, Asn, Gln; (3) acidic: Asp, Glu; (4) basic: His, Lys, Arg; (5) residues that influence chain orientation: Gly, Pro; (6) aromatic: Trp, Tyr, Phe. [0695] In some embodiments, conservative substitutions can involve the exchange of a member of one of these classes for another member of the same class. In some embodiments, non-conservative amino acid substitutions can involve exchanging a member of one of these sf-6059407 22474-20028.40 classes for another class. [0696] As used herein, a composition refers to any mixture of two or more products, substances, or compounds, including cells. It may be a solution, a suspension, liquid, powder, a paste, aqueous, non-aqueous or any combination thereof. [0697] As used herein, a “subject” or an “individual,” which are terms that are used interchangeably, is a mammal. In some embodiments, a “mammal” includes humans, non- human primates, domestic and farm animals, and zoo, sports, or pet animals, such as dogs, horses, rabbits, cattle, pigs, hamsters, gerbils, mice, ferrets, rats, cats, monkeys, etc. In some embodiments, the subject or individual is human. In some embodiments, the subject is a patient that is known or suspected of having a disease, disorder or condition. [0698] As used herein, the term “treating” and “treatment” includes administering to a subject an effective amount of a biological molecule, such as a therapeutic agent, so that the subject has a reduction in at least one symptom of the disease or an improvement in the disease, for example, beneficial or desired clinical results. For instance, a biological molecule may include cells (e.g. T cells), such as cells that have been modified by a DNA-targeting system or polynucleotide(s) encoding the DNA-targeting system described herein. For purposes of this technology, beneficial or desired clinical results include, but are not limited to, alleviation of one or more symptoms, diminishment of extent of disease, stabilized (i.e., not worsening) state of disease, delay or slowing of disease progression, amelioration or palliation of the disease state, and remission (whether partial or total), whether detectable or undetectable. Treating can refer to prolonging survival as compared to expected survival if not receiving treatment. Thus, one of skill in the art realizes that a treatment may improve the disease condition, but may not be a complete cure for the disease. In some embodiments, one or more symptoms of a disease or disorder are alleviated by at least 5%, at least 10%, at least 20%, at least 30%, at least 40%, or at least 50% upon treatment of the disease. [0699] For purposes of this technology, beneficial or desired clinical results of disease treatment include, but are not limited to, alleviation of one or more symptoms, diminishment of extent of disease, stabilized (i.e., not worsening) state of disease, delay or slowing of disease progression, amelioration or palliation of the disease state, and remission (whether partial or total), whether detectable or undetectable. [0700] The term “therapeutically effective amount” refers to the amount of the subject compound that will elicit the biological or medical response of a tissue, system, or subject that is sf-6059407 22474-20028.40 being sought by the researcher, veterinarian, medical doctor or other clinician. The term "therapeutically effective amount" includes that amount of a biological molecule, such as a compound or cells, that, when administered, is sufficient to prevent development of, or alleviate to some extent, one or more of the signs or symptoms of the disorder or disease being treated. The therapeutically effective amount will vary depending on the biological molecule, the disease and its severity and the age, weight, etc., of the subject to be treated. [0701] As used herein, “adoptive cell therapy” (ACT) refers to the administration of T cells targeting a specific antigen to a subject. [0702] As used herein, the term "autologous" is meant to refer to any material derived from the same individual to which it is later to be re-introduced into the individual. [0703] "Allogeneic" refers to a graft derived from a different animal of the same species. VIII. EXEMPLARY EMBODIMENTS [0704] Among the provided embodiments are: 1. An epigenetic-modifying DNA-targeting system comprising a plurality of DNA- targeting modules comprising at least one activator DNA-targeting module and at least one repressor DNA-targeting module, wherein: the at least one activator DNA-targeting module is for increasing transcription of one or more activation genes in a T cell, and each activator DNA-targeting module comprises a fusion protein comprising (i) a first DNA-binding domain for targeting to a target site of one of the one or more activation genes and (ii) at least one transcriptional activator domain; and the at least one repressor DNA-targeting module is for repressing transcription of one or more repression genes in the T cell, and each repressor DNA-targeting module comprises a fusion protein comprising (i) a second DNA-binding domain for targeting to a target site of one of the one or more repression genes and (ii) at least one transcriptional repressor domain; wherein the first DNA-binding domain or domains are all different from the second DNA-binding domain or domains. 2. The epigenetic-modifying DNA-targeting system of embodiment 1, wherein the one or more activation genes are selected from the group consisting of BATF, CD28, EOMES, IL-2, IL-2RB, IRF4, LAT, LCP2, TBX21, and VAV1. 3. The epigenetic-modifying DNA-targeting system of embodiment 1 or embodiment 2, wherein the one or more repression genes are selected from the group consisting sf-6059407 22474-20028.40 of CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and RASA2. 4. An epigenetic-modifying DNA-targeting system comprising a plurality of DNA- targeting modules comprising at least one activator DNA-targeting module and at least one repressor DNA-targeting module, wherein: the at least one activator DNA-targeting module is for increasing transcription of one or more activation genes, and each activator DNA-targeting module comprises a fusion protein comprising (i) a first DNA-binding domain for targeting to a target site of one of the one or more activation genes and (ii) at least one transcriptional activator domain, wherein the one or more activation genes are selected from the group consisting of BATF, CD28, EOMES, IL-2, IL-2RB, IRF4, LAT, LCP2, TBX21, and VAV1; and the at least one repressor DNA-targeting module is for repressing transcription of one or more repression genes, and each repressor DNA-targeting module comprises a fusion protein comprising (i) a second DNA-binding domain for targeting to a target site of one of the one or more repression genes and (ii) at least one transcriptional repressor domain, wherein the one or more repression genes are selected from the group consisting of CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and RASA2; wherein the first DNA-binding domain or domains are all different from the second DNA-binding domain or domains. 5. The epigenetic-modifying DNA-targeting system of any of embodiments 1-4, wherein the epigenetic-modifying DNA-targeting system does not introduce a genetic disruption or a DNA break. 6. The epigenetic-modifying DNA-targeting system of any of embodiments 1-5, wherein each first DNA-binding domain is a Clustered Regularly Interspaced Short Palindromic Repeats associated (Cas) protein; a zinc finger protein (ZFP); a transcription activator-like effector (TALE); a meganuclease; a homing endonuclease; or an I-SceI enzyme. 7. The epigenetic-modifying DNA-targeting system of any of embodiments 1-6, wherein each first DNA-binding domain is catalytically inactive. 8. The epigenetic-modifying DNA-targeting system of any of embodiments 1-7, wherein each second DNA-binding domain is a Cas protein; a ZFP; a TALE; a meganuclease; a homing endonuclease; or an I-SceI enzyme. sf-6059407 22474-20028.40 9. The epigenetic-modifying DNA-targeting system of any of embodiments 1-8, wherein each second DNA-binding domain is catalytically inactive. 10. The epigenetic-modifying DNA-targeting system of any of embodiments 1-9, wherein each first DNA-binding domain is a first Cas protein, and each of the at least one activator DNA-targeting module further comprises a gRNA for targeting the first Cas protein to a target site of one of the one or more activation genes. 11. The epigenetic-modifying DNA-targeting system of any of embodiments 1-10, wherein each second DNA-binding domain is a ZFP. 12. The epigenetic-modifying DNA-targeting system of any of embodiments 1-10, wherein each second DNA-binding domain is a second Cas protein, and each of the at least one repressor DNA-targeting module further comprises a gRNA for targeting the second Cas protein to a target site of one of the one or more repression genes. 13. The epigenetic-modifying DNA-targeting system of any of embodiments 1-9, 11, and 12, wherein each first DNA-binding domain is a ZFP. 14. The epigenetic-modifying DNA-targeting system of any of embodiments 1-10 and 12, wherein: each first DNA-binding domain is a ZFP; and each second DNA-binding domain is a Cas protein, and each of the at least one repressor DNA-targeting module further comprises a gRNA for targeting the second Cas protein to a target site of one of the one or more repression genes. 15. The epigenetic-modifying DNA-targeting system of any of embodiments 1-10 and 12, wherein: each first DNA-binding domain is a first Cas protein, and each of the at least one activator DNA-targeting module further comprises a gRNA for targeting the first Cas protein to a target site of one of the one or more activation genes; and each second DNA-binding domain is a second Cas protein, and each of the at least one repressor DNA-targeting module further comprises a gRNA for targeting the second Cas protein to a target site of one of the one or more repression genes. 16. The epigenetic-modifying DNA-targeting system of embodiment 15, wherein the first Cas protein or proteins are from a different Staphylococcus species than the second Cas protein or proteins. 17. An epigenetic-modifying DNA-targeting system comprising a plurality of DNA- targeting modules comprising at least one activator DNA-targeting module and at least one repressor DNA-targeting module, wherein:the at least one activator DNA-targeting module is sf-6059407 22474-20028.40 for increasing transcription of one or more activation genes, and each activator DNA-targeting module comprises (1) a fusion protein comprising (i) a first Cas protein and (ii) at least one transcriptional activator domain, and (2) a gRNA for targeting the first Cas protein to a target site of one of the one or more activation genes; wherein the one or more activation genes are selected from the group consisting of BATF, CD28, EOMES, IL-2, IL-2RB, IRF4, LAT, LCP2, TBX21, and VAV1; and the at least one repressor DNA-targeting module is for repressing transcription of one or more repression genes, and each repressor DNA-targeting module comprises (1) a fusion protein comprising (i) a second Cas protein and (ii) at least one transcriptional repressor domain, and (2) a gRNA for targeting the second Cas protein to a target site of one of the one or more repression genes; wherein the one or more repression genes are selected from the group consisting of CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and RASA2; wherein the first Cas protein or proteins are from a different Staphylococcus species than the second Cas protein or proteins. 18. The epigenetic-modifying DNA-targeting system of any of embodiments 15-17, wherein the first Cas protein or proteins do not bind to the protospacer-adjacent motifs (PAMs) that the second Cas protein or proteins bind to. 19. An epigenetic-modifying DNA-targeting system comprising a plurality of DNA- targeting modules comprising at least one activator DNA-targeting module and at least one repressor DNA-targeting module, wherein: the at least one activator DNA-targeting module is for increasing transcription of one or more activation genes, and each activator DNA-targeting module comprises (1) a fusion protein comprising (i) a first Cas protein and (ii) at least one transcriptional activator domain, and (2) a gRNA for targeting the first Cas protein to a target site of one of the one or more activation genes; wherein the one or more activation genes are selected from the group consisting of BATF, CD28, EOMES, IL-2, IL-2RB, IRF4, LAT, LCP2, TBX21, and VAV1; and the at least one repressor DNA-targeting module is for repressing transcription of one or more repressor genes, and each repressor DNA-targeting module comprises (1) a fusion protein comprising (i) a second Cas protein and (ii) at least one transcriptional repressor domain, and (2) a gRNA for targeting the second Cas protein to a target site of one of the one or more repression genes; wherein the one or more repression genes are selected from the group consisting of CBLB, CCNC, CD5, CISH, DGKZ, ELOB, FAS, Fli1, GATA3, KDM1A, MED12, MYB, PRDM1, TGFBR2, and RASA2; wherein the first Cas sf-6059407 22474-20028.40 protein or proteins do not bind to the protospacer-adjacent motifs (PAMs) that the second Cas protein or proteins bind to. 20. The epigenetic-modifying DNA-targeting system of embodiment 19, wherein the first Cas protein or proteins are from a different Staphylococcus species than the second Cas protein or proteins. 21. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12 and 15-20, wherein the first Cas protein or proteins bind to a first PAM 5’-NGG-3’ (SEQ ID NO: 224), wherein N is any nucleotide. 22. The epigenetic-modifying DNA-targeting system of any of embodiments 12-21, wherein the second Cas protein or proteins bind to a second PAM 5’- NNGRRT-3’ (SEQ ID NO: 225), wherein N is any nucleotide, and R is G or A. 23. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12 and 15-19, wherein the first Cas protein or proteins bind to a first PAM 5’- NNGRRT-3’ (SEQ ID NO: 225), wherein N is any nucleotide, and R is G or A. 24. The epigenetic-modifying DNA-targeting system of any of embodiments 12-20 and 23, wherein the second Cas protein or proteins bind to a second PAM 5’-NGG-3’ (SEQ ID NO: 224), wherein N is any nucleotide. 25. The epigenetic-modifying DNA-targeting system of any of embodiments 15-24, wherein the first Cas protein or proteins do not bind to the gRNA or gRNAs that target the one or more repression genes, and the second Cas protein or proteins do not bind to the gRNA or gRNAs that target the one or more activation genes. 26. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12 and 15-25, wherein each first Cas protein is a deactivated (dCas) protein. 27. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12 and 15-26, wherein each first Cas protein lacks nuclease activity. 28. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12 and 15-27, wherein each first Cas protein is a dCas9 protein. 29. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-20, and 23-28, wherein each first Cas protein is a Staphylococcus aureus dCas9 (dSaCas9) protein. 30. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-20, and 23-29, wherein each first Cas protein is a dSaCas9 protein comprising at least one sf-6059407 22474-20028.40 amino acid mutation selected from D10A and N580A with reference to numbering of positions of SEQ ID NO:124. 31. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-20, and 23-30, wherein each first Cas protein comprises the sequence set forth in SEQ ID NO:125 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 32. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-20, and 23-31, wherein each first Cas protein is set forth in SEQ ID NO:125. 33. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-22, and 25-28, wherein each first Cas protein is a Streptococcus pyogenes dCas9 (dSpCas9) protein. 34. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-22, 25-28, and 33, wherein each first Cas protein is a dSpCas9 protein comprising at least one amino acid mutation selected from D10A and H840A with reference to numbering of positions of SEQ ID NO:126. 35. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-22, 25-28, 33, and 34, wherein each first Cas protein comprises the sequence set forth in SEQ ID NO:127 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 36. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-22, 25-28, and 33-35, wherein each first Cas protein is set forth in SEQ ID NO:127. 37. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12 and 15-27, wherein each first Cas protein is a dCas12 protein. 38. The epigenetic-modifying DNA-targeting system of any of embodiments 1-37, wherein the at least one activator DNA-targeting module is a single activator DNA-targeting module. 39. The epigenetic-modifying DNA-targeting system of any of embodiments 1-37, wherein the at least one activator DNA-targeting module is a plurality of activator DNA- targeting modules. 40. The epigenetic-modifying DNA-targeting system of any of embodiments 1-9, 12, and 39, wherein at least one of the first DNA-binding domains is a ZFP that targets a target site of one of the one or more activation genes. sf-6059407 22474-20028.40 41. The epigenetic-modfying DNA-targeting system of any of embodiments 1-10 and 39, wherein at least one of the second DNA-binding domains is a ZFP that targets a target site of one of the one or more repression genes. 42. The epigenetic-modifying DNA-targeting system of any of embodiments 1-37 and 39-41, wherein: the first DNA-binding domains of the at least one activator DNA-targeting module are different from one another; and/or each activator DNA-targeting module of the at least one activator DNA-targeting module is different. 43. The epigenetic-modifying DNA-targeting system of any of embodiments 1-37 and 40-42, wherein the at least one activator DNA-targeting module is 2, 3, 4, 5, or 6 activator DNA-targeting modules. 44. The epigenetic-modifying DNA-targeting system of any of embodiments 1-37 and 39-43, wherein the at least one activator DNA-targeting module is 2 activator DNA- targeting modules. 45. The epigenetic-modifying DNA-targeting system of any of embodiments 1-44, wherein the one or more activation genes comprise IL-2. 46. The epigenetic-modifying DNA-targeting system of any of embodiments 1-45, wherein the one or more activation genes is IL-2. 47. The epigenetic-modifying DNA-targeting system of any of embodiments 1-37 and 39-45, wherein the at least one activator DNA-targeting module targets two or more activation genes. 48. The epigenetic-modifying DNA-targeting system of any of embodiments 1-37, 39-45, and 47, wherein the one or more activation genes comprise BATF and IL-2; BATF and VAV1; CD28 and BATF; CD28 and EOMES; CD28 and IL-2; CD28 and LCP2; CD28 and TBX21; CD28 and VAV1; EOMES and BATF; EOMES and LCP2; EOMES and TBX21; EOMES and VAV1; EOMES and IL-2; LCP2 and BATF; LCP2 and IL-2; LCP2 and TBX21; LCP2 and VAV1; TBX21 and BATF; TBX21 and IL-2; TBX21 and TBX21; TBX21 and VAV1; or VAV1 and IL-2. 49. The epigenetic-modifying DNA-targeting system of any of embodiments 1-37, 39-45, 47, and 48, wherein the one or more activation genes comprise IL-2RB and VAV1. 50. The epigenetic-modifying DNA-targeting system of any of embodiments 1-37, 39-45, 47, and 48, wherein the one or more activation genes comprise IL-2 and VAV1. sf-6059407 22474-20028.40 51. The epigenetic-modifying DNA-targeting system of any of embodiments 1-50, wherein the target site for each of the one or more activation genes is in a gene or a regulatory DNA element. 52. The epigenetic-modifying DNA-targeting system of any of embodiments 1-51, wherein the target site for each of the one or more activation genes is in a gene, an enhancer, or a promoter. 53. The epigenetic-modifying DNA-targeting system of any of embodiments 1-52, wherein the target site for each of the one or more activation genes is in an enhancer or a promoter. 54. The epigenetic-modifying DNA-targeting system of any of embodiments 1-53, wherein the target site for each of the one or more activation genes is within 1000 base pairs of a transcriptional start site of one of the one or more activation genes. 55. The epigenetic-modifying DNA-targeting system of any of embodiments 1-54, wherein the target site for each of the one or more activation genes is within 500 base pairs of a transcriptional start site of one of the one or more activation genes. 56. The epigenetic-modifying DNA-targeting system of any of embodiments 1-22, 25-28, 33-36, and 38-55, wherein the target site for at least one of the one or more activation genes is selected from: (a) a target site for VAV1 having the sequence set forth in any one of SEQ ID NOS:7-9, 156, and 170, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (b) a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:78, 388-403, and 451-453, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (c) a target site for BATF having the sequence set forth in any one of SEQ ID NOS:172-174, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (d) a target site for CD28 having the sequence set forth in any one of SEQ ID NOS:144-146 and 189-191, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (e) a target site for EOMES having the sequence set forth in any one of SEQ ID NOS:147-149, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (f) a target site for IRF4 having the sequence set forth in any one of SEQ ID NOS:175-177, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (g) a target site for LAT having the sequence set forth in any one of SEQ ID NOS:184-186, a sf-6059407 22474-20028.40 contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (h) a target site for LCP2 having the sequence set forth in any one of SEQ ID NOS:150-152 and 187-188, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; or (i) a target site for TBX21 having the sequence set forth in any one of SEQ ID NOS:153-155, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. 57. The epigenetic-modifying DNA-targeting system of any of embodiments 1-22, 25-28, 33-36, and 38-56, wherein the target site for at least one of the one or more activation genes is selected from: (a) a target site for VAV1 having the sequence set forth in any one of SEQ ID NOS:7-9, 156, and 170, or a complementary sequence of any of the foregoing; (b) a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:78, 388-403, and 451-453 or a complementary sequence of any of the foregoing; (c) a target site for BATF having the sequence set forth in any one of SEQ ID NOS:172-174, or a complementary sequence of any of the foregoing; (d) a target site for CD28 having the sequence set forth in any one of SEQ ID NOS:144-146 and 189-191, or a complementary sequence of any of the foregoing; (e) a target site for EOMES having the sequence set forth in any one of SEQ ID NOS:147-149, or a complementary sequence of any of the foregoing; (f) a target site for IRF4 having the sequence set forth in any one of SEQ ID NOS:175-177, or a complementary sequence of any of the foregoing; (g) a target site for LAT having the sequence set forth in any one of SEQ ID NOS:184-186, or a complementary sequence of any of the foregoing; (h) a target site for LCP2 having the sequence set forth in any one of SEQ ID NOS:150-152 and 187-188, or a complementary sequence of any of the foregoing; or (i) a target site for TBX21 having the sequence set forth in any one of SEQ ID NOS:153-155, or a complementary sequence of any of the foregoing. 58. The epigenetic-modifying DNA-targeting system of any of embodiments 1-22, 25-28, 33-36, and 38-57, wherein the target site for each of the one or more activation genes is selected from: (a) a target site for VAV1 having the sequence set forth in any one of SEQ ID NOS:7-9, 156, and 170, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (b) a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:78, 388-403, and 452-453 a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (c) a target site for BATF having the sequence set forth in any one of SEQ ID NOS:172-174, a contiguous sf-6059407 22474-20028.40 portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (d) a target site for CD28 having the sequence set forth in any one of SEQ ID NOS:144-146 and 189-191, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (e) a target site for EOMES having the sequence set forth in any one of SEQ ID NOS:147-149, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (f) a target site for IRF4 having the sequence set forth in any one of SEQ ID NOS:175-177, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (g) a target site for LAT having the sequence set forth in any one of SEQ ID NOS:184-186, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (h) a target site for LCP2 having the sequence set forth in any one of SEQ ID NOS:150-152 and 187-188, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; or (i) a target site for TBX21 having the sequence set forth in any one of SEQ ID NOS:153-155, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. 59. The epigenetic-modifying DNA-targeting system of any of embodiments 1-22, 25-28, 33-36, and 38-58, wherein the target site for each of the one or more activation genes is selected from: (a) a target site for VAV1 having the sequence set forth in any one of SEQ ID NOS:7-9, 156, and 170, or a complementary sequence of any of the foregoing; (b) a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:78, 388-403, and 451-453, or a complementary sequence of any of the foregoing; (c) a target site for BATF having the sequence set forth in any one of SEQ ID NOS:172-174, or a complementary sequence of any of the foregoing; (d) a target site for CD28 having the sequence set forth in any one of SEQ ID NOS:144-146 and 189-191, or a complementary sequence of any of the foregoing; (e) a target site for EOMES having the sequence set forth in any one of SEQ ID NOS:147-149, or a complementary sequence of any of the foregoing; (f) a target site for IRF4 having the sequence set forth in any one of SEQ ID NOS:175-177, or a complementary sequence of any of the foregoing; (g) a target site for LAT having the sequence set forth in any one of SEQ ID NOS:184-186, or a complementary sequence of any of the foregoing; (h) a target site for LCP2 having the sequence set forth in any one of SEQ ID NOS:150-152 and 187-188, or a complementary sequence of any of the foregoing; or (i) a target site for TBX21 having the sf-6059407 22474-20028.40 sequence set forth in any one of SEQ ID NOS:153-155, or a complementary sequence of any of the foregoing. 60. The epigenetic-modifying DNA-targeting system of any of embodiments 1-22, 25-28, 33-36, and 38-59, wherein the target site for at least one of the one or more activation genes is a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:78, 388- 403, and 451-453, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. 61. The epigenetic-modifying DNA-targeting system of any of embodiments 1-22, 25-28, 33-36, and 38-60, wherein the target site for at least one of the one or more activation genes is a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:78, 388- 403, and 451-453, or a complementary sequence of any of the foregoing. 62. The epigenetic-modifying DNA-targeting system of any of embodiments 1-22, 25-28, 33-36, and 38-61, wherein the target site for each of the one or more activation genes is a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:78, 388-403, amd 451-453 a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. 63. The epigenetic-modifying DNA-targeting system of any of embodiments 1-22, 25-28, 33-36, and 38-62, wherein the target site for each of the one or more activation genes is a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:78, 388-403, and 451-453, or a complementary sequence of any of the foregoing. 64. The epigenetic-modifying DNA-targeting system of any of embodiments 1-20, 23-32, and 38-55, wherein the target site for at least one of the one or more activation genes is a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:404-412, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. 65. The epigenetic-modifying DNA-targeting system of any of embodiments 1-20, 23-32, 38-55, and 64, wherein the target site for at least one of the one or more activation genes is a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:404-412 or a complementary sequence of any of the foregoing. 66. The epigenetic-modifying DNA-targeting system of any of embodiments 1-20, 23-32, 38-55, 64, and 65, wherein the target site for each of the one or more activation genes is a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:404-412, a sf-6059407 22474-20028.40 contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. 67. The epigenetic-modifying DNA-targeting system of any of embodiments 1-20, 23-32, 38-55, and 64-66, wherein the target site for each of the one or more activation genes is a target site for IL-2 having the sequence set forth in any one of SEQ ID NO:404-412 or a complementary sequence of any of the foregoing. 68. The epigenetic-modifying DNA-targeting system of any of embodiments 1-9, 11- 14, 38-63, wherein the target site for at least one of the one or more activation genes is any one of the sequences set forth in any one of SEQ ID NO:451-453, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. 69. The epigenetic-modifying DNA-targeting system of any of embodiments 1-9, 11- 14, 38-63, and 68, wherein the target site for at least one of the one or more activation genes is any one of the sequences set forth in any one of SEQ ID NO:451-453 or a complementary sequence of any of the foregoing. 70. The epigenetic-modifying DNA-targeting system of any of embodiments 1-9, 11- 14, 38-63, 68, and 69, wherein the target site for each of the one or more activation genes is any one of the sequences set forth in any one of SEQ ID NO:451-453, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. 71. The epigenetic-modifying DNA-targeting system of any of embodiments 1-9, 11- 14, 38-63, and 68-70, wherein the target site for each of the one or more activation genes is any one of the sequences set forth in any one of SEQ ID NO:451-453 or a complementary sequence of any of the foregoing. 72. The epigenetic-modifying DNA-targeting system of any of embodiments 1-9, 11- 14, 38-63, and 68-71, wherein the first DNA-binding domain is a ZFP that targets the target site of one or more activation genes set forth in SEQ ID NO: 451. 73. The epigenetic-modifying DNA-targeting system of embodiment 72, wherein the ZFP comprises a zinc finger recognition region comprising six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, selected from F1-F6 as follows: F1: QNAHRKT (SEQ ID NO: 472), F2: RKYYLAK (SEQ ID NO: 473), F3: RSAHLSR (SEQ ID NO: 474), F4: QSGDLTR (SEQ ID NO: 475), F5: RSDHLTQ (SEQ ID NO: 476), and F6: DSANLSR (SEQ ID NO: 477). sf-6059407 22474-20028.40 74. The epigenetic-modifying DNA-targeting system of embodiment 72 or 73, wherein the ZFP comprises the sequence set forth in SEQ ID NO: 458, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 75. The epigenetic-modifying DNA-targeting system of any of embodiments 72-74, wherein the ZFP comprises the sequence set forth in SEQ ID NO: 458. 76. The epigenetic-modifying DNA-targeting system of any of embodiments 72-75, wherein the ZFP is encoded by the sequence set forth in SEQ ID NO: 465 or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 77. The epigenetic-modifying DNA-targeting system of any of embodiments 72-76, wherein the ZFP is encoded by the sequence set forth in SEQ ID NO: 465. 78. The epigenetic-modifying DNA-targeting system of any of embodiments 1-9, 11- 14, 38-63, and 68-71, wherein the first DNA-binding domain is a ZFP that targets the target site of one or more activation genes set forth in SEQ ID NO: 452. 79. The epigenetic-modifying DNA-targeting system of embodiment 78, wherein the ZFP comprises a zinc finger recognition region comprising six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, selected from F1-F6 as follows: F1: DSSHLEL (SEQ ID NO: 478), F2: DRSNLTR (SEQ ID NO: 479), F3: RSDNLSE (SEQ ID NO: 480), F4: VRRALSS (SEQ ID NO: 481), F5: QSGALAR (SEQ ID NO: 482), and F6: RLDWLPM (SEQ ID NO: 483). 80. The epigenetic-modifying DNA-targeting system of embodiment 78 or 79, wherein the ZFP comprises the sequence set forth in SEQ ID NO: 459, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 81. The epigenetic-modifying DNA-targeting system of any of embodiments 78-80, wherein the ZFP comprises the sequence set forth in SEQ ID NO: 459. 82. The epigenetic-modifying DNA-targeting system of any of embodiments 78-81, wherein the ZFP is encoded by the sequence set forth in SEQ ID NO: 466 or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. sf-6059407 22474-20028.40 83. The epigenetic-modifying DNA-targeting system of any of embodiments 78-82, wherein the ZFP is encoded by the sequence set forth in SEQ ID NO: 466. 84. The epigenetic-modifying DNA-targeting system of any of embodiments 1-9, 11- 14, 38-63, and 68-71, wherein the first DNA-binding domain is a ZFP that targets the target site of one or more activation genes set forth in SEQ ID NO: 453. 85. The epigenetic-modifying DNA-targeting system of embodiment 84, wherein the ZFP comprises a zinc finger recognition region comprising six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, selected from F1-F6 as follows: F1: RSDNLSV (SEQ ID NO: 484), F2: RSAHLSR (SEQ ID NO: 485), F3: QNAHRKT (SEQ ID NO: 486), F4: LRHHLTR (SEQ ID NO: 487), F5: TSSNRKT (SEQ ID NO: 488), and F6: TSSNLSR (SEQ ID NO: 489). 86. The epigenetic-modifying DNA-targeting system of embodiment 84 or 85, wherein the ZFP comprises the sequence set forth in SEQ ID NO: 460, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 87. The epigenetic-modifying DNA-targeting system of any of embodiments 84-86, wherein the ZFP comprises the sequence set forth in SEQ ID NO: 460. 88. The epigenetic-modifying DNA-targeting system of any of embodiments 84-87, wherein the ZFP is encoded by the sequence set forth in SEQ ID NO: 467 or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 89. The epigenetic-modifying DNA-targeting system of any of embodiments 84-88, wherein the ZFP is encoded by the sequence set forth in SEQ ID NO: 467. 90. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12 and 15-67, wherein each gRNA for targeting a target site of one of the one or more activation genes comprises a gRNA spacer sequence that is complementary to a target site of one of the one or more activation genes. 91. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-67, and 90-65, wherein each gRNA for targeting a target site of one of the one or more activation genes comprises a gRNA spacer sequence between 14 nt and 24 nt or between 16 nt and 22 nt in length. sf-6059407 22474-20028.40 92. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-67, 90, and 91, wherein each gRNA for targeting a target site of one of the one or more activation genes comprises a gRNA spacer sequence that is 18 nt, 19 nt, 20 nt, 21 nt, or 22 nt in length. 93. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-22, 25-28, 33-36, 38-67, and 90-92, wherein each gRNA for targeting a target site of one of the one or more activation genes further comprises a gRNA scaffold sequence set forth in SEQ ID NO:69. 94. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-20, 23-32, 38-56, 64-67, and 90-92, wherein each gRNA for targeting a target site of one or the one or more activation genes further comprises a gRNA scaffold sequence set forth in SEQ ID NO:387. 95. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-22, 25-28, 33-36, 38-63, and 68-93, wherein at least one of the gRNA for targeting a target site of one or the one or more activation genes is selected from: (a) a gRNA targeting a target site for VAV1 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:41-43, 169, and 171, or a contiguous portion thereof of at least 14 nt; (b) a gRNA targeting a target site for IL-2 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NO:79 and 413-428, or a contiguous portion thereof of at least 14 nt; (c) a gRNA targeting a target site for BATF and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:178-180, or a contiguous portion thereof of at least 14 nt; (d) a gRNA targeting a target site for CD28 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:157-159 and 197-199, or a contiguous portion thereof of at least 14 nt; (e) a gRNA targeting a target site for EOMES and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:160-162, or a contiguous portion thereof of at least 14 nt; (f) a gRNA targeting a target site for IRF4 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:181-183, or a contiguous portion thereof of at least 14 nt; (g) a gRNA targeting a target site for LAT and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:192-194, or a contiguous portion thereof of at least 14 nt; (h) a gRNA targeting a target site for LCP2 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID sf-6059407 22474-20028.40 NOS:163-165 and 195-196, or a contiguous portion thereof of at least 14 nt; or (i) a gRNA targeting a target site for TBX21 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:166-168, or a contiguous portion thereof of at least 14 nt. 96. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-22, 25-28, 33-36, 38-63, 68-93, and 95, wherein at least one of the gRNA for targeting a target site of one or the one or more activation genes is selected from: (a) a gRNA targeting a target site for VAV1 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:41-43, 169, and 171; (b) a gRNA targeting a target site for IL-2 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NO:79 and 413-428; (c) a gRNA targeting a target site for BATF and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:178-180; (d) a gRNA targeting a target site for CD28 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:157-159 and 197-199; (e) a gRNA targeting a target site for EOMES and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:160-162; (f) a gRNA targeting a target site for IRF4 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:181-183; (g) a gRNA targeting a target site for LAT and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:192-194; (h) a gRNA targeting a target site for LCP2 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:163-165 and 195-196; or (i) a gRNA targeting a target site for TBX21 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:166-168. 97. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-22, 25-28, 33-36, 38-63, 68-93, 95, and 96, wherein each gRNA for targeting a target site of one or the one or more activation genes is selected from: (a) a gRNA targeting a target site for VAV1 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:41-43, 169, and 171, or a contiguous portion thereof of at least 14 nt; (b) a gRNA targeting a target site for IL-2 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NO:79 and 413-428, or a contiguous portion thereof of at least 14 nt; (c) a gRNA targeting a target site for BATF and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:178-180, or a contiguous portion thereof of at least 14 nt; (d) a gRNA targeting a target site for CD28 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:157-159 and 197- 199, or a contiguous portion thereof of at least 14 nt; (e) a gRNA targeting a target site for sf-6059407 22474-20028.40 EOMES and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:160-162, or a contiguous portion thereof of at least 14 nt; (f) a gRNA targeting a target site for IRF4 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:181-183, or a contiguous portion thereof of at least 14 nt; (g) a gRNA targeting a target site for LAT and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:192-194, or a contiguous portion thereof of at least 14 nt; (h) a gRNA targeting a target site for LCP2 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:163-165 and 195-196, or a contiguous portion thereof of at least 14 nt; or (i) a gRNA targeting a target site for TBX21 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:166-168, or a contiguous portion thereof of at least 14 nt. 98. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-22, 25-28, 33-36, 38-63, 68-93, and 95-97, wherein each gRNA for targeting a target site of one or the one or more activation genes is selected from: (a) a gRNA targeting a target site for VAV1 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:41-43, 169, and 171; (b) a gRNA targeting a target site for IL-2 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NO:79 and 413-428; (c) a gRNA targeting a target site for BATF and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:178- 180; (d) a gRNA targeting a target site for CD28 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:157-159 and 197-199; (e) a gRNA targeting a target site for EOMES and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:160-162; (f) a gRNA targeting a target site for IRF4 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:181-183; (g) a gRNA targeting a target site for LAT and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:192-194; (h) a gRNA targeting a target site for LCP2 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:163-165 and 195-196; or (i) a gRNA targeting a target site for TBX21 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:166-168. 99. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-22, 25-28, 33-36, 38-63, and 68-98, wherein at least one of the gRNA for targeting a target site of one or the one or more activation genes is a gRNA targeting a target site for IL-2 and comprises a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NO:79 and 413-428, or a contiguous portion thereof of at least 14 nt. sf-6059407 22474-20028.40 100. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-22, 25-28, 33-36, 38-63, 68-99, wherein at least one of the gRNA for targeting a target site of one or the one or more activation genes is a gRNA targeting a target site for IL-2 and comprises a gRNA spacer sequence set forth in any one of SEQ ID NO:79 and 413-428. 101. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-22, 25-28, 33-36, 38-63, 90-93, and 95-98, wherein each gRNA for targeting a target site of one or the one or more activation genes is a gRNA targeting a target site for IL-2 and comprises a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NO:79 and 413-428, or a contiguous portion thereof of at least 14 nt. 102. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-22, 25-28, 33-36, 38-63, 90-93, and 95-101, wherein each gRNA for targeting a target site of one or the one or more activation genes is a gRNA targeting a target site for IL-2 and comprises a gRNA spacer sequence set forth in any one of SEQ ID NO:79 and 413-428. 103. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-20, 23-32, 38-55, 64-92, and 94, wherein at least one of the gRNA for targeting a target site of one or the one or more activation genes is a gRNA targeting a target site for IL-2 and comprises a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NO:429-437, or a contiguous portion thereof of at least 14 nt. 104. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-20, 23-32, 38-55, 64-92, 94, and 103, wherein at least one of the gRNA for targeting a target site of one or the one or more activation genes is a gRNA targeting a target site for IL-2 and comprises a gRNA spacer sequence set forth in any one of SEQ ID NO:429-437. 105. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-20, 23-32, 38-55, 64-67, 90-92, 94, 103, and 104, wherein each gRNA for targeting a target site of one or the one or more activation genes is a gRNA targeting a target site for IL-2 and comprises a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NO:429-437, or a contiguous portion thereof of at least 14 nt. 106. The epigenetic-modifying DNA-targeting system of any of embodiments 10-12, 15-20, 23-32, 38-55, 64-67, 90-92, 94, and 103-105, wherein each gRNA targeting a target site of one or the one or more activation genes is a gRNA targeting a target site for IL-2 and comprises a gRNA spacer sequence set forth in any one of SEQ ID NO:429-437. sf-6059407 22474-20028.40 107. The epigenetic-modifying DNA-targeting system of any of embodiments 1-106, wherein each transcriptional activator domain epigenetically modifies a target site of one of the one or more activation genes to promote increased transcription of one of the one or more activation genes. 108. The epigenetic-modifying DNA-targeting system of any of embodiments 1-107, wherein each transcriptional activator domain is a NCOA3 domain, a FOXO3 domain, a VP64 domain, a p65 activation domain, a p300 domain, an Rta domain, a CBP domain, a VPR domain, a VPH domain, an HSF1 domain, a TET protein domain, optionally wherein the TET protein is TET1, a SunTag domain, or a domain, portion, variant, or truncation of any of the foregoing. 109. The epigenetic-modifying DNA-targeting system of any of embodiments 1-108, wherein each transcriptional activator domain comprises at least one VP16 domain or a variant or portion thereof that exhibits transcriptional activation activity. 110. The epigenetic-modifying DNA-targeting system of any of embodiments 1-109, wherein each transcriptional activator domain comprises a VP16 tetramer (VP64) domain or a variant or portion thereof that exhibits transcriptional activation activity. 111. The epigenetic-modifying DNA-targeting system of any of embodiments 1-110, wherein each transcriptional activator domain is a VP64 domain. 112. The epigenetic-modifying DNA-targeting system of any of embodiments 1-110, wherein each transcriptional activator domain comprises a NCOA3 domain or a variant or portion thereof that exhibits transcriptional activation activity. 113. The epigenetic-modifying DNA-targeting system of any of embodiments 1-110 and 112, wherein each transcriptional activator domain comprises a FOXO3 domain or a variant or portion thereof that exhibits transcriptional activation activity. 114. The epigenetic-modifying DNA-targeting system of any of embodiments 1-110, 112, and 113, wherein each transcriptional activator domain comprises a NCOA3-FOXO3- NCOA3 domain. 115. The epigenetic-modifying DNA-targeting system of any of embodiments 1-110 and 112-114, wherein each transcriptional activator domain comprises a NCOA3-FOXO3- NCOA3 domain. sf-6059407 22474-20028.40 116. The epigenetic-modifying DNA-targeting system of any of embodiments 1-108 and 112-115, wherein each transcriptional activator domain is a NCOA3-FOXO3-NCOA3 domain. 117. The epigenetic-modifying DNA-targeting system of any of embodiments 1-110 and 112-115, wherein each transcriptional activator domain comprises a VP16 tetramer (VP64) domain and a NCOA3-FOXO3-NCOA3 domain. 118. The epigenetic-modifying DNA-targeting system of any of embodiments 1-110, 112-115, and 117, wherein each transcriptional activator domain is a VP16 tetramer (VP64) domain and a NCOA3-FOXO3-NCOA3 domain. 119. The epigenetic-modifying DNA-targeting system of any of embodiments 1-111, 117, and 118, wherein each transcriptional activator domain comprises the sequence set forth in SEQ ID NO:142, a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. 120. The epigenetic-modifying DNA-targeting system of any of embodiments 1-12, 15-22, 25-28, 33-36, 38, 39, 41-63, 90-93, 95-97, and 107-111, and 119, wherein each fusion protein of the at least one activator DNA-targeting module comprises the sequence set forth in SEQ ID NO:77 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 121. The epigenetic-modifying DNA-targeting system of any of embodiments 1-12, 15-20, 23-32, 38, 39, 41-55, 64-67, 90-92, 94, and 103-111, and 119, wherein each fusion protein of the at least one activator DNA-targeting module comprises the sequence set forth in SEQ ID NO:386 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 122. The epigenetic-modifying DNA-targeting system of any of embodiments 1-110 and 112-118, wherein each fusion protein of the at least one activator DNA-targeting module comprises the sequence set forth in SEQ ID NO:528 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 123. The epigenetic-modifying DNA-targeting system of any of embodiments 1-12, 15-20, 23-32, 38, 39, 41-55, 64-67, 90-92, 94, and 103-110, 112-115, 117, 118, and 122, wherein each fusion protein of the at least one activator DNA-targeting module comprises the sequence set forth in SEQ ID NO:529 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. sf-6059407 22474-20028.40 124. The epigenetic-modifying DNA-targeting system of any of embodiments 1-9, 11- 14, 38-63, 68-77, 107, 108, 112-116, and 122, wherein each fusion protein of the at least one activator DNA-targeting module comprises the sequence set forth in SEQ ID NO:514 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 125. The epigenetic-modifying DNA-targeting system of any of embodiments 1-9, 11- 14, 38-63, 68-71, 78-83, 107, 108, 112-116, and 122, wherein each fusion protein of the at least one activator DNA-targeting module comprises the sequence set forth in SEQ ID NO:515 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 126. The epigenetic-modifying DNA-targeting system of any of embodiments 1-9, 11- 14, 38-63, 68-71, 84-89, 107, 108, 112-116, and 122, wherein each fusion protein of the at least one activator DNA-targeting module comprises the sequence set forth in SEQ ID NO:516 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 127. The epigenetic-modifying DNA-targeting system of any of embodiments 12-126, wherein each second Cas protein is a deactivated (dCas) protein. 128. The epigenetic-modifying DNA-targeting system of any of embodiments 12-127, wherein each second Cas protein lacks nuclease activity. 129. The epigenetic-modifying DNA-targeting system of any of embodiments 12-128, wherein each second Cas protein is a dCas9 protein. 130. The epigenetic-modifying DNA-targeting system of any of embodiments 12-22, 25-28, 33-63, 68-93, 95-102, 107-120, and 127-129, wherein each second Cas protein is a dSaCas9 protein. 131. The epigenetic-modifying DNA-targeting system of any of embodiments 12-22, 25-28, 33-63, 68-93, 95-102, 107-120, and 127-130, wherein each second Cas protein is a dSaCas9 protein comprising at least one amino acid mutation selected from D10A and N580A with reference to numbering of positions of SEQ ID NO:124. 132. The epigenetic-modifying DNA-targeting system of any of embodiments 12-22, 25-28, 33-63, 68-93, 95-102, 107-120, and 127-131, wherein each second Cas protein comprises the sequence set forth in SEQ ID NO:125 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. sf-6059407 22474-20028.40 133. The epigenetic-modifying DNA-targeting system of any of embodiments 12-22, 25-28, 33-63, 68-93, 95-102, 107-120, and 127-132, wherein each second Cas protein is set forth in SEQ ID NO:125. 134. The epigenetic-modifying DNA-targeting system of any of embodiments 12-20, 23-32, 37-55, 64-92, 94, 103-119, and 121-129, wherein each second Cas protein is a dSpCas9 protein. 135. The epigenetic-modifying DNA-targeting system of any of embodiments 12-20, 23-32, 37-55, 64-92, 94, 103-119, 121-129, and 134, wherein each second Cas protein is a dSpCas9 protein comprising at least one amino acid mutation selected from D10A and H840A with reference to numbering of positions of SEQ ID NO:126. 136. The epigenetic-modifying DNA-targeting system of any of embodiments 12-20, 23-32, 37-55, 64-92, 94, 103-119, 121-129, 134, and 135, wherein each second Cas protein comprises the sequence set forth in SEQ ID NO:127 or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 137. The epigenetic-modifying DNA-targeting system of any of embodiments 12-20, 23-32, 37-55, 64-92, 94, 103-119, 121-129, and 134-136, wherein each second Cas protein is set forth in SEQ ID NO:127. 138. The epigenetic-modifying DNA-targeting system of any of embodiments 12-128, wherein each second Cas protein is a dCas12 protein. 139. The epigenetic-modifying DNA-targeting system of any of embodiments 1-138, wherein the at least one repressor DNA-targeting module is a single repressor DNA-targeting module. 140. The epigenetic-modifying DNA-targeting system of any of embodiments 1-138, wherein the at least one repressor DNA-targeting module is a plurality of repressor DNA- targeting modules. 141. The epigenetic-modifying DNA-targeting system of any of embodiments 1-138 and 140, wherein: the second DNA-binding domains of the at least one repressor DNA-targeting module are different from one another; and/or each repressor DNA-targeting module of the at least one repressor DNA-targeting module is different. 142. The epigenetic-modifying DNA-targeting system of any of embodiments 1-138, 140, and 141, wherein the at least one repressor DNA-targeting module is 2, 3, 4, 5, or 6 repressor DNA-targeting modules. sf-6059407 22474-20028.40 143. The epigenetic-modifying DNA-targeting system of any of embodiments 1-138 and 140-142, wherein the at least one repressor DNA-targeting module is 2 repressor DNA- targeting modules. 144. The epigenetic-modifying DNA-targeting system of any of embodiments 1-143, wherein the one or more repression genes comprise MED12. 145. The epigenetic-modifying DNA-targeting system of any of embodiments 1-144, wherein the one or more repression genes is MED12. 146. The epigenetic-modifying DNA-targeting system of any of embodiments 1-138 and 140-144, wherein the at least one repressor DNA-targeting modules targets two or more repression genes. 147. The epigenetic-modifying DNA-targeting system of any of embodiments 1-138, 140-144, and 146, wherein the one or more repression genes comprise CBLB and CCNC; CBLB and CD5; CBLB and CISH; CBLB and DGKZ; CBLB and ELOB; CBLB and FAS; CBLB and Fli1; CBLB and GATA3; CBLB and KDM1A; CBLB and MED12; CBLB and MYB; CBLB and PRDM1; CBLB and RASA2; CD5 and CISH; CD5 and MYB; CISH and DGKZ; CISH and MYB; CISH and RASA2; GATA3 and CD5; GATA3 and CISH; GATA3 and MYB; MED12 and CBLB; MED12 and CD5; MED12 and CISH; MED12 and DGKZ; MED12 and ELOB; MED12 and GATA3; MED12 and MYB; MED12 and PRDM1; MED12 and RASA2; MYB and RASA2; MED12 and TGFBR2; PRDM1 and CISH; PRDM1 and GATA3; PRDM1 and MYB; PRDM1 and RASA2; CD5, CISH, and MYB; GATA3, CBLB, and MYB; GATA3, CD5, and MYB; or PRDM1, GATA3, and CISH. 148. The epigenetic modifying DNA-targeting system of any of embodiments 1-138, 140-144, 146, and 147, wherein: the two or more repression genes comprise CBLB and MYB; the two or more repression genes comprise CBLB and MED12; the two or more repression genes comprise CBLB and CCNC; or the two or more repression genes comprise MED12 and TGFBR2. 149. The epigenetic-modifying DNA-targeting system of any of embodiments 1-138 and 140-144, wherein the at least one repressor DNA-targeting modules targets three or more repression genes. 150. The epigenetic-modifying DNA-targeting system of any of embodiments 1-138, 140-144, and 149, wherein the one or more repression genes comprise MED12, TGFBR2 and CISH. sf-6059407 22474-20028.40 151. The epigenetic-modifying DNA-targeting system of any of embodiments 1-150, wherein the target site for each of the one or more repression genes is in a gene or a regulatory DNA element. 152. The epigenetic-modifying DNA-targeting system of any of embodiments 1-151, wherein the target site for each of the one or more repression genes is in a gene, an enhancer, or a promoter. 153. The epigenetic-modifying DNA-targeting system of any of embodiments 1-152, wherein the target site for each of the one or more repression genes is in an enhancer or a promoter. 154. The epigenetic-modifying DNA-targeting system of any of embodiments 1-153, wherein the target site for each of the one or more repression genes is within 1000 base pairs of a transcriptional start site of one of the one or more repression genes. 155. The epigenetic-modifying DNA-targeting system of any of embodiments 1-154, wherein the target site for each of the one or more repression genes is within 500 base pairs of a transcriptional start site of one of the one or more repression genes. 156. The epigenetic-modifying DNA-targeting system of any of embodiments 1-20, 23-32, 37-55, 64-89, 94, 103-119, 121-129, 134-137, and 139-155, wherein the target site for at least one of the one or more repression genes is selected from: (a) a target site for CD5 having the sequence set forth in any one of SEQ ID NOS:1-3, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (b) a target site for KDM1A having the sequence set forth in any one of SEQ ID NOS:4-6, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (c) a target site for CBLB having the sequence set forth in any one of SEQ ID NOS:10-12, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (d) a target site for DGKZ having the sequence set forth in any one of SEQ ID NOS:13-15, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (e) a target site for MYB having the sequence set forth in any one of SEQ ID NOS:16-18, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (f) a target site for RASA2 having the sequence set forth in any one of SEQ ID NOS:19-21, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (g) a target site for ELOB having the sequence set forth in any one of SEQ ID NOS:22-24, a contiguous portion sf-6059407 22474-20028.40 thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (h) a target site for GATA3 having the sequence set forth in any one of SEQ ID NOS:25-27, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (i) a target site for CISH having the sequence set forth in any one of SEQ ID NOS:28-30, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (j) a target site for PRDM1 having the sequence set forth in any one of SEQ ID NOS:31-33, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (k) a target site for MED12 having the sequence set forth in any one of SEQ ID NOS:80-90 and 454-456, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (l) a target site for CCNC having the sequence set forth in any one of SEQ ID NOS:102-112, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (m) a target site for FAS having the sequence set forth in any one of SEQ ID NOS:200-205 and 292-295, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (n) a target site for Fli1 having the sequence set forth in any one of SEQ ID NOS:206-211, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; and (o) a target site for TGFBR2 having the sequence set forth in any one of SEQ ID NOS:300-302, 306-308 and 457, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. 157. The epigenetic-modifying DNA-targeting system of any of embodiments 1-20, 23-32, 37-55, 64-89, 94, 103-119, 121-129, 134-137, and 139-156, wherein the target site for at least one of the one or more repression genes is selected from: (a) a target site for CD5 having the sequence set forth in any one of SEQ ID NOS:1-3 or a complementary sequence thereof; (b) a target site for KDM1A having the sequence set forth in any one of SEQ ID NOS:4-6 or a complementary sequence thereof; (c) a target site for CBLB having the sequence set forth in any one of SEQ ID NOS:10-12 or a complementary sequence thereof; (d) a target site for DGKZ having the sequence set forth in any one of SEQ ID NOS:13-15 or a complementary sequence of any of the foregoing; (e) a target site for MYB having the sequence set forth in any one of SEQ ID NOS:16-18 or a complementary sequence of any of the foregoing; (f) a target site for RASA2 having the sequence set forth in any one of SEQ ID NOS:19-21 or a complementary sequence of any of the foregoing; (g) a target site for ELOB having the sequence set forth in any one of SEQ sf-6059407 22474-20028.40 ID NOS:22-24 or a complementary sequence of any of the foregoing; (h) a target site for GATA3 having the sequence set forth in any one of SEQ ID NOS:25-27 or a complementary sequence of any of the foregoing; (i) a target site for CISH having the sequence set forth in any one of SEQ ID NOS:28-30 or a complementary sequence of any of the foregoing; (j) a target site for PRDM1 having the sequence set forth in any one of SEQ ID NOS:31-33 or a complementary sequence of any of the foregoing; (k) a target site for MED12 having the sequence set forth in any one of SEQ ID NOS:80-90 and 454-456 or a complementary sequence of any of the foregoing; (l) a target site for CCNC having the sequence set forth in any one of SEQ ID NOS:102-112 or a complementary sequence of any of the foregoing; (m) a target site for FAS having the sequence set forth in any one of SEQ ID NOS:200-205 and 292-295 or a complementary sequence of any of the foregoing; (n) a target site for Fli1 having the sequence set forth in any one of SEQ ID NOS:206-211 or a complementary sequence of any of the foregoing; and (o) a target site for TGFBR2 having the sequence set forth in any one of SEQ ID NOS:300-302, 306-308, and 457, or a complementary sequence of any of the foregoing. 158. The epigenetic-modifying DNA-targeting system of any of embodiments 1-20, 23-32, 37-55, 64-89, 94, 103-119, 121-129, 134-137, and 139-157, wherein the target site for each of the one or more repression genes is selected from: (a) a target site for CD5 having the sequence set forth in any one of SEQ ID NOS:1-3, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (b) a target site for KDM1A having the sequence set forth in any one of SEQ ID NOS:4-6, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (c) a target site for CBLB having the sequence set forth in any one of SEQ ID NOS:10-12, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (d) a target site for DGKZ having the sequence set forth in any one of SEQ ID NOS:13-15, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (e) a target site for MYB having the sequence set forth in any one of SEQ ID NOS:16-18, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (f) a target site for RASA2 having the sequence set forth in any one of SEQ ID NOS:19-21, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (g) a target site for ELOB having the sequence set forth in any one of SEQ ID NOS:22-24, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (h) sf-6059407 22474-20028.40 a target site for GATA3 having the sequence set forth in any one of SEQ ID NOS:25-27, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (i) a target site for CISH having the sequence set forth in any one of SEQ ID NOS:28-30, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (j) a target site for PRDM1 having the sequence set forth in any one of SEQ ID NOS:31-33, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (k) a target site for MED12 having the sequence set forth in any one of SEQ ID NOS:80-90 and 454-456, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (l) a target site for CCNC having the sequence set forth in any one of SEQ ID NOS:102-112, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (m) a target site for FAS having the sequence set forth in any one of SEQ ID NOS:200-205 and 292-295, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; (n) a target site for Fli1 having the sequence set forth in any one of SEQ ID NOS:206-211, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing; and (o) a target site for TGFBR2 having the sequence set forth in any one of SEQ ID NOS:300-302, 306-308, and 557, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. 159. The epigenetic-modifying DNA-targeting system of any of embodiments 1-20, 23-32, 37-55, 64-89, 94, 103-119, 121-129, 134-137, and 139-158, wherein the target site for each of the one or more repression genes is selected from: (a) a target site for CD5 having the sequence set forth in any one of SEQ ID NOS:1-3 or a complementary sequence thereof; (b) a target site for KDM1A having the sequence set forth in any one of SEQ ID NOS:4-6 or a complementary sequence thereof; (c) a target site for CBLB having the sequence set forth in any one of SEQ ID NOS:10-12 or a complementary sequence thereof; (d) a target site for DGKZ having the sequence set forth in any one of SEQ ID NOS:13-15 or a complementary sequence of any of the foregoing; (e) a target site for MYB having the sequence set forth in any one of SEQ ID NOS:16-18 or a complementary sequence of any of the foregoing; (f) a target site for RASA2 having the sequence set forth in any one of SEQ ID NOS:19-21 or a complementary sequence of any of the sf-6059407 22474-20028.40 foregoing; (g) a target site for ELOB having the sequence set forth in any one of SEQ ID NOS:22-24 or a complementary sequence of any of the foregoing; (h) a target site for GATA3 having the sequence set forth in any one of SEQ ID NOS:25-27 or a complementary sequence of any of the foregoing; (i) a target site for CISH having the sequence set forth in any one of SEQ ID NOS:28-30 or a complementary sequence of any of the foregoing; (j) a target site for PRDM1 having the sequence set forth in any one of SEQ ID NOS:31-33 or a complementary sequence of any of the foregoing; (k) a target site for MED12 having the sequence set forth in any one of SEQ ID NOS:80-90 and 454-456 or a complementary sequence of any of the foregoing; (l) a target site for CCNC having the sequence set forth in any one of SEQ ID NOS:102-112 or a complementary sequence of any of the foregoing; (m) a target site for FAS having the sequence set forth in any one of SEQ ID NOS:200-205 and 292-295 or a complementary sequence of any of the foregoing; (n) a target site for Fli1 having the sequence set forth in any one of SEQ ID NOS:206-211 or a complementary sequence of any of the foregoing; and (o) a target site for TGFBR2 having the sequence set forth in any one of SEQ ID NOS:300-302, 306-308, and 457, or a complementary sequence of any of the foregoing. 160. The epigenetic-modifying DNA-targeting system of any of embodiments 1-20, 23-32, 37-55, 64-89, 94, 103-119, 121-129, 134-137, and 139-159, wherein the target site for at least one of the one or more repression genes is a target site for MED12 having the sequence set forth in any one of SEQ ID NOS:80-90 and 454-456, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. 161. The epigenetic-modifying DNA-targeting system of any of embodiments 1-20, 23-32, 37-55, 64-89, 94, 103-109, 121-129, 134-137, and 139-160, wherein the target site for at least one of the one or more repression genes is a target site for MED12 having the sequence set forth in any one of SEQ ID NOS:80-90 and 454-456, or a complementary sequence of any of the foregoing. 162. The epigenetic-modifying DNA-targeting system of any of embodiments 1-20, 23-32, 37-55, 64-89, 94, 103-119, 121-129, 134-137, and 139-161, wherein the target site for each of the one or more repression genes is a target site for MED12 having the sequence set forth in any one of SEQ ID NOS:80-90 and 454-456, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. 163. The epigenetic-modifying DNA-targeting system of any of embodiments 20, 23- 32, 37-55, 64-89, 94, 103-119, 121-129, 134-137, and 139-162, wherein the target site for each sf-6059407 22474-20028.40 of the one or more repression genes is a target site for MED12 having the sequence set forth in any one of SEQ ID NOS:80-90 and 454-456, or a complementary sequence of any of the foregoing. 164. The epigenetic-modifying DNA-targeting system of any of embodiments 1-11, 13, and 37-126, and 139-163, wherein the target site of one of the one or more repression genes is any one of the sequences set forth in any one of SEQ ID NO:454-457, a contiguous portion thereof of at least 14 nucleotides (nt), or a complementary sequence of any of the foregoing. 165. The epigenetic-modifying DNA-targeting system of any of embodiments 1-11, 13, and 37-126, and 139-164, wherein the target site of one of the one or more repression genes is any one of the sequences set forth in any one of SEQ ID NO:454-457 or a complementary sequence of any of the foregoing. 166. The epigenetic-modifying DNA-targeting system of any of embodiments 1-11, 13, and 37-126, and 139-165, wherein the second DNA-binding domain is a ZFP that targets the target site of one or more repression genes set forth in SEQ ID NO: 454. 167. The epigenetic-modifying DNA-targeting system of embodiment 166, wherein the ZFP comprises a zinc finger recognition region comprising six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, selected from F1-F6 as follows: F1: DRSHLTR (SEQ ID NO: 490), F2: DRSYRNT (SEQ ID NO: 491), F3: QRRSLPH (SEQ ID NO: 492), F4: RSADLSR (SEQ ID NO: 493), F5: RSDTLSE (SEQ ID NO: 494), and F6: NRRGRWS (SEQ ID NO: 495). 168. The epigenetic-modifying DNA-targeting system of embodiment 166 or 167, wherein the ZFP comprises the sequence set forth in SEQ ID NO: 461, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 169. The epigenetic-modifying DNA-targeting system of any of embodiments 166- 168, wherein the ZFP comprises the sequence set forth in SEQ ID NO: 461. 170. The epigenetic-modifying DNA-targeting system of any of embodiments 166- 169, wherein the ZFP is encoded by the sequence set forth in SEQ ID NO: 468 or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 171. The epigenetic-modifying DNA-targeting system of any of embodiments 166- 170, wherein the ZFP is encoded by the sequence set forth in SEQ ID NO: 468. sf-6059407 22474-20028.40 172. The epigenetic-modifying DNA-targeting system of any of embodiments 1-11, 13, and 37-126, and 139-165, wherein the second DNA-binding domain is a ZFP that targets the target site of one or more repression genes set forth in SEQ ID NO: 455. 173. The epigenetic-modifying DNA-targeting system of embodiment 172, wherein the ZFP comprises a zinc finger recognition region comprising six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, selected from F1-F6 as follows: F1: RSANLAR (SEQ ID NO: 496), F2: DRSALAR (SEQ ID NO: 497), F3: RSDALST (SEQ ID NO: 498), F4: QSATRTK (SEQ ID NO: 499), F5: RSDTLSE (SEQ ID NO: 500), and F6: FRYARQY (SEQ ID NO: 501). 174. The epigenetic-modifying DNA-targeting system of embodiment 172 or 173, wherein the ZFP comprises the sequence set forth in SEQ ID NO: 462, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 175. The epigenetic-modifying DNA-targeting system of any of embodiments 172- 174, wherein the ZFP comprises the sequence set forth in SEQ ID NO: 462. 176. The epigenetic-modifying DNA-targeting system of any of embodiments 172- 175, wherein the ZFP is encoded by the sequence set forth in SEQ ID NO: 469 or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 177. The epigenetic-modifying DNA-targeting system of any of embodiments 172- 176, wherein the ZFP is encoded by the sequence set forth in SEQ ID NO: 469. 178. The epigenetic-modifying DNA-targeting system of any of embodiments 1-11, 13, and 37-126, and 139-165, wherein the second DNA-binding domain is a ZFP that targets the target site of one or more repression genes set forth in in SEQ ID NO: 456. 179. The epigenetic-modifying DNA-targeting system of embodiment 178, wherein the ZFP comprises a zinc finger recognition region comprising six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, selected from F1-F6 as follows: F1: DQSNLRA (SEQ ID NO: 502), F2: QNAHRKT (SEQ ID NO: 503), F3: TSGSLSR (SEQ ID NO: 504), F4: DRSNLSS (SEQ ID NO: 505), F5: RSAHLSR (SEQ ID NO: 506), and F6: RSDHLTQ (SEQ ID NO: 507). 180. The epigenetic-modifying DNA-targeting system of embodiment 178 or 179, wherein the ZFP comprises the sequence set forth in SEQ ID NO: 463, or a portion thereof, or sf-6059407 22474-20028.40 an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 181. The epigenetic-modifying DNA-targeting system of any of embodiments 178- 180, wherein the ZFP comprises the sequence set forth in SEQ ID NO: 463. 182. The epigenetic-modifying DNA-targeting system of any of embodiments 178- 181, wherein the ZFP is encoded by the sequence set forth in SEQ ID NO: 470 or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 183. The epigenetic-modifying DNA-targeting system of any of embodiments 178- 182, wherein the ZFP is encoded by the sequence set forth in SEQ ID NO: 470. 184. The epigenetic-modifying DNA-targeting system of any of embodiments 1-11, 13, and 37-126, and 139-159, wherein the second DNA-binding domain is a ZFP that targets the target site of one or more repression genes set forth in SEQ ID NO: 457. 185. The epigenetic-modifying DNA-targeting system of embodiment 184, wherein the ZFP comprises a zinc finger recognition region comprising six zinc fingers denoted F1 through F6 in order from N-terminus to C-terminus, selected from F1-F6 as follows: F1: RSDHLSA (SEQ ID NO: 508), F2: QSSDLRR (SEQ ID NO: 509), F3: HHNNRTH (SEQ ID NO: 510), F4: RNASRTR (SEQ ID NO: 511), F5: RSDHLSA (SEQ ID NO: 512), and F6: RSANLTR (SEQ ID NO: 513). 186. The epigenetic-modifying DNA-targeting system of embodiment 185, wherein the ZFP comprises the sequence set forth in SEQ ID NO: 464, or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 187. The epigenetic-modifying DNA-targeting system of embodiment 185 or 186, wherein the ZFP comprises the sequence set forth in SEQ ID NO: 464. 188. The epigenetic-modifying DNA-targeting system of any of embodiments 185- 187, wherein the ZFP is encoded by the sequence set forth in SEQ ID NO: 471 or a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 189. The epigenetic-modifying DNA-targeting system of any of embodiments 185- 188, wherein the ZFP is encoded by the sequence set forth in SEQ ID NO: 471. sf-6059407 22474-20028.40 190. The epigenetic-modifying DNA-targeting system of any of embodiments 12-163, wherein each gRNA for targeting a target site of one of the one or more repression genes comprises a gRNA spacer sequence that is complementary to a target site of one of the one or more repression genes. 191. The epigenetic-modifying DNA-targeting system of any of embodiments 12-163 and 190, wherein each gRNA for targeting a target site of one of the one or more repression genes comprises a gRNA spacer sequence between 14 nt and 24 nt or between 16 nt and 22 nt in length. 192. The epigenetic-modifying DNA-targeting system of any of embodiments 12-163, 190, and 191, wherein each gRNA for targeting a target site of one of the one or more repression genes comprises a gRNA spacer sequence that is 18 nt, 19 nt, 20 nt, 21 nt, or 22 nt in length. 193. The epigenetic-modifying DNA-targeting system of any of embodiments 12-20, 23-32, 37-55, 64-67, 90-92, 94, 103-119, 121-1239, 134-137, 139-163, and 190-192, wherein each gRNA for targeting a target site of one of the one or more repression genes further comprises a gRNA scaffold sequence set forth in SEQ ID NO:69. 194. The epigenetic-modifying DNA-targeting system of any of embodiments 12-22, 25-28, 33-63, 90-93, 95-102, 107-120, 122, , 124-133, 139-155, and 190-191, wherein each gRNA for targeting a target site of one of the one or more repression genes further comprises a gRNA scaffold sequence set forth in SEQ ID NO:387. 195. The epigenetic-modifying DNA-targeting system of any of embodiments 12-20, 23-32, 37-55, 64-67, 90-92, 94, 103-119, 121-123, 134-137, 139-163, and 190-192, wherein at least one of the gRNA for targeting a target site of one of the one or more repression genes is selected from: (a) a gRNA targeting a target site for CD5 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:35-37, or a contiguous portion thereof of at least 14 nt; (b) a gRNA targeting a target site for KDM1A and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:38-40, or a contiguous portion thereof of at least 14 nt; (c) a gRNA targeting a target site for CBLB and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:44-46, or a contiguous portion thereof of at least 14 nt; (d) a gRNA targeting a target site for DGKZ and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:47-49, or a contiguous portion thereof of at least 14 nt; (e) a gRNA targeting a target site for MYB and comprising a gRNA spacer sequence comprising the sf-6059407 22474-20028.40 sequence set forth in any one of SEQ ID NOS:50-52, or a contiguous portion thereof of at least 14 nt; (f) a gRNA targeting a target site for RASA2 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:53-55, or a contiguous portion thereof of at least 14 nt; (g) a gRNA targeting a target site for ELOB and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:56-58, or a contiguous portion thereof of at least 14 nt; (h) a gRNA targeting a target site for GATA3 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:59-61, or a contiguous portion thereof of at least 14 nt; (i) a gRNA targeting a target site for CISH and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:62-64, or a contiguous portion thereof of at least 14 nt; (j) a gRNA targeting a target site for PRDM1 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:65-67, or a contiguous portion thereof of at least 14 nt; (k) a gRNA targeting a target site for MED12 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:91-101, or a contiguous portion thereof of at least 14 nt; (l) a gRNA targeting a target site for CCNC and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:113-123, or a contiguous portion thereof of at least 14 nt; (m) a gRNA targeting a target site for FAS and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:212-217 and 296-299, or a contiguous portion thereof of at least 14 nt; (n) a gRNA targeting a target site for Fli1 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:218-223, or a contiguous portion thereof of at least 14 nt; and (o) a gRNA targeting a target site for TGFBR2 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:303-305 and 309-311, or a contiguous portion thereof of at least 14 nt. 196. The epigenetic-modifying DNA-targeting system of any of embodiments 20, 23- 32, 37-55, 64-67, 90-92, 94, 103-119, 121-123, 134-137, 139-163, 190-193, and 195, wherein at least one of the gRNA for targeting a target site of one of the one or more repression genes is selected from: (a) a gRNA targeting a target site for CD5 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:35-37; (b) a gRNA targeting a target site for KDM1A and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:38-40; (c) a gRNA targeting a target site for CBLB and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:44-46; (d) a gRNA targeting a target site for DGKZ and comprising sf-6059407 22474-20028.40 a gRNA spacer sequence set forth in any one of SEQ ID NOS:47-49; (e) a gRNA targeting a target site for MYB and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:50-52; (f) a gRNA targeting a target site for RASA2 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:53-55; (g) a gRNA targeting a target site for ELOB and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:56-58; (h) a gRNA targeting a target site for GATA3 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:59-61; (i) a gRNA targeting a target site for CISH and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:62-64; (j) a gRNA targeting a target site for PRDM1 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:65-67; (k) a gRNA targeting a target site for MED12 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:91-101; (l) a gRNA targeting a target site for CCNC and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:113-123; (m) a gRNA targeting a target site for FAS and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:212-217 and 296-299; (n) a gRNA targeting a target site for Fli1 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:218-223; and (o) a gRNA targeting a target site for TGFBR2 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:303-305 and 309-311. 197. The epigenetic-modifying DNA-targeting system of any of embodiments 20, 23- 32, 37-55, 64-67, 90-92, 94, 103-119, 121-1239, 134-137, 139-163, 190-193195, and 196, wherein each gRNA for targeting a target site of one of the one or more repression genes is selected from: (a) a gRNA targeting a target site for CD5 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:35-37, or a contiguous portion thereof of at least 14 nt; (b) a gRNA targeting a target site for KDM1A and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:38-40, or a contiguous portion thereof of at least 14 nt; (c) a gRNA targeting a target site for CBLB and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:44-46, or a contiguous portion thereof of at least 14 nt; (d) a gRNA targeting a target site for DGKZ and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:47-49, or a contiguous portion thereof of at least 14 nt; (e) a gRNA targeting a target site for MYB and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:50-52, or a contiguous portion thereof of at least 14 nt; (f) a gRNA targeting a target site for RASA2 and comprising a gRNA spacer sequence sf-6059407 22474-20028.40 comprising the sequence set forth in any one of SEQ ID NOS:53-55, or a contiguous portion thereof of at least 14 nt; (g) a gRNA targeting a target site for ELOB and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:56-58, or a contiguous portion thereof of at least 14 nt; (h) a gRNA targeting a target site for GATA3 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:59-61, or a contiguous portion thereof of at least 14 nt; (i) a gRNA targeting a target site for CISH and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:62-64, or a contiguous portion thereof of at least 14 nt; (j) a gRNA targeting a target site for PRDM1 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:65-67, or a contiguous portion thereof of at least 14 nt; (k) a gRNA targeting a target site for MED12 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:91-101, or a contiguous portion thereof of at least 14 nt; (l) a gRNA targeting a target site for CCNC and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:113-123, or a contiguous portion thereof of at least 14 nt; (m) a gRNA targeting a target site for FAS and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:212-217 and 296-299, or a contiguous portion thereof of at least 14 nt; (n) a gRNA targeting a target site for Fli1 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:218-223, or a contiguous portion thereof of at least 14 nt; and (o) a gRNA targeting a target site for TGFBR2 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:303-305 and 309-311, or a contiguous portion thereof of at least 14 nt. 198. The epigenetic-modifying DNA-targeting system of any of embodiments 20, 23- 32, 37-55, 64-67, 90-92, 94, 103-119, 121-1239, 134-137, 139-163, and 190-193, and 195-197, wherein each gRNA for targeting a target site of one of the one or more repression genes is selected from: (a) a gRNA targeting a target site for CD5 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:35-37; (b) a gRNA targeting a target site for KDM1A and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:38-40; (c) a gRNA targeting a target site for CBLB and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:44-46; (d) a gRNA targeting a target site for DGKZ and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:47-49; (e) a gRNA targeting a target site for MYB and comprising a gRNA spacer sequence set forth in any one of SEQ ID sf-6059407 22474-20028.40 NOS:50-52; (f) a gRNA targeting a target site for RASA2 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:53-55; (g) a gRNA targeting a target site for ELOB and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:56-58; (h) a gRNA targeting a target site for GATA3 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:59-61; (i) a gRNA targeting a target site for CISH and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:62-64; (j) a gRNA targeting a target site for PRDM1 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:65-67; (k) a gRNA targeting a target site for MED12 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:91-101; (l) a gRNA targeting a target site for CCNC and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:113-123; (m) a gRNA targeting a target site for FAS and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:212-217 and 296-299; (n) a gRNA targeting a target site for Fli1 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:218-223; and (o) a gRNA targeting a target site for TGFBR2 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:303-305 and 309-311. 199. The epigenetic-modifying DNA-targeting system of any of embodiments 20, 23- 32, 37-55, 64-67, 90-92, 94, 103-119, 121-1239, 134-137, 139-163, and 190-193, and 195-198, wherein at least one of the gRNA for targeting a target site of one of the one or more repression genes is a gRNA targeting a target site for MED12 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:91-101, or a contiguous portion thereof of at least 14 nt. 200. The epigenetic-modifying DNA-targeting system of any of embodiments 20, 23- 32, 37-55, 64-67, 90-92, 94, 103-119, 121-1239, 134-137, 139-163, and 190-193, and 195-199, wherein at least one of the gRNA for targeting a target site of one of the one or more repression genes is a gRNA targeting a target site for MED12 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:91-101. 201. The epigenetic-modifying DNA-targeting system of any of embodiments 20, 23- 32, 37-55, 64-67, 90-92, 94, 103-119, 121-1239, 134-137, 139-163, and 190-193, and 195-200, wherein each gRNA for targeting a target site of one of the one or more repression genes is a gRNA targeting a target site for MED12 and comprising a gRNA spacer sequence comprising the sequence set forth in any one of SEQ ID NOS:91-101, or a contiguous portion thereof of at least 14 nt. sf-6059407 22474-20028.40 202. The epigenetic-modifying DNA-targeting system of any of embodiments 20, 23- 32, 37-55, 64-67, 90-92, 94, 103-119, 121-1239, 134-137, 139-163, and 190-193, and 195-201, wherein each gRNA for targeting a target site of one of the one or more repression genes is a gRNA targeting a target site for MED12 and comprising a gRNA spacer sequence set forth in any one of SEQ ID NOS:91-101. 203. The epigenetic-modifying DNA-targeting system of any of embodiments 1-202, wherein each transcriptional repressor domain epigenetically modifies a target site of one of the one or more repression genes to promote reduced transcription of one of the one or more repression genes. 204. The epigenetic-modifying DNA-targeting system of any of embodiments 1-203, wherein each transcriptional repressor domain is a KRAB domain, a DNMT3A domain, a DNMT3L domain, a DNMT3B domain, a DNMT3A-DNMT3L fusion protein domain, an ERF repressor domain, an Mxi1 repressor domain, a SID4X repressor domain, a Mad-SID repressor domain, an LSD1 repressor domain, an EZH2 repressor domain, a SunTag domain, a variant or portion of any of the foregoing, or a combination of any of the foregoing. 205. The epigenetic-modifying DNA-targeting system of any of embodiments 1-204, wherein each transcriptional repressor domain is a KRAB domain, a DNMT3A domain, a DNMT3L domain, or a combination of any of the foregoing. 206. The epigenetic-modifying DNA-targeting system of any of embodiments 1-205, wherein each transcriptional repressor domain comprises a KRAB domain or a variant or portion thereof that exhibits transcriptional repressor activity. 207. The epigenetic-modifying DNA-targeting system of any of embodiments 1-206, wherein each transcriptional repressor domain comprises the sequence set forth in any one of SEQ ID NOS:70, 235, and 355-358, a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. 208. The epigenetic-modifying DNA-targeting system of any of embodiments 1-205, wherein each transcriptional repressor domain comprises a DNMT3A domain or a variant or portion thereof that exhibits transcriptional repressor activity. 209. The epigenetic-modifying DNA-targeting system of any of embodiments 1-205 and 208, wherein each transcriptional repressor domain comprises the sequence set forth in SEQ sf-6059407 22474-20028.40 ID NO:131 or 238, a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. 210. The epigenetic-modifying DNA-targeting system of any of embodiments 1-205, wherein each transcriptional repressor domain comprises a DNMT3L domain or a variant or portion thereof that exhibits transcriptional repressor activity. 211. The epigenetic-modifying DNA-targeting system of any of embodiments 1-205 and 210, wherein each transcriptional repressor domain comprises the sequence set forth in any one of SEQ ID NOS:133 and 240-242, a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. 212. The epigenetic-modifying DNA-targeting system of any of embodiments 1-205, wherein each transcriptional repressor domain is a DNMT3A-DNMT3L fusion protein domain, a DNMT3B-DNMT3L fusion protein domain, or a variant thereof that exhibits transcriptional repressor activity. 213. The epigenetic-modifying DNA-targeting system of any of embodiments 1-205 and 212, wherein each transcriptional repressor domain comprises the sequence set forth in any one of SEQ ID NOS:135, 137, or 363, a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity to any of the foregoing. 214. The epigenetic-modifying DNA-targeting system of any of embodiments 1-20, 23-32, 37-55, 64-67, 90-92, 94, 103-119, 121-123, 134-137, 139-163, 190-192, and 195-213, wherein each fusion protein of the at least one repressor DNA-targeting module comprises the sequence set forth in any one of SEQ ID NOS:138-141, 332-351, and 365-384, a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 215. The epigenetic-modifying DNA-targeting system of any of embodiments 1-11, 13, and 37-126, 139-189, and 203-213, wherein each fusion protein of the at least one repressor DNA-targeting module comprises the sequence set forth in any one of SEQ ID NOS:517-520, a portion thereof, or an amino acid sequence that has at least 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% sequence identity thereto. 216. The epigenetic-modifying DNA-targeting system of any of embodiments 1-215, wherein delivery of the epigenetic-modifying DNA-targeting system to the T cell increases sf-6059407 22474-20028.40 transcription of the one or more activation genes and decreases transcription of the one or more repression genes, compared to a T cell that has not been delivered the epigenetic-modifying DNA-targeting system. 217. The epigenetic-modifying DNA-targeting system of any of embodiments 1-216, wherein delivery of the epigenetic-modifying DNA-targeting system to the T cell promotes increased T cell effector function upon T cell stimulation, compared to a T cell that has not been delivered the epigenetic-modifying DNA-targeting system. 218. The epigenetic-modifying DNA-targeting system of embodiment 216 or embodiment 217, wherein the delivery is transient delivery of the epigenetic-modifying DNA- targeting system to the T cell. 219. The epigenetic-modifying DNA-targeting system of embodiment 217 or embodiment 218, wherein the T cell effector function is selected from the group consisting of IL-2 production, IFN-gamma production, TNF-alpha production, T cell proliferation, and a combination of any of the foregoing. 220. The epigenetic-modifying DNA-targeting system of any of embodiments 217- 219, wherein the T cell effector function is IL-2 production. 221. The epigenetic-modifying DNA-targeting system of any of embodiments 217- 219, wherein the T cell effector function is IFN-gamma production. 222. The epigenetic-modifying DNA-targeting system of any of embodiments 217- 219, wherein the T cell effector function is IL-2 production and IFN-gamma production. 223. The epigenetic-modifying DNA-targeting system of any of embodiments 217- 219, wherein the T cell effector function is polyfunctional production of IL-2, IFN-gamma, and TNF-alpha. 224. The epigenetic-modifying DNA-targeting system of any of embodiments 217- 223, wherein the T cell effector function further comprises T cell proliferation. 225. The epigenetic modifying DNA-targeting system of any of embodiments 217- 224, wherein the T cell effector function further comprises killing of target cells. 226. The epigenetic-modifying DNA-targeting system of any of embodiments 217- 225, wherein the T cell effector function further comprises T cell persistence. 227. The epigenetic-modifying DNA-targeting system of any of embodiments 217- 226, wherein the increased T cell effector function occurs 48 hours or more after the delivery of the epigenetic-modifying DNA-targeting system to the T cell. sf-6059407 22474-20028.40 228. The epigenetic-modifying DNA-targeting system of any of embodiments 217- 227, wherein the increased T cell effector function occurs up to 6 days, up to 9 days, up to 12 days, up to 15 days, up to 21 days, up to 28 days, up to 35 days, up to 42 days, up to 49 days, up to 56 days, up to 63 days, or up to 71 days after the delivery of the epigenetic-modifying DNA- targeting system to the T cell. 229. The epigenetic-modifying DNA-targeting system of any of embodiments 217- 228, wherein the T cell stimulation is with an anti-CD3 and anti-CD28 activation reagent. 230. The epigenetic-modifying DNA-targeting system of any of embodiments 1-229, wherein the T cell expresses a recombinant receptor. 231. The epigenetic-modifying DNA-targeting system of embodiment 230, wherein the recombinant receptor is a chimeric antigen receptor (CAR) or a T cell receptor (eTCR). 232. The epigenetic-modifying DNA-targeting system of embodiment 230 or embodiment 231, wherein the recombinant receptor is a CAR. 233. The epigenetic-modifying DNA-targeting system of any of embodiments 230- 232, wherein the recombinant receptor is directed against an antigen, and the T cell stimulation is an antigen-specific stimulation of the recombinant receptor. 234. The epigenetic-modifying DNA-targeting system of any of embodiments 217- 228 and 233, wherein the T cell stimulation is with antigen-expressing target cells. 235. The epigenetic-modifying DNA-targeting system of any of embodiments 217- 234, wherein the T cell stimulation is a restimulation after at least one prior T cell stimulation of the T cells. 236. A polynucleotide encoding the epigenetic-modifying DNA-targeting system of any of embodiments 1-235. 237. Two or more polynucleotides together encoding the epigenetic-modifying DNA- targeting system of any of embodiments 1-235. 238. A polynucleotide comprising (a) a promoter sequence; (b) a first nucleic acid sequence encoding at least one activator DNA-targeting module comprising a fusion protein comprising (i) a first zinc finger protein (ZFP) for targeting to a target site of one of the one or more activation genes and (ii) at least one transcriptional activator domain; (c) a second nucleic acid encoding at least one repressor DNA-targeting module comprising a fusion protein comprising (i) a second zinc finger protein (ZFP) for targeting to a target site of one of the one or more repression genes and (ii) at least one transcriptional repressor domain; and (d) a sf-6059407 22474-20028.40 cleavable linker sequence located between the first and second nucleic acid sequence, wherein the promoter is operably linked to the first and second nucleic acid to control expression their expression. 239. The polynucleotide of embodiment 238, wherein the cleavable linker sequence encodes a self-cleaving peptide, optionally P2A or T2A. 240. The polynucleotide of embodiment 238 or embodiment 239, wherein the first ZFP targets the target site of one or more activation genes set forth in any one of SEQ ID NOs: 451-453; and the second ZFP targets the target site set forth in any one of SEQ ID NOs: 454-457. 241. The polynucleotide of any of embodiments 238-240, wherein the first ZFP comprises the sequence set forth in any one of SEQ ID NOs: 458-460; and the second ZFP comprises the sequence set forth in any one of SEQ ID NOs: 461-464. 242. The polynucleotide of any of embodiments 238-241, wherein the at least one transcriptional activator domain comprises a sequence set forth in SEQ ID NO: 549; and the at least one transcriptional repressor domain comprises sequences set forth in SEQ ID NO: 70 and SEQ ID NO: 135. 243. The polynucleotide of any of embodiments 238-242, wherein the at least one activator DNA-targeting module comprises the sequence set forth in any one of SEQ ID NOs: 514-516; and the at least one repressor DNA-targeting module comprises the sequence set forth in any one of SEQ ID NOs: 517-520. 244. The polynucleotide of any of embodiments 238-243, wherein the at least one activator DNA-targeting module comprises SEQ ID NO: 516; and the at least one repressor DNA-targeting module comprises SEQ ID NO: 517. 245. The polynucleotide of any of embodiments 238-244, wherein the polynucleotide, from N-terminus to C-terminus, comprises :(a) the promoter sequence; (b) the first nucleic acid encoding the at least one activator DNA-targeting module, (c) the cleavable linker sequence, and (d) the second nucleic acid encoding the at least one repressor DNA-targeting module. 246. The polynucleotide of any of embodiments 238-244, wherein the polynucleotide, from N-terminus to C-terminus, comprises: (a) the promoter sequence; (b) the second nucleic acid encoding the at least one repressor DNA-targeting module, (c) the cleavable linker sequence, and (d) the first nucleic acid encoding the at least one activator DNA-targeting module. sf-6059407 22474-20028.40 247. The polynucleotide of any of embodiments 238-246, wherein the self-cleaving peptide comprises the sequence set forth in SEQ ID NO: 352. 248. A vector comprising the polynucleotide of any of embodiments 236 and 238-247. 249. A vector comprising the two or more polynucleotides of embodiment 237. 250. The vector of embodiment 248 or embodiment 249, wherein the vector is a viral vector. 251. The vector of any of embodiments 248-250, wherein the vector is an adeno- associated virus (AAV) vector. 252. The vector of any of embodiments 248-251, wherein the vector is an AAV1, AAV2, AAV3, AAV4, AAV5, AAV6, AAV7, AAV8, or AAV9 vector. 253. The vector of embodiment 248 or embodiment 249, wherein the vector is a non- viral vector. 254. The vector of any of embodiments 248, 249, and 253, wherein the vector is a lipid nanoparticle, a liposome, an exosome, or a cell penetrating peptide. 255. The vector of any of embodiments 248-249, 253, and 254, wherein the vector is a lipid nanoparticle. 256. The vector of any of embodiments 248-255, wherein the vector exhibits immune cell tropism, optionally wherein the vector exhibits T cell tropism. 257. Two or more vectors together comprising the two or more polynucleotides of embodiment 237. 258. The two or more vectors of embodiment 257, wherein the two or more vectors are viral vectors. 259. The two or more vectors of embodiment 257 or embodiment 258, wherein the two or more vectors are AAV vectors. 260. The two or more vectors of any of embodiments 257-259, wherein each vector is an AAV1, AAV2, AAV3, AAV4, AAV5, AAV6, AAV7, AAV8, or AAV9 vector. 261. The two or more vectors of embodiment 257, wherein the two or more vectors are non-viral vectors. 262. The two or more vectors of embodiment 257 or embodiment 261, wherein each vector is a lipid nanoparticle, a liposome, an exosome, or a cell penetrating peptide. 263. The two or more vectors of any of embodiments 257, 261, and 262, wherein the two or more vectors are lipid nanoparticles. sf-6059407 22474-20028.40 264. The two or more vectors of any of embodiments 257-263, wherein the two or more vectors exhibit immune cell tropism. 265. The two or more vectors of any of embodiments 257-264, wherein the two or more vectors exhibit T cell tropism. 266. A modified T cell comprising the DNA-targeting system of any of embodiments 1-235, the polynucleotide of any of embodiments 236 and 238-247, or the two or more polynucleotides of embodiment 237. 267. A modified T cell comprising an epigenetic or phenotypic modification resulting from being contacted by the DNA-targeting system of any of embodiments 1-235, the polynucleotide of any of embodiments 236 and 238-247, the two or more polynucleotides of embodiment 237, the vector of any of embodiments 248-256, or the two or more vectors of any of embodiments 257-265. 268. The modified T cell of embodiment 266 or embodiment 267, wherein the modified T cell is derived from a cell from a subject. 269. The modified T cell of any of embodiments 266-268, wherein the modified T cell is derived from a primary T cell. 270. The modified T cell of any of embodiments 266-268, wherein the modified T cell is derived from a T cell progenitor, a pluripotent stem cell, or an induced pluripotent stem cell. 271. The modified T cell of any of embodiments 266-268, wherein the modified T cell is a tumor infiltrating lymphocyte (TIL) or is an engineered T cell that further comprises an eTCR or CAR. 272. A method of modulating transcription in a T cell, the method comprising introducing into a T cell the DNA-targeting system of any of embodiments 1-235, the polynucleotide of any of embodiments 236 and 238-247, the two or more polynucleotides of embodiment 237, the vector of any of embodiments 248-256, or the two or more vectors of any of embodiments 257-265. 273. A method of increasing T cell effector function, the method comprising introducing into a T cell the DNA-targeting system of any of embodiments 1-235, the polynucleotide of any of embodiments 236 and 238-247, the two or more polynucleotides of embodiment 237, the vector of any of embodiments 248-256, or the two or more vectors of any of embodiments 257-265. sf-6059407 22474-20028.40 274. The method of embodiment 272 or embodiment 273, wherein T cell effector function of the T cell is increased upon T cell stimulation compared to a T cell that has not been introduced to the DNA-targeting system, the polynucleotide, the two or more polynucleotides, the vector, or the two or more vectors. 275. The method of any of embodiments 272-274, further comprising stimulating the T cell. 276. The method of embodiment 274 or embodiment 275, wherein the T cell effector function is selected from the group consisting of IL-2 production, IFN-gamma production, TNF- alpha production, T cell proliferation, and a combination of any of the foregoing. 277. The method of any of embodiments 274-276, wherein the T cell effector function is IL-2 production. 278. The method of any of embodiments 274-276, wherein the T cell effector function is IFN-gamma production. 279. The method of any of embodiments 274-276, wherein the T cell effector function is IL-2 production and IFN-gamma production. 280. The method of any of embodiments 274-276, wherein the T cell effector function is polyfunctional production of IL-2, IFN-gamma, and TNF-alpha. 281. The method of any of embodiments 274-280, wherein the T cell effector function further comprises T cell proliferation. 282. The method of any of embodiments 274-281, wherein the T cell effector function further comprises killing of target cells. 283. The method of any of embodiments 274-282, wherein the T cell effector function further comprises T cell persistence. 284. The method of any of embodiments 274-283, wherein the increased T cell effector function occurs 48 hours or more after the introducing to the T cell. 285. The method of any of embodiments 274-284, wherein the increased T cell effector function occurs up to 6 days, up to 9 days, up to 12 days, up to 15 days, up to 21 days, up to 28 days, up to 35 days, up to 42 days, up to 49 days, up to 56 days, up to 63 days, or up to 71 days after the introducing. 286. The method of any of embodiments 274-285, wherein the T cell stimulation is with an anti-CD3 and anti-CD28 activation reagent. sf-6059407 22474-20028.40 287. The method of any of embodiments 274-286, wherein the T cell is a tumor infiltrating lymphocyte (TIL) therapy. 288. The method of any of embodiments 274-286, wherein the T cell expresses a recombinant receptor. 289. The method of embodiment 288, wherein the recombinant receptor is a CAR or a eTCR. 290. The method of embodiment 288 or embodiment 289, wherein the recombinant receptor is directed against an antigen, and the T cell stimulation is an antigen-specific stimulation of the recombinant receptor. 291. The method of any of embodiments 274-285 and 290, wherein the T cell stimulation is with antigen-expressing target cells. 292. The method of any of embodiments 274-291, wherein the T cell stimulation is a restimulation after at least one prior T cell stimulation of the T cells. 293. The method of any of embodiments 272-292, wherein the T cell is a T cell in a subject, and the method is carried out in vivo. 294. The method of any of embodiments 272-292, wherein the T cell is a T cell from a subject or derived from a cell from the subject, and the method is carried out ex vivo. 295. The method of embodiment 294, wherein the T cell is a primary T cell. 296. The method of embodiment 294, wherein the T cell is derived from a T cell progenitor, a pluripotent stem cell, or an induced pluripotent stem cell. 297. The method of any of embodiments 272-296, wherein the introducing is by transient delivery into the T cell. 298. The method of any of embodiments 272-297, wherein the introducing comprises electroporation, transfection, or transduction. 299. The method of any of embodiments 272-298, wherein the DNA-targeting system, the polynucleotide, the two or more polynucleotides, the vector, or the two or more vectors are transiently present in the T cell. 300. The method of any of embodiments 272-299, wherein the introducing increases transcription of the one or more activation genes in the T cell. 301. The method of any of any of embodiments 272-300, wherein the introducing represses transcription of the one or more repression genes in the T cell. 302. A modified T cell produced by the method of any of embodiments 272-301. sf-6059407 22474-20028.40 303. A method of treating a disease or condition in a subject, the method comprising administering to the subject the modified T cell of any of embodiments 266-271 and 302. 304. A method of increasing T cell persistence in T cells of a subject, the method comprising administering to the subject the DNA-targeting system of any of embodiments 1- 235, the polynucleotide of any of embodiments 236 and 238-247, the two or more polynucleotides of embodiment 237, the vector of any of embodiments 248-256, or the two or more vectors of any of embodiments 257-265. 305. The method of embodiment 304, wherein the T cells are from an adoptive T cell therapy for treating a disease or condition in the subject. 306. The method of embodiment 305, wherein the adoptive T cell therapy comprises T cells expressing a recombinant receptor directed against an antigen associated with the disease or condition. 307. The method of embodiment 305 or embodiment 306, wherein the administration is carried out prior to, concurrently with, or after administration of the adoptive T cell therapy. 308. The method of any of embodiments 305-307, wherein the administration is carried out after administration of the adoptive T cell therapy to the subject and at a time after the numbers or effector function of T cells of the adoptive T cell therapy are reduced, or are suspected of being reduced, in the subject. 309. A method of treating a disease or condition in a subject, the method comprising administering to a subject: an adoptive T cell therapy for treating the disease or condition; and the DNA-targeting system of any of embodiments 1-235, the polynucleotide of any of embodiments 236 and 238-247, the two or more polynucleotides of embodiment 237, the vector of any of embodiments 248-256, or the two or more vectors of any of embodiments 257-265. 310. The method of any of embodiments 305 and 307-309, wherein the adoptive T cell therapy is a tumor infiltrating lymphocyte (TIL) therapy. 311. The method of any of embodiments 305-309, wherein the adoptive T cell therapy comprises T cells expressing a recombinant receptor directed against an antigen associated with the disease or condition. 312. The method of any of embodiments 306-308 and 311, wherein the recombinant receptor is an eTCR or CAR. sf-6059407 22474-20028.40 313. The method of any of embodiments 306-308, 311, and 312, wherein the antigen is a tumor antigen. 314. The method of any of embodiments 303 and 305-313, wherein the disease or condition is a cancer. 315. The method of embodiment 314, wherein the cancer is a hematological cancer or is a solid tumor. 316. The method of any of embodiments 303 and 305-313, wherein the disease or condition is an autoimmune condition and/or an inflammatory condition. 317. The method of any of embodiments 304-316, wherein the administering of the DNA-targeting system, the polynucleotide, the two or more polynucleotides, the vector, or the two or more vectors results in transient delivery to the T cells of the DNA-targeting system, the polynucleotide, the two or more polynucleotides, the vector, or the two or more vectors. 318. The method of any of embodiments 304-317, wherein the administering of the DNA-targeting system, the polynucleotide, the two or more polynucleotides, the vector, or the two or more vectors increases transcription of the one or more activation genes in the T cells. 319. The method of any of any of embodiments 304-318, wherein the administering of the DNA-targeting system, the polynucleotide, the two or more polynucleotides, the vector, or the two or more vectors represses transcription of the one or more repression genes in the T cells. IX. EXAMPLES [0705] The following examples are included for illustrative purposes only and are not intended to limit the scope of the invention. Example 1: A CRISPRi screen for gRNAs targeting genes affecting T-cell phenotype [0706] A library of gRNAs targeting genes involved in T cell regulation was screened in a pooled format in primary human T-cells transiently expressing an exemplary dCas9- transcriptional repressor fusion protein to identify gRNAs that upregulate or downregulate cytokines upon T-cell stimulation. A. Screen of gRNA library for gRNAs that regulate cytokines via CRISPR-based transcriptional interference (CRISPRi) sf-6059407 22474-20028.40 [0707] A library of approximately 13,500 gRNAs was generated. The library was composed of gRNAs targeted to 77 human genes, with approximately 150 gRNAs per transcription start site (TSS), and approximately 500 control gRNAs with spacers not aligned to the human genome. gRNAs were designed according to the protospacer adjacent motif (PAM) sequence for SpCas9 (5’-NGG-3’). The gRNAs were tiled around the TSS of the 77 target genes, generally within 1 kb and closer to the TSS. [0708] The library was screened to identify gRNAs that upregulate or downregulate cytokine production upon T-cell receptor stimulation in cells transiently expressing the transcriptional repressor domain dSpCas9-KRAB (SEQ ID NO:72, encoding SEQ ID NO:73) or dSpCas9-KRAB-DNMT3A/3L (SEQ ID NO:74, encoding SEQ ID NO:75), two exemplary RNA-guided DNA-targeting nuclease-inactivated Cas9 fusion proteins for transcriptional repression of gRNA-targeted genes. FIG.1A and FIG.1B show the screen workflow, which is further described below. [0709] On day 0, primary human CD4+T cells and CD8+T cells (at a ratio of 1:1 CD4+ to CD8+ T cells) were activated with an anti-CD3 and anti-CD28 T cell activation reagent (e.g. T cell TransAct™, Miltenyi Biotec). Experiments were performed with two different donors in parallel. [0710] On day 1, 24 hours post stimulation, T cells were transduced with lentiviral constructs encoding the gRNA library. To be able to enrich for gRNA+ cells, the cells were cotransfected with CD90.1 (Thy1.1) and mCherry as reporters. Cells were incubated overnight at 37°C, 5% CO
2, and then fresh media with cytokine was added 24-30 hours after transduction. At days 3-5, transduction efficiency of the gRNA library was monitored based on mCherry fluorescent signal. On day 5, CD90+ cells were enriched by positive selection. [0711] On day 6, gRNA-enriched cells were electroporated with 0.5 µg mRNA encoding dSpCas9-KRAB, or 1 µg mRNA encoding dSpCas9-KRAB-DNMT3A/3L, per 1E6 cells. Cells were then cultured in fresh media until day 8, at which point the cells were stimulated with the anti-CD3/anti-CD28 T cell activation reagent. T cells were harvested for analysis on days 9 or 12. Cells that were harvested on day 12 were restimulated overnight on day 11 with the anti- CD3/anti-CD28 T cell activation reagent prior to their harvest on day 12. [0712] On day 9 or 12, approximately 15 hours after stimulation or restimulation, cells were stained by intracellular cytokine staining (ICS) for IL-2 production and IFN-gamma (IFNg) production. Cells were analyzed by flow cytometry for cells that were positive or negative for sf-6059407 22474-20028.40 IL-2 and IFN-g, and the following populations of cells were collected: (1) an unsorted population, (2) an IL-2+ population, (3) an IFNg+/IL-2- population, and (4) an IL-2-/IFNg- double-negative (DN) population. An exemplary flow cytometry expression plot and sorted populations are shown in FIG.1B. Screening for an IL-2+ phenotype was chosen as it was expected it may be associated with T cells that are less differentiated, have higher potential for sustained homeostasis, and longevity. Screening for an IFNg+/IL-2- phenotype was chosen as it was expected it may be associated with robust effector T cells with more limited homeostatic potential. gRNAs that facilitate upregulation or downregulation of IL-2 and/or IFNg were expected to be enriched in the respective sorted populations, for example in comparison to the double-negative population. gRNAs that facilitate increased proliferation of transduced cells were expected to be enriched in unsorted cells at day 12 in comparison to day 6 before dCas9 expression. gRNA enrichment was analyzed in order to identify targets whose modulation results in enrichment of cells that express IL-2 and/or IFNg and exhibit increased proliferation, as described below. [0713] To identify enriched gRNAs, genomic DNA was isolated from the populations, amplified, and sequenced. Sequencing was performed to compare the abundance of each gRNA between the respective sorted populations and the unsorted or double-negative population, as described below. B. Identification of gRNAs and genes affecting proliferation, IL-2 expression, and/or IFNg expression. [0714] gRNAs enriched in cells with a negative or positive impact on IL-2 and/or IFN- gamma production, or on proliferation, were identified based on sequencing analysis. gRNA enrichment analysis based on sequencing of sorted cell populations was used to identify gRNAs and corresponding genes that affect proliferation, IL-2 expression, and/or IFN-gamma expression. [0715] gRNAs targeting genes that affect IL-2 and/or IFNg expression were expected to be enriched in the respective FACS-sorted populations, for example in comparison to the double- negative population. For example, a gRNA targeting the transcriptional repressor dCas9-KRAB to a gene whose downregulation leads to upregulation of IL-2 (i.e. a gene that inhibits IL-2 expression) would be expected to be enriched in the IL-2+ population in comparison to the double-negative population. In contrast, a gRNA targeting the transcriptional repressor dCas9- KRAB to a gene whose downregulation leads to downregulation of IL-2 (i.e. a gene that sf-6059407 22474-20028.40 promotes IL-2 expression) would be expected to be depleted from the IL-2+ population in comparison to the double-negative population. [0716] gRNAs targeting genes that affect proliferation were expected to be enriched or depleted from unsorted cells following electroporation with the targeted transcriptional repressor in comparison to unsorted cells pre-electroporation. For example, a gRNA targeting the transcriptional repressor dCas9-KRAB to a gene whose downregulation leads to increased proliferation (i.e. a gene that inhibits proliferation) would be expected to be enriched in unsorted cells at Day 12 (6 days after electroporation) in comparison to Day 6 (before electroporation). In contrast, a gRNA targeting the transcriptional repressor dCas9-KRAB to a gene whose downregulation leads to reduced proliferation (i.e. a gene that promotes proliferation) would be expected to be depleted from unsorted cells at Day 12 (6 days after electroporation) in comparison to Day 6 (before electroporation). [0717] Exemplary results depicted in FIG.2 show hits for gRNAs targeting genes that, when inhibited, either decrease (negative hits) or increase (positive hits) IL-2 expression as determined using a cutoff with an abs(log2 FC) >1 and FDR <0.05. None of the non-targeting control gRNAs were identified as a hit. An exemplary non-targeting control gRNA (non- targeting_sp_1) targeted the sequence of SEQ ID NO: 34, and contained the gRNA spacer sequence SEQ ID NO: 68. Exemplary results depicted in FIG.3 show hits for gRNAs targeting genes that, when inhibited, either decrease (negative hits) or increase (positive hits) proliferation. [0718] FIG.4 shows the number of gRNAs that were hits based on similar analysis at day 9 only, day 12 only, or both day 9 and day 12, for the indicated conditions. Fewer hits were obtained at day 12 than at day 9. Without wishing to be bound by theory, it is believed that hits from day 12 represent gRNAs capable of facilitating extended persistence of the relevant phenotype. [0719] Table E1 summarizes gene targets whose repression by CRISPRi upregulated IL-2 and/or IFN-gamma expression in T cells from one or both donors using either transcriptional repressor domain (KRAB or KRAB-DNMT3A/3L).“IL-2 & IFNg hits” shows genes targeted by gRNAs that were enriched in both the IL-2+ population and in the IFNg+/IL-2- population. “IL- 2 hits” shows genes targeted by gRNAs that were enriched in the IL-2+ population but not in the IFNg+/IL-2- population. “IFNg hits” shows genes targeted by gRNAs that were enriched in the IFNg+/IL-2- population but not in the IL-2+ population. Enriched gRNAs targeting the genes sf-6059407 22474-20028.40 CD5, RASA2, MYB, CBLB and ELOB (underlined in the table) were also found as proliferation hits (e.g. as shown and described for FIG.3). Additional exemplary gene targets whose repression by CRISPRi lead to increased proliferation included PRDM1 and CISH (not shown in Table E1). Exemplary gene targets whose repression by CRISPRi downregulated IL-2 and/or IFN-gamma (i.e. negative hits) included VAV1, LAT, LCP2, and CD28 (not shown in Table E1). Table E1: Summary of Repression Screen Gene Hits

[0720] Table E2 shows exemplary enriched gRNAs for the gene hits described above. Exemplary designed target site (protospacer) sequences are set forth in SEQ ID NOs: 1-33, 150, and 184-191, and gRNA spacer sequences for gRNAs targeting each site are set forth in SEQ ID NOS: 35-67, 163, and 192-199, as shown in Table E2. Each gRNA further comprised a scaffold sequence for SpCas9, comprising the sequence: GUUUAAGAGCUAUGCUGGAAACAGCAUAGCAAGUUUAAAUAAGGCUAGUCCGUU AUCAACUUGAAAAAGUGGCACCGAGUCGGUGC (SEQ ID NO: 69). Table E2: Enriched gRNAs from Repression Screen
sf-6059407 22474-20028.40
sf-6059407 22474-20028.40

Example 2: A CRISPRa screen for gRNAs targeting genes affecting T-cell phenotype [0721] A library of gRNAs targeting genes involved in T cell regulation was screened in a pooled format in primary human T-cells transiently expressing an exemplary dCas9- transcriptional activator fusion protein to identify gRNAs that upregulate or downregulate cytokines upon T-cell stimulation. [0722] The gRNA library described above for Example 1 was screened in a similar format, this time using an exemplary dCas9 fusion protein for transcriptional activation of gRNA- targeted genes, dSpCas9-2xVP64 (SEQ ID NO:76, encoding SEQ ID NO:77). [0723] gRNAs enriched in the sorted populations were identified, revealing gRNAs and corresponding target genes that, when transcriptionally activated, promoted the respective phenotypes. FIG.5 shows an exemplary plot of gRNA abundance in the IL-2+ population versus the IL-2-/IFNg- double negative (DN) population from the activation screen. Positive hits are enriched in the IL-2+ population in comparison to the DN population. [0724] Table E3 shows gene targets whose activation by CRISPRa led to increased IL-2 expression and/or increased IFNg expression, or increased proliferation. Table E4 shows exemplary gRNAs for target genes identified in the activation screen. Table E3: Summary of Activation Screen Gene Hits Gene

Phenotype enrichment when activated
sf-6059407 22474-20028.40
Table E4: Exemplary gRNAs for target genes identified in activation screen
sf-6059407 22474-20028.40
Example 3: Identification of additional genes and gRNAs for modulating T cell phenotypes [0725] Additional genes and gRNAs were identified for modulating T cell phenotypes. [0726] Additional gRNAs were designed for targeted repression of genes including MED12, CCNC, FAS, TGFBR2, and Fli1, as shown in Table E5.A. Additional gRNAs were designed for targeted activation of genes including VAV1 and IL-2, as shown in Table E5.B. Table E5.A: gRNAs for targeted gene repression for modulating T cell phenotypes
sf-6059407 22474-20028.40
sf-6059407 22474-20028.40
Table E5.B: gRNAs for targeted gene activation for modulating T cell phenotypes

Example 4: Validation of gRNAs for epigenetic repression of target genes by CRISPRi [0727] DNA-targeting systems composed of an exemplary dCas9-effector fusion protein for transcriptional repression and guide RNAs (gRNAs) were tested for ability to repress expression of target genes including MED12, CCNC, and FAS in T cells. [0728] Multiple gRNAs targeting the genes were designed by selecting target sites according to the protospacer adjacent motif (PAM) sequence for SpCas9 (5’-NGG-3’). Exemplary designed target site (protospacer) sequences for MED12 are set forth in SEQ ID NOs: 80-90 and gRNA spacer sequences for gRNAs targeting each site are set forth in SEQ ID NOS: 91-101 (Table E5.A). Exemplary designed target site (protospacer) sequences for CCNC are set forth in SEQ ID NOs: 102-112 and gRNA spacer sequences for gRNAs targeting each site are set sf-6059407 22474-20028.40 forth in SEQ ID NOS: 113-123 (Table E5.A). Exemplary designed target site (protospacer) sequences for FAS are set forth in SEQ ID NOs:200-205 and 292-295 and gRNA spacer sequences for gRNAs targeting each site are set forth in SEQ ID NOS:212-217 and 296-299. Each gRNA further comprised a scaffold sequence for SpCas9, comprising the sequence: GUUUAAGAGCUAUGCUGGAAACAGCAUAGCAAGUUUAAAUAAGGCUAGUCCGUU AUCAACUUGAAAAAGUGGCACCGAGUCGGUGC (SEQ ID NO: 69). [0729] Human primary T cells from two different donors were transiently transfected with the generated gRNAs and dSpCas9-KRAB (SEQ ID NO:72, encoding SEQ ID NO:73), and assessed for knockdown of the targeted genes. [0730] For MED12, mRNA was isolated from the transfected cells at 48 hours, 72 hours, 6 days, and 7 days after transfection, and assessed for knockdown of the target gene by qRT-PCR. Exemplary qRT-PCR results for MED12 are depicted in FIG.6A-6B, which show that expression of dSpCas9-KRAB and the indicated MED12-targeting gRNAs led to transcriptional repression of MED12 to varying levels, with some gRNAs demonstrating 90% or greater knock down at 48 hours. Several gRNAs facilitated sustained knockdown at 6 days after transfection. As shown in FIG.6B, delivery of dSpCas9-KRAB and the gRNA MED12_2 (targeting SEQ ID NO:81) led to robust and sustained knockdown of MED12 for at least 7 days following delivery. [0731] For CCNC, mRNA was isolated from the transfected cells at 48 hours and 6 days after transfection, and assessed for knockdown of the target gene by qRT-PCR. Exemplary qRT- PCR results for CCNC are depicted in FIG.6C, which shows that expression of dSpCas9- KRAB and the indicated CCNC-targeting gRNAs led to transcriptional repression of CCNC to varying levels, with some gRNAs demonstrating sustained knockdown of CCNC at 6 days after transfection. [0732] To assess FAS knockdown, cells were assessed by flow cytometry for expression of FAS at 72 hours and 7 days after transduction. Exemplary results for FAS are shown in FIG. 6D, which shows that expression of dSpCas9-KRAB and the indicated FAS gRNAs led to transcriptional repression of FAS to varying levels. Several gRNAs facilitated knockdown at both 72 hours and 7 days after transfection. Example 5: Modulation of T cell phenotypes with DNA-targeting systems sf-6059407 22474-20028.40 [0733] DNA-targeting systems for epigenetic modulation of target genes containing gRNAs identified in the preceding Examples and dCas9 effector fusion proteins for repression or activation were tested for ability to modulate phenotypes in CAR T cells. [0734] For CAR T cell experiments, CD4 and CD8 T cells from healthy donors were thawed and activated on day 0 with the anti-CD3/anti-CD28 T cell activation reagents as described above, and transduced with a polynucleotide encoding a chimeric antigen receptor (CAR) 24 hours after CD3/CD28 activation (day 1). CAR T cells were derived from various donors, and a referenced donor, first donor, or second donor in any given experiment or corresponding figure does not necessarily correspond to the same donor, first donor, or second donor from another experiment or figure. In these experiments, the polynucleotide encoded an exemplary Her2- targeted chimeric antigen receptor (Her2 CAR), although any other suitable CARs could be used. Further, in this experiment, the polynucleotide encoding the CAR also included a truncated EGFR (EGFRt) marker as a surrogate marker for T cells expressing the CAR (i.e. Her2 CAR T cells). Her2 CAR T cells were electroporated, generally on day 4, with mRNA encoding the dCas9 effector fusion protein and pre-transcribed gRNAs to achieve transient expression of the DNA-targeting systems. For transcriptional repression, experiments were carried out with dSpCas9-KRAB (SEQ ID NO:72, encoding SEQ ID NO:73) or dSpCas9- KRAB-DNMT3A/L (SEQ ID NO:74, encoding SEQ ID NO:75). For transcriptional activation, experiments were carried out with dSpCas9-2xVP64 (SEQ ID NO:76, encoding SEQ ID NO:77). In some cases, Her2 CAR T cells were expanded until day 9 and cryopreserved prior to thawing and further functional characterization. Her2 CAR T cells were stimulated by incubation with Her2 antigen-expressing tumor cell lines (i.e. target cells) such as SKOV3, 143B (ATCC CRL-8303), or NCI-H1975 (ATCC CRL-5908) at various time points and CAR T cell : target cell ratios to mimic physiological conditions. In some experiments, Her2 CAR T cells underwent multiple rounds of stimulation (i.e. serial stimulation) by co-culture with target cells. Serial stimulations were performed by replating CAR T cells with fresh target cells every three to four days, at ratios indicated in the Examples below, generally using a ratio of approximately 1:4 CAR T cells:target cells, depending on the experiment. DNA-targeting systems were delivered only once (generally at day 4, as described above), and not re-delivered during re-stimulations. Experiments where data was collected after serial stimulations are indicated in the text and figures. sf-6059407 22474-20028.40 [0735] Resulting phenotypes of the Her2 CAR T cells were assessed based on multiple criteria, including cytokine expression, proliferation, and ability to kill target cells, as described below. [0736] For intracellular cytokine staining (ICS), cells were labeled with antibodies for cytokines (including for IL-2, IFNg, and TNF-alpha (TNFa)), and assessed for intracellular expression of the cytokines by flow cytometry. [0737] Secreted cytokines were measured in the cell culture medium, using immunoassays from Meso Scale Discovery (MSD) (e.g. as described at https://www.mesoscale.com/en/technical_resources/our_technology/our_immunoassays). MSD immunoassays combine electrochemiluminescence and multiarray technology for detection of multiple proteins in a single sample. MSD Immunoassays are sandwich-based and involve a Multi-Spot microplate, where each spot is coated with unique capture antibodies. [0738] To measure proliferation (i.e. fold expansion), CAR T cell numbers were determined and compared before and after stimulations with target cells. To count CAR T cells in co-culture with target cells, total cell counts from the co-culture were multiplied by the proportion of total cells that were CAR T cells (e.g. multiplied by 0.5 if the proportion of total cells that were CAR T cells was 50%), based on expression of the transgenic EGFRt marker as assessed flow cytometry using an anti-EGFR antibody. [0739] To measure ability of CAR T cells to kill target cells (i.e. cytotoxic activity), numbers of target cells were tracked in co-culture with the CAR T cells using an Incucyte automated imaging system. Her2 antigen-expressing target cells expressed a fluorescent tag (Nuclight Red), allowing imaging and quantification of target cell numbers over time to generate a growth curve for the target cells. Reduced fluorescence over time was indicative of decreased target cell numbers, and increased killing. The area under the growth curve was used to calculate a killing index to compare conditions, with a higher killing index reflecting a lower area under the curve and greater killing (i.e. cytotoxic activity) by the CAR T cells. [0740] As shown in FIG.7A, activation of IL-2 with a transiently expressed DNA-targeting system containing dSpCas9-2xVP64 and the IL-2-targeting gRNA targeting SEQ ID NO:78 (IL- 2_1) led to increased IL-2 cytokine production (as assessed by ICS and flow cytometry) in stimulated Her2 CAR T cells from two different donors at day 3 post-electroporation (EP). The Her2 CAR T cells were stimulated with Her2 antigen-expressing cells. IL-2 expression was increased in comparison to control cells lacking the CAR (mock), lacking the gRNA (effector sf-6059407 22474-20028.40 alone), or lacking stimulation with a Her2-expressing cell line (no stim). IL-2 expression was increased in CAR T cells from both donors, and in CAR T cells stimulated with either of two different target cell lines (SKOV3 or 143B). As shown in FIG.7B, the DNA-targeting system for IL-2 activation led to increased IL-2 expression (shown as percentage of cells positive for IL-2) in comparison to control cells at both day 3 and day 7 post-EP. [0741] Her2 CAR T cells were electroporated with a transiently expressed DNA-targeting system for IL-2 activation containing dSpCas9-2xVP64 and the IL-2-targeting gRNA targeting SEQ ID NO:78 (IL-2_1), and assessed for IL-2 mRNA expression levels by RT-qPCR at day 3 and day 7 post-EP. Control cells included cells lacking the CAR (mock), and cells not delivered with the DNA-targeting system (CAR only). IL-2 mRNA expression levels were normalized to the mock control cells. As shown in FIG.7C, electroporation of the DNA-targeting system for IL-2 activation increased IL-2 mRNA expression levels by greater than 60-fold at 3 days (left panel), and greater than 2-fold at 7 days (middle panel) post-EP. At 7 days post-EP, CAR T cells were stimulated with Her2 antigen-expressing tumor cells (NCI H1975), and the cells were further assessed by ICS and flow cytometry for IL-2 protein expression one day later. As shown in FIG.7C (right panel), cells delivered with the DNA-targeting system for IL-2 activation exhibited increased expression of IL-2 as compared to CAR T cells not delivered with the DNA- targeting system. [0742] As shown in FIGS. 8A-8C, activation of VAV1 using a transiently expressed DNA- targeting system containing dSpCas9-2xVP64 and the VAV1-targeting gRNA targeting SEQ ID NO:170 (VAV1_5) led to increased IL-2 (FIG.8A), IFNg (FIG.8B), and TNFa (FIG.8C) intracellular cytokine production (as assessed by ICS and flow cytometry) in stimulated Her2 CAR T cells, as compared to control cells. [0743] As shown in FIG.9, activation of IL-2 or VAV1 using a transiently expressed DNA- targeting system containing dSpCas9-2xVP64 and gRNA IL-2_1 or gRNA VAV1_5, respectively, led to an increase in the percentage of polyfunctional T cells (i.e. triple-positive IL- 2+/IFNg+/TNFa+ T cells) at both day 3 and day 7 post-electroporation (post-EP), in comparison to control cells (as assessed by ICS and flow cytometry). [0744] As shown in FIG.10, repression of CBLB using a transiently expressed DNA- targeting system containing dCas9-KRAB and the indicated CBLB-targeting gRNAs (gRNA CBLB_2, targeting SEQ ID NO:11 or gRNA CBLB_3, targeting SEQ ID NO:12) led to increased IL-2 expression in CD4+ CAR T cells, and increased IFNg expression in CD8+ CAR sf-6059407 22474-20028.40 T cells, in comparison to control CAR T cells electroporated with a non-targeting gRNA (as assessed by ICS and flow cytometry). Expression was increased based on both the percentage of cells quantified as positive, and by mean fluorescence intensity (MFI) of the indicated cytokines, as assessed by FACS. [0745] As shown in FIG.11A, DNA-targeting systems containing dSpCas9-2xVP64 and gRNAs for activation of target genes identified in the preceding Examples and as shown in Table E4 and Table E5.B were tested to identify systems that promote advantageous phenotypes in CAR T cells. DNA-targeting systems containing dSpCas9-2xVP64 and the individual gRNAs were electroporated into Her2 CAR T cells for transient expression, and the Her2 CAR T cells were stimulated by co-culture with the Her2 expressing tumor cell line, 143B. Each condition was assessed (by ICS) based on the percentage of cells with specific intracellular cytokine expression profiles. Cytokine expression was quantified according to % Her2 CAR T cells with the indicated phenotypes (e.g. CD4+/IL-2+, CD4+/IFNg+/TNFa+) as assessed by flow cytometry, and log 2 fold change was calculated with respect to the control condition with a non-targeting gRNA (non-targeting_sp_1). Darker shades correspond to higher cytokine expression, as indicated in the legend. Each condition was assigned a cumulative score according to all cytokine expression measurements and ranked from low to high cytokine expression. As shown in FIG.11A, multiple DNA-targeting systems with different gRNAs increased cytokine expression in comparison to a control non-targeting gRNA, including with gRNAs targeting the genes EOMES, IL-2, TBX21, VAV1, BATF, LCP2, or CD28. [0746] As shown in FIG.11B, transiently expressed DNA-targeting systems for activation of IL-2 or repression of MED12 in CAR T cells led to increased and sustained killing of antigen- expressing target cells in serial stimulation assays (performed as described above in Example 5). For activation of IL-2, CAR T cells were electroporated with a transiently expressed DNA- targeting system containing dSpCas9-2xVP64 and a gRNA targeting IL-2 (IL-2_1, targeting SEQ ID NO:78). For repression of MED12, CAR T cells were electroporated with a transiently expressed DNA-targeting system containing dSpCas9-KRAB and a gRNA targeting MED12 (MED12_2, targeting SEQ ID NO:81). Control cells did not receive the DNA-targeting system (CAR alone) or did not express a CAR (Mock). FIG.11B shows quantification of growth of fluorescent antigen-expressing target cells as determined using the Incucyte automated tracking system. Target cell growth was quantified based on fluorescence every 2 hours and throughout the serial stimulations. After a first stimulation, as expected, control (Mock) cells without a CAR sf-6059407 22474-20028.40 failed to kill antigen-expressing cells, which proliferated. In contrast, CAR T cells effectively killed target antigen-expressing cells, with target cell numbers decreasing over time. After a second and third stimulation, CAR T cells not delivered with a DNA-targeting system (CAR alone) exhibited reduced killing, indicating exhaustion of the T cells. In contrast, CAR T cells delivered with a DNA-targeting system for IL-2 activation or MED12 repression retained killing activity after a second and third stimulation. Fold expansion of the CAR T cells was quantified after the second stimulation, based on counts of CAR T cells at the onset and end of the second stimulation period. As shown in FIG.11C (left panel), CAR T cells delivered with the DNA- targeting system for IL-2 activation or the DNA-targeting system for MED12 repression expanded by approximately 2-fold during the second stimulation, as compared to control cells, which exhibited reduced numbers (mock), or failed to expand (CAR alone). CAR T cell IL-2 secretion was also quantified 24 hours after the second stimulation by MSD immunoassay. As shown in FIG.11C (right panel), CAR T cells delivered with the DNA-targeting system for IL- 2 activation or the DNA-targeting system for MED12 repression exhibited a dramatic increase in secreted IL-2 levels in comparison to both CAR alone and Mock control cells. [0747] The results support using the DNA-targeting systems for activation or repression of target genes for driving advantageous cellular phenotypes in T cells, including increased cytokine expression and cytotoxic activity, for example in cellular therapies. Example 6: Modulation of T cell phenotypes with DNA-targeting systems including multiplexed DNA-targeting systems [0748] DNA-targeting systems for individual or multiplexed gene activation or individual or multiplexed gene repression were tested to identify systems that modulate phenotypes related to T cell effector function. The DNA-targeting systems contained gRNAs or combinations thereof identified in the previous Examples. A. DNA-targeting systems for individual or multiplexed gene activation of IL-2 and VAV1 for promoting polyfunctional cytokine production [0749] DNA-targeting systems containing gRNAs for targeting a target site in VAV1 (gRNA VAV1_5, targeting SEQ ID NO:170), IL-2 (gRNA IL-2_1, targeting SEQ ID NO:78), or both VAV1 and IL-2, and a dSpCas9-2xVP64 effector fusion protein (SEQ ID NO:76, encoding SEQ ID NO:77) were transiently expressed in Her2 CAR T cells, which were stimulated by co- culture with antigen-expressing tumor cell lines, as described in Example 5 above. About 48 sf-6059407 22474-20028.40 hours after transient transfection, IL-2 and IFNg expression were assessed by ICS. ICS flow cytometry plots are shown in FIG.12A for control dSpCas9-2xVP64 effector only (without targeting gRNA), and for DNA-targeting systems targeting either VAV1 or IL-2 individually. FIG.12B shows the results of multiplexed targeting of both VAV1 and IL-2 with the gRNAs targeting VAV1 and IL-2 in combination with dSpCas9-2xVP64. The percent increase in IL-2 production or in polyfunctional cytokines IL-2, IFNg and TNF-alpha is shown in FIG.12C. The results from this experiment show a synergistic increase in IL-2+ and IL-2+IFNg+ cells compared to cells in which only one of the genes was targeted by the DNA-targeting systems. B. DNA-targeting systems for individual or multiplexed gene activation or repression for modulating CAR T cell phenotypes [0750] As shown in FIG.13A and FIG.13B, DNA-targeting systems for repression (using dSpCas9-KRAB) or activation (using dSpCas9-2xVP64) of target genes were tested to identify systems that modulate phenotypes in CAR T cells related to T cell effector function. Included were DNA-targeting systems with a single gRNA (e.g. shown in Table E2, Table E5.A, and Table E5.B), as well as multiplexed DNA-targeting systems for multiplexed activation of IL-2 and VAV-1 (containing gRNAs IL-2_1 and VAV1_5 as shown in Table E5.B), or multiplexed repression of CBLB and MYB (containing gRNAs CBLB_2 and MYB_3, as shown in Table E2). The DNA-targeting systems were electroporated for transient expression into Her2 CAR T cells stimulated by co-culture with a Her2 expressing tumor cell line. Multiple rounds of stimulation were performed, and cells electroporated with the different DNA-targeting systems were assessed based on multiple readouts of T cell effector function after the stimulations, including intracellular cytokine expression (by ICS), cytokine secretion, proliferation, and killing of target cells. The readouts of T cell effector function were assessed after a first, second, and/or third stimulation (shown as stim 1, stim 2, or stim 3 in figures). ICS was performed after a first stimulation only. Assays were performed using CAR T cells derived from a first donor (FIG.13A) and a second donor (FIG.13B). The results are shown as Log2 fold change in comparison to control conditions with a non-targeting gRNA (NT). Darker shades correspond to increased measured T cell effector functions, as indicated in the legend. Each condition was assigned a cumulative score according to all measurements and ranked from low to high T cell effector function. As shown in FIG.13A and FIG.13B, multiple DNA-targeting systems containing different gRNAs increased T cell effector functions, including with gRNAs targeting the genes IL-2 or VAV1 for activation, or CBLB, MED12, MYB, or CCNC for repression. In sf-6059407 22474-20028.40 addition, in this experiment, the multiplexed DNA-targeting systems outperformed many of the non-multiplexed DNA-targeting systems, resulting in increased cytokine production, proliferation, and/or killing. In addition, multiplexed repression of CBLB and MYB resulted in higer cytokine production, proliferation, and/or killing than repression of either CBLB or MYB alone in CAR T cells from the second donor. [0751] As shown in FIG.14, additional DNA-targeting systems for multiplexed activation of combinations of individual genes using gRNAs identified in the preceding Examples and dSpCas9-2xVP64 were assessed for ability to modulate cytokine production in stimulated Her2 CAR T cells. Each DNA-targeting system included dSpCas9-2xVP64 and two gRNAs as indicated for each condition in Table E6. Where the two gRNAs for a condition are the same, the gRNA was delivered at twice the concentration. Control non-multiplexed DNA-targeting systems included one gene-targeting gRNA and one non-targeting gRNA (NT). A control non- targeting system with non-targeting gRNA only was also included (NT + NT). Each condition was assessed for cytokine expression (by ICS) for a number of cytokine expression profiles (e.g. CD4+/TNFa+). The results are shown as Log2 fold change in comparison to control conditions with non-targeting gRNA only (NT + NT). Darker shades correspond to higher cytokine expression, as indicated in the legend. Each condition was assigned a cumulative score according to all cytokine expression measurements and ranked from low to high cytokine expression. As shown in FIG.14, in this experiment, several of the DNA-targeting systems, including multiplexed DNA-targeting systems, increased cytokine production in comparison to the non-targeting system. Furthermore, in this experiment, DNA-targeting systems for multiplexed activation of a first gene and second gene using two different gene-targeting gRNAs led to higher cytokine production than DNA-targeting systems including only one of the gene- targeting gRNAs and optionally a second non-targeting gRNA, for several of the combinations. [0752] Table E6 shows target genes and gRNAs for each condition shown in FIG.14. Table E6: Target genes and gRNAs corresponding to FIG.14

sf-6059407 22474-20028.40

[0753] As shown in FIG.15A and FIG.15B, additional DNA-targeting systems for multiplexed activation or multiplexed repression of combinations of individual genes using gRNAs identified in the preceding Examples and dSpCas9-2xVP64 (for activation) or dSpCas9- KRAB (for repression), were assessed for ability to modulate phenotypes in stimulated Her2 CAR T cells in comparison to control conditions with a single gRNA, a non-targeting gRNA, or no gRNA (dCas only). Target genes and gRNAs for each condition in FIG.15A are shown in Table E7. Target genes and gRNAs for each condition in FIG.15B are shown in Table E8. Each condition was assessed for cytokine expression (by ICS) and proliferation. Results are shown as Log2 fold change in comparison to control conditions (non-targeting gRNA in FIG. 15A; dCas only in FIG.15B). Darker shades correspond to increased proliferation and cytokine expression, as indicated in the legend. Each condition was assigned a cumulative score according to all cytokine and proliferation measurements and ranked from low to high cytokine expression and proliferation. As shown in FIG.15A and FIG.15B, in this experiment, several sf-6059407 22474-20028.40 of the DNA-targeting systems, including multiplexed DNA-targeting systems, increased CAR T cell proliferation and/or cytokine production in comparison to control conditions. Furthermore, DNA-targeting systems for multiplexed activation or multiplexed repression of at least a first gene and second gene using different gene-targeting gRNAs led to higher proliferation and/or cytokine production than DNA-targeting systems including only one of the gene-targeting gRNAs, for several of the combinations. Table E7: Target genes and gRNAs corresponding to FIG.15A

sf-6059407 22474-20028.40
Table E8: Target genes and gRNAs corresponding to FIG.15B

C. DNA-targeting systems for multiplexed gene activation of IL-2 and VAV1, or multiplexed repression of combinations of CBLB, CCNC, MED12, and MYB [0754] DNA-targeting systems for individual or multiplexed gene activation (with dSpCas9- 2xVP64) of IL-2, VAV1, or a combination of IL-2 and VAV-1, or individual or multiplexed gene repression (with dSpCas9-KRAB) of CBLB, CCNC, MED12, MYB, or a combination thereof, were further characterized for ability to modulate phenotypes in CAR T cells related to sf-6059407 22474-20028.40 T cell effector function. The DNA-targeting systems containing gene-targeting gRNAs and dSpCas9 effector fusion proteins for targeted activation or repression were electroporated for transient expression in Her2 CAR T cells that were stimulated with antigen expressing target cells, generally as described above. The CAR T cells were assessed based on proliferation (fold expansion), cytokine expression (both ICS and cytokine secretion) and ability to kill antigen- expressing target cells in co-culture. Negative control conditions included CAR T cells delivered with a dCas-effector and no gRNA, CAR T cells not delivered with a DNA-targeting system (CAR only), and cells not expressing a CAR (mock). Target genes and corresponding gRNAs for these experiments are shown in Table E9. Table E9. Target genes and gRNAs corresponding to FIGS.16-19.

[0755] As shown in FIG.16, in this experiment, DNA-targeting systems, including multiplexed DNA-targeting systems, led to increased proliferation (fold expansion) of CAR T cells in comparison to negative control conditions in a first donor and a second donor. Proliferation data in FIG.16 was measured after a first stimulation with target cells (stim 1). Multiplexed repression of CBLB and MED12 dramatically increased proliferation by 5-10 fold in comparison to controls. [0756] As shown in FIG.17, the DNA-targeting systems for individual or multiplexed gene modulation were further characterized based on ability to modulate secreted cytokines IL-2 and IFNg in CAR T cells. Cytokine secretion was measured by MSD immunoassays, as described above in Example 5. Cyokine data in FIG.17 was measured after a first stimulation with target cells (stim 1). In this experiment, DNA-targeting systems, including multiplexed DNA-targeting systems, increased cytokine secretion in comparison to negative control conditions. Multiplexed activation of IL-2 and VAV1 led to higher secretion of IL-2 than activation of either gene alone. Multiplexed repression of CBLB and MED12 led to higher secretion of both IL-2 and IFNg than repression of either gene alone. sf-6059407 22474-20028.40 [0757] As shown in FIG.18, the DNA-targeting systems for individual or multiplexed gene modulation were further characterized based on ability of electroporated CAR T cells to kill antigen expressing target cells after a third round of stimulation (a model for T cell exhaustion), generally as described above in Example 5. A killing index based on the growth curve of antigen-expressing target cells (area under the curve; AUC) was determined for each condition, as shown in FIG.18. In this experiment, DNA-targeting systems, including multiplexed DNA- targeting systems, increased killing activity in comparison to negative control conditions. Multiplexed activation of IL-2 and VAV1 led to greater killing activity than activation of either gene alone. Multiplexed repression of CBLB and MED12 led to greater killing activity than repression of either gene alone. These results are consistent with a finding that the DNA- targeting systems may improve persistence of antigen-targeted T cells following exposure and activation by target cells. [0758] As shown in FIGS. 19A-19B, the DNA-targeting systems for individual or multiplexed gene modulation were further characterized based on multiple phenotypic readouts of T cell effector function in electroporated CAR T cells derived from a first donor (FIG.19A) or second donor (FIG.19B). Cells were assessed for intracellular cytokine expression (by ICS), secreted cytokine expression, proliferation, and killing, after a first, second, and/or third stimulation, as indicated in the figures (shown as stim 1, stim 2, stim 3). Results are shown as Log2 fold change in comparison to control conditions (CAR alone). Darker shades correspond to increased measured T cell effector functions, as indicated in the legend. Each condition was assigned a cumulative score according to all measurements and ranked from low to high T cell effector function. In this experiment, DNA-targeting systems, including multiplexed DNA- targeting systems, increased T cell effector function in comparison to control conditions. Multiplexed repression of CBLB and MED12 led to increased T cell effector function in comparison to repression of either gene alone in both donors, and multiplexed repression of CBLB and MYB led to increased T cell effector function in comparison repression of either gene alone in the second donor. Multiplexed activation of IL-2 and VAV1 led to increased T cell effector function in comparison to repression of either gene alone in the second donor. [0759] These results demonstrate that simultaneous modifications create synergy and expand options for driving cell phenotype without the potential negative impact of DNA breaks. sf-6059407 22474-20028.40 D. Expansion, Killing, and Cytokine Expression in CAR T cells delivered with DNA-target systems for repression of one or more genes or activation of one or more genes [0760] CAR T cells delivered with DNA-targeting systems for transcriptional repression of one or more genes, or DNA-targeting systems for transcriptional activation of one or more genes, were assessed for cell expansion, target cell killing, and cytokine expression. [0761] T cells from two different donors were thawed, activated, transduced with a Her2 CAR, electroporated with DNA-targeting systems, and serially stimulated by co-culture with Her2 positive NCI-H1975 cells, generally as described above. DNA-targeting systems for repression included dSpCas9-KRAB and one or more gene-targeting gRNAs shown in Table E10. DNA-targeting systems for activation included dSpCas9-2xVP64 and one or more gRNAs shown in Table E10. Table E10. Target genes and gRNAs corresponding to FIGS. 20A-C

[0762] The CAR T cells were assessed for expansion (fold change in CAR T cell numbers; i.e. proliferation) during 4 different time periods, as shown in FIG.20A. The first time period (shown as Production in FIG.20A) was following transduction of the CAR and electroporation of the DNA-targeting system, and prior to stimulation with Her2 antigen-expressing cells. The next three time periods were during a first, second, and third stimulation with Her2 antigen- expressing cells (shown as Round 1 Round 2 and Round 3 in FIG.20), with cell numbers being compared between the onset and the end of each stimulation. As shown in FIG.20A, CAR T cells delivered with DNA-targeting systems for transcriptional repression or transcriptional activation exhibited increased expansion in comparison to control cells (CAR alone; not delivered with a DNA-targeting system) after one or multiple rounds of stimulation with the Her2 antigen-expressing cells. For each condition, expansion results are plotted for each of the two donors (triangles), with average value indicated by vertical line. sf-6059407 22474-20028.40 [0763] The CAR T cells were assessed for ability to kill Her2 antigen-expressing target cells after the second and third rounds of stimulation. A target killing index for each experimental condition with a DNA-targeting system was calculated based on the area under the growth curve (AUC) of antigen-expressing target cells in the experimental condition and mock control cells (no CAR), according to the formula: (Mock AUC – Experimental condition AUC) / Mock AUC. FIG.20B shows killing index for CAR T cells delivered with DNA-targeting systems for activation or repression of genes with the indicated gRNAs. For each experimental condition, results are plotted for each of the two donors (triangles), with average value indicated by vertical line. As shown in FIG.20B, CAR T cells delivered with DNA-targeting systems for transcriptional repression or transcriptional activation exhibited increased killing in comparison to control cells (CAR alone) during the second and third round of stimulation. [0764] The CAR T cells were assessed for cytokine expression by ICS and flow cytometry at Day 9 post-electroporation with the DNA-targeting systems, and after exposure to antigen- expressing target cells. The percentage of CAR T cells that were IL-2+, IFNg+, TNF-alpha+, or IL-2+/IFNg+/TNF-alpha+ (polyfunctional) was assessed. As shown in FIG.20C, CAR T cells delivered with DNA-targeting systems for transcriptional repression or transcriptional activation exhibited increased cytokine expression in comparison to control cells (CAR alone). For each experimental condition, results are plotted for each of the two donors (triangles), with average value indicated by vertical line. [0765] The results support using the DNA-targeting systems for activation or repression of target genes for driving advantageous cellular phenotypes in T cells, including increased proliferation, cytokine expression, cytotoxic activity, for example in cellular therapies. Example 7: Enhanced TGFBR2 and T cell effector function with an alternative fusion protein for targeted transcriptional repression [0766] DNA-targeting systems with TGFBR2-targeting gRNAs and dSpCas9 fusion proteins with different transcriptional repressor effector domains were tested for ability to repress TGFBR2 and modulate T cell effector function. [0767] CD4 and CD8 T cells underwent CD3/CD28 activation and were transduced with Her2 CAR 24 hours later.4 days post-transduction, CAR T cells were electroporated with DNA- targeting systems containing a TGFBR2-targeting gRNA (as shown in Table E5.A) and either a dSpCas9-KRAB fusion protein (mRNA encoding SEQ ID NO:332) or a DNMT3A/L-XTEN80- sf-6059407 22474-20028.40 dSpCas9-KRAB fusion protein (mRNA encoding SEQ ID NO:337). Control cells included CAR T cells delivered with dSpCas9 alone (dSpCas9 Only), mock cells not transduced with a CAR (Mock), and CAR T cells not delivered with a DNA-targeting system (CAR alone).48 hours post-electroporation, TGFBR2 expression was assessed by RT-qPCR, normalized to dSpCas9 Only controls. As shown in FIG.21A, each of the tested TGFBR2-targeting DNA-targeting systems repressed TGFBR2. DNMT3A/L-XTEN80-dSpCas9-KRAB enhanced repression of TGFBR2 in comparison to dSpCas9-KRAB for all 7 of the tested TGFBR2-targeting gRNAs. [0768] The CAR T cells delivered with the transiently expressed TGFBR2-targeting DNA- targeting systems with 3 of the TGFBR2-targeting gRNAs (TGFBR2_1, targeting SEQ ID NO:300; TGFBR2_2, targeting SEQ ID NO:301; and TGBR2_3, targeting SEQ ID NO:302) were serially stimulated with Her2-positive NCI H1975 tumor cells at a ratio of 1:5 CAR T:tumor cells, with serial stimulations occurring 4 days apart, in the presence of 10ng/mL TGFb. 24 hours after the second stimulation, secreted IFN-gamma was measured by MSD immunoassay (as described above), and compared to CAR alone control cells. As shown in FIG. 21B, secreted IFNg was increased in CAR T cells delivered with each of the DNA-targetings systems, including for each of the three tested TGFBR2-targeting gRNAs and with both dSpCas9-KRAB and DNMT3A/L-XTEN80-dSpCas9-KRAB fusion proteins. In addition, DNA- targeting systems with the DNMT3A/L-XTEN80-dSpCas9-KRAB fusion protein dramatically enhanced secretion of IFNg by about 10-fold or more, in comparison to DNA-targeting systems with the dSpCas9-KRAB fusion protein. CAR T cell proliferation was measured after the second stimulation (based on live CAR T cell counts, as described above), and proliferation was normalized to CAR alone control cells. As shown in FIG.21C, proliferation was at least modestly increased in each of the TGFBR2-targeting DNA-targeting systems. In addition, DNA- targeting systems with the DNMT3A/L-XTEN80-dSpCas9-KRAB fusion protein dramatically enhanced proliferation in comparison to DNA-targeting systems with the dSpCas9-KRAB fusion protein. Example 8: TGFBR2 knockdown modulates T cell function [0769] DNA-targeting systems for repression of TGF-beta receptor 2 (TGFBR2) were identified and tested for ability to modulate phenotypes in CAR T cells. [0770] DNA-targeting systems for repression of TGFBR2 containing DNMT3A/L- XTEN80-dSpCas9-KRAB and a gRNA targeting TGFBR2 were delivered to CAR T cells by sf-6059407 22474-20028.40 electroporation for transient expression, generally as described above in Example 5. TGFBR2- targeting gRNA target site (protospacer) sequences are set forth in SEQ ID NOS:300-302 and gRNA spacer sequences for gRNAs targeting each site are set forth in SEQ ID NOS:303-305, as shown in Table E5.A. Negative controls included cells that were CAR T cells delivered with DNMT3A/L-XTEN80-dSpCas9-KRAB (SEQ ID NO:337) and a non-targeting gRNA (NT), or cells that did not express a CAR (mock). [0771] Cell surface expression of TGFBR2 was measured by flow cytometry at multiple time points post-electroporation with the DNA-targeting systems, and the percentage of cells negative for TFGBR2 was quantified. As shown in FIG.21D, the DNA-targeting systems for repression of TGFBR2 led to sustained knockdown of TGFBR2 expression for up to 2 weeks post-electroporation, in comparison to negative control cells. [0772] The CAR T cells delivered with DNA-targeting systems for repression of TGFBR2 were assessed for proliferation in the presence of titrating concentrations of TGF-beta (TGFb) ranging from 0 ng/mL to 10 ng/mL. Cell numbers of CAR T cells electroporated with the DNA- targeting systems were determined at 0 hours and then plated at 10,000 cells per well. Proliferation was monitored at 96 hours post activation with plate-bound anti-CD3/anti-CD28 T cell activation reagents to determine fold expansion. Fold expansion was normalized to 0 ng/mL TGFb conditions. As shown in FIG.21E, negative control cells delivered with DNMT3A/L- XTEN80-dSpCas9-KRAB and a non-targeting gRNA exhibited markedly reduced proliferation upon exposure to TGFb. In contrast, DNA-targeting systems for repression of TGFBR2 facilitated proliferation of CAR T cells even in the presence of TGFb. [0773] The CAR T cells were also assessed for secreted cytokine production when exposed to target antigen-expressing Her2 positive NCI-H1975 tumor cells in the presence of 10 ng/mL TGFb. NCI-H1975 cells were plated at 50,000 cells per well and T cells were plated at 10,000 cells per well for an effector cell to target cell ratio of 5 to 1. At 24 hours post co-culture of CAR T cells with target cells, supernatant was obtained from the wells and IFNg production was measured by MSD immunoassays, generally as described above in Example 5. As shown in FIG.21F, in the presence of TGF-beta at a concentration of 10ng/mL, CAR T cells electroporated with DNA-targeting systems for repression of TGFBR2 exhibited increased secreted IFNg when exposed to antigen-expressing target cells. [0774] The CAR T cells were also assessed for secreted cytokine expression at 96 hours post activation with plate-bound anti-CD3/anti-CD28 T cell activation reagents in the presence of sf-6059407 22474-20028.40 titrating amounts of TGFb. For each concentration of TGFb, secreted cytokine expression was normalized to expression after exposure to 0ng/mL TGFb. As shown in FIG.21G, control activated CAR T cells with a non-targeting gRNA exhibited markedly reduced secretion of IFNg, IL-2, and TNFa upon exposure to TGFb. In contrast, activated CAR T cells delivered with DNA-targeting systems for repression of TGFBR2 exhibited sustained levels of secretion of IFNg, IL-2, and TNFa upon exposure to TGFBR2 at the various concentrations. [0775] The results support using the DNA-targeting systems for repression of TGFBR2 for driving advantageous cellular phenotypes in T cells, such as increased T cell effector function, for example in cellular therapies involving CAR T cells. Example 9: Transient delivery of DNA-targeting systems for activation of IL-2 or repression of MED12 to CAR T cells improves function in vivo [0776] DNA-targeting systems for activation of IL-2 or repression of MED12 were transiently delivered to CAR T cells. The CAR T cells were transplanted to a mouse model with Her2 antigen-expressing tumor cells to assess CAR T cell function in vivo. [0777] Her2 positive NCI-H1975 cells were implanted subcutaneously into the flank of NSG MHC KO mice (immunodeficient NOD scid gamma, major histocompatibility complex knockout mice). Five days after tumor implant, 1 million Her2 CAR T cells were injected intravenously into the tail vein. Experimental mice were injected with CAR T cells previously delivered with a transiently expressed DNA-targeting system for IL-2 activation (dSpCas9- 2xVP64 and gRNA IL-2_1, targeting SEQ ID NO:78), or CAR T cells previously delivered with a transiently expressed DNA-targeting system for MED12 repression (dSpCas9-KRAB and gRNA MED12_2, targeting SEQ ID NO:81). Control mice were injected with CAR T cells not delivered with a DNA-targeting system (CAR alone), were injected with T cells not expressing a CAR (Mock T Cells), or were not injected with T cells (Tumor Alone). Six mice were included in each group. Mice were assessed for survival, tumor volume (measured every 2-3 days), and levels of circulating CAR T cells in the blood after T cell transfusion. The timecourse for the in vivo experiment is shown in FIG.22A. [0778] As shown in FIG.22B and FIG.22C (left), mice injected with CAR T cells delivered with the DNA-targeting system for IL-2 activation or the DNA-targeting system for MED12 repression exhibited better survival than control conditions, including mice injected with CAR T cells not delivered with a DNA-targeting system (CAR Alone). sf-6059407 22474-20028.40 [0779] As shown in FIG.22B and FIG.22C (middle), mice injected with CAR T cells delivered with the DNA-targeting system for IL-2 activation or the DNA-targeting system for MED12 repression exhibited delayed growth or elimination of the tumor. Control mice generally exhibited rapid tumor growth. [0780] As shown in FIG.22B and FIG.22C (right), mice injected with CAR T cells delivered with the DNA-targeting system for IL-2 activation or the DNA-targeting system for MED12 repression exhibited higher levels of the CAR T cells than mice injected with CAR T cells not delivered with a DNA-targeting system (CAR Alone). [0781] The results support using the DNA-targeting systems for activation or repression of target genes for improving T cell function in vivo, including increased circulating CAR T cells, greater tumor killing, and improved survival outcomes in adoptive cell therapies with CAR T cells. Example 10: Enhanced repression, durability, and T cell function with alternative fusion protein for targeted transcriptional repression [0782] Transiently expressed DNA-targeting systems comprising dSpCas9 fusion proteins with different transcriptional repressor effector domains were tested for ability to repress MED12 expression and modulate T cell effector function. [0783] First, CD4 and CD8 T cells were thawed and activated with anti-CD3/anti-CD28 reagents. At 5 days after CD3/CD28 activation, T cells were electroporated with a MED12- targeting gRNA (MED12_2, targeting SEQ ID NO:81), and one of: a control dSpCas9 not fused to a transcriptional repressor effector domain (dSpCas9 control), a dSpCas9-KRAB fusion protein (mRNA encoding SEQ ID NO:332), or DNMT3A/L-XTEN80-dSpCas9-KRAB (mRNA encoding SEQ ID NO:337). Electroporated mRNAs encoding the dSpCas9 fusion proteins further encoded an N-terminal FLAG epitope (SEQ ID NO:364) and a C-terminal P2A-mCherry domain (SEQ ID NO:354) for assessing expression of the transduced fusion proteins. The full mRNA for the dSpCas9-KRAB encoded SEQ ID NO:312, and the full mRNA for the DNMT3A/L-XTEN80-dSpCas9-KRAB encoded SEQ ID NO:317. Expression of MED12 was assessed by RT-qPCR at day 4 and day 21 post-electroporation with the DNA-targeting systems, and expression levels were normalized to expression levels in T cells electroporated with the same fusion protein but with a non-targeting gRNA. As shown in FIG.23, DNA-targeting systems with both dSpCas9-KRAB and DNMT3A/L-XTEN80-dSpCas9-KRAB fusion proteins sf-6059407 22474-20028.40 induced strong repression of MED12 at 4 days post-EP in comparison to dSpCas9 condition, with MED12 expression being reduced by greater than 90% with both fusion proteins. However, at day 21 post-EP, cells delivered with dSpCas9-KRAB and gRNA MED12_2 recovered full MED12 expression in comparison to controls. In contrast, cells delivered with DNMT3A/L- XTEN80-dSpCas9-KRAB and gRNA MED12_2 exhibited sustained repression at greater than 90% knockdown in comparison to controls. The results showed that the transiently expressed DNA-targeting system with DNMT3A/L-XTEN80-dSpCas9-KRAB and MED12-targeting gRNA mediated lasting epigenetic effects as evidenced by robust and sustained repression. [0784] CD4 and CD8 T cells were thawed and activated with anti-CD3/anti-CD28 reagents, and transduced with a Her2 CAR at 24 hours post-activation. Mock T cells not transduced with the Her2 CAR were also included as control cells. At four days after transduction with the Her2 CAR, the CAR T cells were electroporated with DNA-targeting systems containing a MED12- targeting gRNA (selected from: MED12_2 targeting SEQ ID NO:81; MED12_3 targeting SEQ ID NO:82, MED12_4 targeting SEQ ID NO:83, and MED12_7 targeting SEQ ID NO:86), and with mRNA encoding either a dSpCas9-KRAB fusion protein (encoding SEQ ID NO:332) or a DNMT3A/L-XTEN80-dSpCas9-KRAB fusion protein (encoding SEQ ID NO:337). mRNAs encoding the dSpCas9 fusion proteins further comprised an N-terminal FLAG epitope (SEQ ID NO:364) and a C-terminal P2A-mCherry domain (SEQ ID NO:354) for assessing expression of the transduced fusion proteins. The full mRNA for the dSpCas9-KRAB encoded SEQ ID NO:312, and the full mRNA for DNMT3A/L-XTEN80-dSpCas9-KRAB encoded SEQ ID NO:317. Control cells included CAR T cells electroporated with DNMT3A/L-XTEN80- dSpCas9-KRAB alone, CAR T cells electroporated with DNMT3A/L-XTEN80-dSpCas9- KRAB and a non-targeting gRNA (NTg), or T cells not expressing a CAR (Mock). Cells were analyzed at various time points post-electroporation (post-EP) with the DNA-targeting systems to assess targeted gene repression and T cell effector function phenotypes. [0785] First, knockdown of MED12 expression was measured by qRT-PCR. FIG.24A shows MED12 expression at day 10 post-electroporation. As shown in the figure, dSpCas9- KRAB and DNMT3A/L-XTEN80-dSpCas9-KRAB both mediated knockdown of MED12 repression with the various indicated MED12 gRNAs, which was sustained at least until Day 10. DNMT3A/L-XTEN80-dSpCas9-KRAB mediated stronger repression of MED12 at day 10 than dSpCas9-KRAB, including with each of the four tested MED12-targeting gRNAs. The results sf-6059407 22474-20028.40 showed that the DNA-targeting systems induced sustained transcriptional repression of targeted genes, and that DNMT3A/L-XTEN80-dSpCas9-KRAB mediated an enhanced effect. [0786] MED12 expression was measured in the CAR T cells electroporated with DNMT3A/L-XTEN80-dSpCas9-KRAB and MED12-targeting gRNAs, in comparison to control cells, at 2, 7, 10, and 14 days post-delivery. As shown in FIG.24B, MED12 expression was strongly repressed for at least 14 days post-delivery including with each of the MED12-targeting gRNAs. With 3 of the 4 tested gRNAs, MED12 expression was reduced to at or about 10% or less of control levels at day 14. [0787] CD25 cell surface expression, a measure of IL-2 sensitivity, was also measured by flow cytometry in the CAR T cells electroporated with DNMT3A/L-XTEN80-dSpCas9-KRAB and MED12-targeting gRNAs, in comparison to control cells, at 2, 7, 10, and 14 days post- delivery. As shown in FIG.24C, MED12 repression with the transiently expressed DNA- targeting systems led to increased CD25 expression, including a dramatic increase in CD25 expression at 2 weeks post-EP in comparison to control cells. [0788] The CAR T cells electroporated with the transiently expressed DNA-targeting systems containing dSpCas9-KRAB or DNMT3A/L-XTEN80-dSpCas9-KRAB and one of the MED12-targeting gRNAs were serially stimulated with Her2-positive NCI H1975 tumor cells at a ratio of 1:5 CAR T:tumor cells, with serial stimulations occurring 4 days apart.24 hours after the second stimulation, secreted IFN-gamma and IL-2 were measured by MSD immunoassay (as described above), normalized to CAR T cells electroporated with a non-targeting gRNA (CAR NTg). CAR T cell proliferation was measured after the second stimulation (based on live CAR T cell counts, as described above), and was also normalized to CAR T cells electroporated with a non-targeting gRNA (CAR NTg). As shown in FIG.24D, dSpCas9-KRAB with gRNA MED12_2 increased secreted IFNg in the CAR T cells by approximately 2-fold at the assayed timepoint. MED12-targeting systems with DNMT3A/L-XTEN80-dSpCas9-KRAB dramatically increased secreted IFNg in comparison to control cells, by a factor of 30-fold to 50-fold, with all four tested MED12-targeting gRNAs, demonstrating an enhanced effect on CAR T cell effector function in comparison to the DNA-targeting systems with dSpCas9-KRAB. As shown in FIG. 24E, DNA-targeting systems with DNMT3A/L-XTEN80-dSpCas9-KRAB also dramatically increased secreted IL-2 by 20-fold to 30-fold in comparison to control cells with all four tested MED12-targeting gRNAs. IL-2 secretion was also enhanced in comparison to cells delivered with DNA-targeting systems with dSpCas9-KRAB. As shown in FIG.24F, CAR T cells sf-6059407 22474-20028.40 delivered with DNA-targeting systems with both dSpCas9-KRAB and DNMT3A/L-XTEN80- dSpCas9-KRAB exhibited increased proliferation in comparison to control cells by up to 2-fold increased proliferation. Example 11: Transient expression of DNA-targeting systems [0789] An exemplary DNA-targeting system was delivered by electroporation to T cells in accordance with the methods provided herein, and the transient nature of expression of the DNA-targeting system was assessed. [0790] FIG.25 demonstrates transient expression in T cells of an exemplary dSpCas9 protein following delivery by electroporation of mRNA encoding the protein and a non-targeting gRNA or gene-targeting gRNA, in accordance with the transient delivery methods used herein and as described the above Examples. The mRNA encoding the dSpCas9 further comprised a sequence encoding GFP, which was used as a surrogate to detect expression of the dSpCas9 protein in electroporated cells. The cells were assessed at timepoints following electroporation by flow cytometry to determine the percentage of cells expressing the dSpCas9 protein (i.e. % GFP+ cells). Control cells were not electroporated. FIG.25 shows robust expression of the dSpCas9 protein at 3 days post-electroporation, followed by a decline to no detectable dSpCas9 expression after Day 7 post-electroporation. These results confirm transient expression of the DNA-targeting systems described herein, and suggest that the resulting enduring phenotypes result from sustained and/or heritable epigenetic effects in dividing T cells, including gene repression and T cell effector function. Example 12: Identification of fusion proteins for targeted transcriptional repression [0791] Fusion proteins comprising dSpCas9 and transcriptional repressor effector domains in different arrangements and combinations were tested for ability to repress and maintain repression of gene expression when delivered with a gene-targeting gRNA. [0792] 20 different dSpCas9 fusion proteins for transcriptional repression were designed. The 20 different dSpCas9 fusion proteins comprised four general arrangements of components, shown as Fusion Arrangement 1 through 4, in FIG.26A. In the first arrangement (Fusion Arrangement 1), the dSpCas9 fusion protein comprised from N-terminus to C-terminus: dSpCas9, and a variable repression domain. In Fusion Arrangement 2, the dSpCas9 fusion protein comprised from N-terminus to C-terminus: a DNMT3A domain, a DNMT3L domain, an sf-6059407 22474-20028.40 XTEN80 linker, dSpCas9, and a variable repression domain. In Fusion Arrangement 3, the dSpCas9 fusion protein comprised from N-terminus to C-terminus: a DNMT3A domain, a DNMT3L domain, an XTEN80 linker, a variable repression domain, and dSpCas9. In Fusion Arrangement 4, the dSpCas9 fusion protein comprised from N-terminus to C-terminus: a DNMT3B domain, a DNMT3L domain, an XTEN80 linker, dSpCas9, and a variable repression domain. For each of the 4 Fusion Arrangements, 5 different fusion proteins were designed, with each fusion protein comprising a different variable repression domain, selected from a KRAB domain from KOX1 (KOX1(2-99)) (SEQ ID NO:355; KRAB domain used in dSpCas9 fusion proteins of preceding Examples), a KRAB domain from KOX1 (KOX1(1-72)) (SEQ ID NO:356), a KRAB domain from ZIM3 (SEQ ID NO:357), a KRAB domain from ZNF324 (SEQ ID NO:358), and an EZH2 domain (SEQ ID NO:359). Fusion proteins further comprised components such as linkers and NLS sequences, including those shown in Table E12. mRNA encoding each of the fusion proteins was prepared for transient delivery by electroporation, as described above, with mRNA further encoding an N-terminal FLAG epitope (SEQ ID NO:364) and a C-terminal P2A-mCherry domain (SEQ ID NO:354) for assessing expression of the transduced fusion proteins. SEQ ID NOs of the fusion protein sequences without, and with the FLAG and P2A-mCherry domains are shown in Table E11 below. Various components of the fusion proteins from Table E11 are shown in Table E12. Table E11. Fusion proteins for transcriptional repression

sf-6059407 22474-20028.40
Table E12. Components of Fusion Proteins from Table E11 / FIG.26A
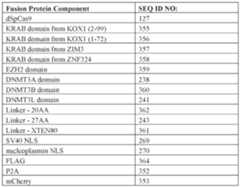
[0793] CD4 and CD8 T cells were thawed and activated with anti-CD3/anti-CD28 reagents. 5 days after T cell activation, cells were electroporated with DNA-targeting systems comprising a MED12-targeting gRNA (MED12_2, targeting SEQ ID NO:81), and the different dSpCas9 fusion proteins with different repressor domains. The fusion proteins were delivered as mRNA encoding the fusion proteins, with mRNAs further encoding P2A-mCherry to confirm expression of the fusion proteins in the electroporated T cells. Control cells were electroporated sf-6059407 22474-20028.40 with mRNA encoding dSpCas9 without a transcriptional repressor effector domain. At day 4 and day 20 post-electroporation with the DNA-targeting systems, MED12 expression was assessed by RT-qPCR.For each experimental condition, results were normalized to expression levels in T cells electroporated with the same fusion protein but with a non-targeting gRNA. [0794] As shown in FIG.26B, as expected, dSpCas9 alone did not repress the targeted gene MED12, whereas the dSpCas9-KRAB fusion protein tested in the previous examples (having KRAB domain KOX1(2-99)) robustly repressed expression of MED12 at 4 days post- electroporation. Several of the other dSpCas9 fusion proteins with different Fusion Arrangements and variable repression domains also repressed expression of MED12 at day 4. Cells electroporated with dSpCas9-KRAB(KOX1(2-99)) did not exhibit sustained repression of MED12 at day 21. In contrast, several of the dSpCas9 fusion proteins induced sustained repression of MED12 at day 21 post-EP. [0795] Taken together, the results show that the exemplary DNA-targeting systems comprising dSpCas9 transcriptional repressor effector domain fusion proteins are capable of inducing heritable epigenetic modifications, including gene repression, to modulate T cell effector function in a durable manner. Example 13: Design of guide RNAs, Screening and Validation A. Identification of Genes and Design of gRNAs Targeting IL-2 Loci Affecting IL-2 Modulation [0796] Guide RNAs (gRNAs) targeted to sites across a selected set of putative regulatory regions of a human IL-2 locus were designed, and packaged for delivery in a lentiviral (LV) library. The sequence of the IL-2 gene body is set forth in SEQ ID NO:452 and the full sequence of the targeted IL-2 locus including the putative regulatory regions is set forth in SEQ ID NO:451. The targeted IL-2 locus spans a region of approximately 150 kb, within human genome assembly GRCh38 (hg38) genomic coordinates chr4:122,451,261-122,593,946 (SEQ ID NO: 539), as shown in FIG.27A. gRNAs were primarily targeted to 7 distinct regions as identified in FIG.27A that exhibit characteristics (e.g. epigenetic marks, regulatory features and relevant transcription factor (TF) motifs) indicative of regulatory regions. [0797] An IL-2 minipool library composed of 1755 guide RNAs (gRNAs), 243 non- targeting guides and two control guides was generated (total of 2000 guides). Proposed gRNAs were cross-referenced against assay for transposase-accessible chromatin (ATAC) and DNAse datasets. Additionally, guides were filtered by epigenome browser criteria, which included sf-6059407 22474-20028.40 examining DNAse and ENCODE regulatory notation. CHOPCHOP, a web tool that facilitates identification and scoring of gRNA targets was used for guide QC. Guides with >0 or >“0MM” were removed. “0MM” refers to a stretch of DNA with zero sequence mis-match (i.e. a perfect alignment). That is, any guides with any 0 MM predicted alignments were removed. In addition, guides with >10 or >“1MM” were also removed. That is, guides with more than 10 predicted alignments with one mis-match were also removed. Moreover, guides with >3 self- complementarity regions were also removed. [0798] The designed gRNAs targeted putative regulatory elements in the IL-2 gene locus for enhancing IL-2 activation. gRNAs were designed for targeting of SpCas9 proteins, and accordingly targeted sites with protospacer adjacent motifs (PAMs) having the sequence NGG, where N is any nucleotide. Each gRNA further comprised a scaffold sequence for SpCas9, comprising the sequence: GUUUAAGAGCUAUGCUGGAAACAGCAUAGCAAGUUUAAAUAAGGCUAGUCCGUU AUCAACUUGAAAAAGUGGCACCGAGUCGGUGC (SEQ ID NO: 69). [0799] A previously validated IL-2-targeting gRNA (also referred to as “IL-2 gRNA-1”) targeted to a region proximal to the transcriptional start site of IL-2 was included as a positive control guide. IL-2 gRNA-1 targeted the sequence set forth in SEQ ID NO:78 (GAGAGCTATCACCTAAGTGT), and comprised a spacer having the sequence set forth in SEQ ID NO:79 (GAGAGCUAUCACCUAAGUGU). B. Screening and Validation of gRNAs Targeting IL-2 Loci Affecting IL-2 Modulation [0800] The minipool library was screened to identify gRNAs that upregulate IL-2 production upon T-cell receptor stimulation in cells transiently expressing an exemplary dCas9 fusion protein for transcriptional activation of gRNA-targeted genes, dSpCas9-2xVP64 (SEQ ID NO:76, encoding SEQ ID NO:77). [0801] Experiments were performed with two different donors (3 replicates). On day 0, primary human CD4+T cells and CD8+T cells (at a ratio of 1:1 CD4+ to CD8+ T cells) were activated with an anti-CD3 and anti-CD28 T cell activation reagent (e.g. T cell TransAct™, Miltenyi Biotec). On day 1, 24 hours post stimulation, T cells were transduced with lentiviral constructs encoding the gRNA library. To be able to enrich for gRNA+ cells, the cells were cotransfected with CD90.1 (Thy1.1) and mCherry as reporters. Cells were incubated overnight at 37°C, 5% CO
2, and then fresh media with cytokine was added 24-30 hours after transduction. sf-6059407 22474-20028.40 [0802] gRNA-enriched cells were electroporated with mRNA encoding dSpCas9-2xVP64, per 1E6 cells. Cells were then cultured and then were stimulated with the anti-CD3/anti-CD28 T cell activation reagent. T cells were harvested for analysis on days 3 (72 hours) or 7. At 3 days (72 hours) and 7 days post electroporation (post-EP), T cells were harvested, sorted by flow cytometry, and assessed for enrichment of gRNAs in an IL-2+ sorted population (compared to cells with no effector and no guide RNAs). [0803] A number of enriched guides that enhanced the expression of IL-2 when transcriptionally activated were found in various regions 50-150 kb upstream of the IL-2 gene. In particular, the initial assessment of hits identified gRNA IL-2_H targeting Region 4 as facilitating strong activation of IL-2, with several other gRNAs with activity exceeding that of the previously validated IL-2 gRNA-1. Four gRNAs were identified to facilitate strong activation of IL-2; they are are (presented from the highest ranked to the fourth ranked): IL-2_H, IL-2_J, IL-2_K, and IL-2_B. An exemplary non-targeting gRNA, SpNT (referred to also as “NT control” in Figures), which was specifically found to be depleted from IL-2+ sorted cells. SpNT targeted the sequence set forth in SEQ ID NO:440 (GAAACATCGACCGAAAGCGT), and comprised a spacer having the sequence set forth in SEQ ID NO:441 (GAAACAUCGACCGAAAGCGU). [0804] Table E13 shows exemplary spCas9 gRNAs for IL-2 activation identified in the activation screen. Table E13: Summary of exemplary spCas9 guide RNAs

sf-6059407 22474-20028.40

[0805] The gRNAs identified above were verified in independent donors. The verified hits were consistent between the two donors, as demonstrated by FIG.27B. Additionally, the hits were similarly distributed across proximal and distal regulatory regions between the two donors, as detailed by FIG.27C. Example 14: Modulation of T cell IL-2 Activation with DNA-targeting systems [0806] DNA-targeting systems for epigenetic modulation of IL-2 containing gRNAs identified in the preceding Example and dCas9 effector fusion proteins for activation were tested for ability to modulate expression of IL-2 in CAR T cells. [0807] For CAR T cell experiments, CD4 and CD8 T cells from three healthy donors were thawed and activated on day 0 with the anti-CD3/anti-CD28 T cell activation reagents, and transduced with a polynucleotide encoding a chimeric antigen receptor (CAR) 24 hours after CD3/CD28 activation (day 1). In these experiments, the polynucleotide encoded an exemplary Her2-targeted chimeric antigen receptor (Her2 CAR), although any other suitable CARs could be used. Further, in this experiment, the polynucleotide encoding the CAR also included a truncated EGFR (EGFRt) marker as a surrogate marker for T cells expressing the CAR (i.e. Her2CAR T cells). Her2 CAR T cells were electroporated, generally on day 4, with mRNA encoding the dCas9 effector fusion protein and pre-transcribed gRNAs to achieve transient expression of the DNA-targeting systems. Experiments were carried out with dSpCas9-2xVP64 (SEQ ID NO:76, encoding SEQ ID NO:77). sf-6059407 22474-20028.40 [0808] In some cases, Her2 CAR T cells were expanded until day 9 and cryopreserved prior to thawing and further functional characterization. Her2 CAR T cells were stimulated by incubation with Her2 antigen-expressing tumor cell lines (i.e. target cells) such as SKOV3, 143B (ATCC CRL-8303), or NCI-H1975 (ATCC CRL-5908) at various time points and CAR T cell : target cell ratios to mimic physiological conditions. In some experiments, Her2 CAR T cells underwent multiple rounds of stimulation (i.e. serial stimulation) by co-culture with target cells. Serial stimulations were performed by replating CAR T cells with fresh target cells every three to four days, generally using a ratio of approximately 1:4 CAR T cells:target cells, depending on the experiment. DNA-targeting systems were delivered only once (generally at day 4, as described above), and not re-delivered during re-stimulations. [0809] Cells were labeled with antibodies for IL-2 and assessed for intracellular expression of IL-2 by flow cytometry. As shown in FIGS.28A, 28B and 28C, transiently expressed DNA- targeting systems for activation of IL-2 in CAR T cells with specific guide RNAs led to sustained expression of IL-2 despite repeated stimulation in serial stimulation assays. For example, guide RNAs IL-2_H and IL-2_J showed a higher percentage of IL-2 at the second stimulation when compared to the percentage of IL-2 using control guide RNA IL-2 gRNA-1, as shown by FIG.28B (control demarcated by solid, horizontal line). As another example, guide RNAs IL-2_H, IL-2_I and IL-2_J showed a higher percentage of IL-2 at the third stimulation when compared to the percentage of IL-2 using control guide RNA IL-2 gRNA-1, as shown by FIG.28C (control demarcated by solid, horizontal line). [0810] FIG.29A and FIG.29B show the percentage of IL2+ T cells and the fold increase in IL2+ T cells, respectively, at the first stimulation and third stimulation for guide RNA IL-2_H as well as other guide hits identified in the screen compared to the control guide RNA IL-2 gRNA- 1. The results demonstrated, for example, that targeting dCas9 effector fusion proteins for activation using guide RNA IL-2_H resulted in a higher percentage of cells being IL2+and a nearly 8-fold increase in IL2+CAR T cells, as compared to the IL-2 gRNA-2 control. In addition, there was a higher IL-2 expression in Her2 CAR T cells that transiently expressed dCas9 effector fusion proteins for activation of IL-2 targeted by guide RNA IL-2_H compared to other guides (FIG.30). [0811] The results support using the identified guide RNAs in conjunction with DNA- targeting systems for activation of IL-2 for driving enhanced expression of IL-2 in T cells. sf-6059407 22474-20028.40 Example 15: Multiplexing of IL-2 Activator Guides to Tune Activity [0812] DNA-targeting systems for individual or multiplexed IL-2 activation were tested to assess multiplexing of IL-2 activator guides across the IL-2 gene locus to tune IL-2 activity. The DNA-targeting systems contained gRNAs or combinations thereof identified in the previous Examples. [0813] Serial stimulation of Her2 CAR T cells was carried out as described in Example 2. Her2 CAR T cells were activated and then electroporated with mRNA encoding the dCas9 effector fusion protein (e.g., dSpCas9-2xVP64; set forth in SEQ ID NO:76 and encoding SEQ ID NO:77) and a single pre-transcribed gRNA or combinations of pre-transcribed gRNAs. Her2 CAR T cells were expanded and then were incubated with Her2 antigen-expressing cells in multiple rounds of serial stimulation. Cells were labeled with antibodies for IL-2 and assessed for intracellular expression of IL-2 by flow cytometry. Table E14 sets forth exemplary paired combinations of gRNAs that were tested, directed to the same or different regions of the IL-2 locus. The combinations of gRNAs also included a combination of five gRNAs, directed to different regions of the IL-2 locus. The five gRNAs used together in a combination were gRNA IL-2_B, gRNA IL2_H, IL-2_I, IL-2_J, and IL-2_O. Table E14: Exemplary paired combinations of spCas9 guide RNAs

[0814] The serial stimulation experiments were further analyzed for IL-2 activation durability. IL-2 activation durability was defined as the retention of IL-2 producing per cell capability. The index is the ratio of the percentage of IL-2+ cells in the third round of serial killing to the percentage of IL-2+ cells in the first round of serial killing. sf-6059407 22474-20028.40 [0815] FIG.31A plots the fold increase in IL-2+ cells (which is the ratio of the absolute count of edited CAR+ IL-2+ cells to the absolute count of NT control, CAR+ IL-2+ cells at the third round of serial killing) against the IL-2 activation durability in an exemplary donor. As shown in FIG.31A, multiplexing of IL-2-targeting gRNAs with the dCas9 effector fusion protein led to both enhanced fold increase in IL2+ cells as well as improved IL-2 activation durability. Select combinations of gRNAs are highlighted. Further targeting dCas9 effector fusion proteins for activation using twice the amount (2X) of pre-transcribed guide RNA IL-2_H also demonstrated potent, durable activation of IL-2. This was superior to targeting dCas9 effector fusion proteins for activation using 2X the control gRNA IL-2 gRNA-1. In all results, multiplexing by using at least 2 gRNAs or a 2X gRNA was better than targeting with single-plex gRNAs, although single-plex targeting with gRNA IL-2_H achieved a relatively high IL-2 activation durability and fold increase in IL-2+ cells compared to single-plex targeting with other IL-2 gRNAs. Similar results were observed with other donors as shown in FIGS.31B and 31C. [0816] The results demonstrate that IL-2 activation can be tuned by combining different IL- 2 activator guides across the IL-2 gene locus. Example 16: Testing IL-2 Activator Guides with Staphylococcus aureus Cas9 Fusion Protein [0817] Guide RNAs (gRNAs) targeting the IL-2 locus were designed and screened for transcriptional activation of IL-2 substantially as described in Example 1. Each gRNA further comprised a scaffold sequence for SaCas9, comprising the sequence: GUUUUAGUACUCUGGAAACAGAAUCUACUAAAACAAGGCAAAAUGCCGUGUUUA UCUCGUCAACUUGUUGGCGAGA (SEQ ID NO: 387). The gRNAs were screened for the ability to increase expression of IL-2 when co-expressed with dSaCas9-2xVP64 (set forth in SEQ ID NO:385 and encoding SEQ ID NO:386) in T cells activated with an anti-CD3 and anti- CD28 T cell activation reagent. Table E15 shows several SaCas9 gRNA candidates identified in the screen. Table E15: Summary of exemplary spCas9 guide RNAs

sf-6059407 22474-20028.40
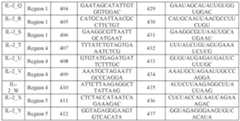
[0818] Serial stimulation experiments were performed as described in Example 2 using Her2 CAR T cells, and ability to increase IL-2 expression was compared to SpCas9 gRNA and systems described in Example 2. In addition to SpNT, another exemplary non-targeting gRNA, SaNT was also used. SaNT targeted the sequence set forth in SEQ ID NO:438 (CCGAATAATTGGTGTAGTGAG), and comprised a spacer having the sequence set forth in SEQ ID NO:439 (CCGAAUAAUUGGUGUAGUGAG). [0819] As shown in FIG.32A and 32B, the SaCas9 gRNAs enhanced IL-2 expression in antigen-stimulated CAR T cells, when compared to IL-2 expression in cells that had not been transiently transfected with the DNA-targeting systems (CAR only cells). The increase in IL-2 expression was particularly pronounced after the second stimulation as shown in FIG.32B for particular exemplary tested gRNAs using either the dSpCas9 or dSpCas9 gRNAs and fusion proteins. Example 17: Combined activation and repression of different genes with DNA-targeting systems [0820] DNA-targeting systems for epigenetic modulation of target genes for combined activation and repression of different genes were tested to identify systems that modulate phenotypes related to T cell effector function. [0821] CD4+ and CD8+ T cells from healthy donors were thawed and activated on day 0 with an anti-CD3/anti-CD28 T cell activation reagent, in this case T cell TransAct™ (Miltenyi sf-6059407 22474-20028.40 Biotec), and transduced with a polynucleotide encoding a CAR 24 hours after CD3/CD28 activation (day 1). The polynucleotide encoded an exemplary Her2-targeted CAR (Her2 CAR), though any other suitable CARs could be used. The polynucleotide encoding the CAR also included a truncated EGFR (EGFRt) marker as a surrogate marker for T cells expressing the CAR (i.e., Her2 CAR T cells). Her2 CAR T cells were electroporated, generally on day 4, with mRNA encoding a dCas9 effector fusion protein for transcriptional activation, mRNA encoding a dCas9 effector fusion protein for transcriptional repression, and pre-transcribed gRNAs to achieve transient expression of the DNA-targeting systems. [0822] For transcriptional activation, a dSpCas9-2xVP64 effector fusion protein (SEQ ID NO:76, encoding SEQ ID NO:77) was used with one of the SpCas9 IL-2-targeting gRNAs shown in Table E16. Each SpCas9 IL-2 targeting gRNA further comprised an SpCas9 scaffold sequence comprising the sequence GUUUAAGAGCUAUGCUGGAAACAGCAUAGCAAGUUUAAAUAAGGCUAGUCCGUU AUCAACUUGAAAAAGUGGCACCGAGUCGGUGC (SEQ ID NO:69). Alternatively for transcriptional activation, a dSaCas9-2xVP64 effector fusion protein (SEQ ID NO:385, encoding SEQ ID NO:386) was used with the SaCas9 IL-2-targeting gRNA shown in Table E16. The SaCas9 IL-2 targeting gRNA further comprised an SaCas9 scaffold sequence comprising the sequence GUUUUAGUACUCUGGAAACAGAAUCUACUAAAACAAGGCAAAAUGCCGUGUUUA UCUCGUCAACUUGUUGGCGAGA (SEQ ID NO:387). [0823] For transcriptional repression, a dSpCas9-KRAB-DNMT3A/L effector fusion protein (SEQ ID NO:74, encoding SEQ ID NO:75) was used with one of the MED12-targeting gRNAs shown in Table E16. Each SpCas9 MED12-targeting gRNA further comprised an SpCas9 scaffold sequence comprising the sequence GUUUAAGAGCUAUGCUGGAAACAGCAUAGCAAGUUUAAAUAAGGCUAGUCCGUU AUCAACUUGAAAAAGUGGCACCGAGUCGGUGC (SEQ ID NO:69). [0824] For assessing T cell effector function, Her2 CAR T cells expressing the DNA- targeting systems were stimulated by incubation with Her2 antigen-expressing, NCI-H1975 (ATCC CRL-5908) tumor cells (i.e., target cells) using a ratio of approximately 1:4 CAR T cells:target cells, after which for intracellular cytokine staining (ICS), cells were labeled with antibodies for IL-2 and assessed for intracellular expression of IL-2 by flow cytometry. [0825] Table E16: gRNAs for combined gene activation and repression sf-6059407 22474-20028.40

[0826] First, the feasibility of using dSpCas9 effector fusion proteins for both IL-2 activation and MED12 repression was tested. For the results shown in FIG.33A, Her2 CAR T cells were delivered mRNA encoding the dSpCas9-KRAB-DNMT3A/L effector fusion protein, a SpCas9 MED12-targeting gRNA, and mRNA encoding the dSpCas9-2xVP64 effector fusion protein. For the results shown in FIG.33B, Her2 CAR T cells were delivered mRNA encoding the dSpCas9-KRAB-DNMT3A/L effector fusion protein, an SpCas9 IL-2-targeting gRNA, and mRNA encoding the dSpCas9-2xVP64 effector fusion protein. As a control, an SpCas9 non- target guide (SpNT, GAAACAUCGACCGAAAGCGU, SEQ ID NO:441) was used. For these experiments, only one gRNA was provided. [0827] As shown in FIG.33A, increased day-8 IL-2 expression was observed with MED12 repression mediated by the dSpCas9-KRAB-DNMT3A/L effector fusion protein, but not when the dSpCas9-2xVP64 effector fusion protein was simultaneously present. Likewise, and as shown in FIG.33B, increased day-8 IL-2 expression was observed with IL-2 activation mediated by the dSpCas9-2xVP64 effector fusion protein, but not when the dSpCas9-KRAB- DNMT3A/L effector fusion protein was simultaneously present. [0828] Thus, increased T cell effector function was not achieved when using dSpCas9 effector fusion proteins for both activation and repression. [0829] Next, the feasibility of using dCas9 effector fusion proteins from different Staphylococcus species for IL-2 activation and MED12 repression was tested (see FIG.34A and FIG.34B). For the results shown in FIG.34C and FIG.34D, Her2 CAR T cells were delivered sf-6059407 22474-20028.40 mRNA encoding the dSpCas9-KRAB-DNMT3A/L effector fusion protein, an SpCas9 MED12- targeting gRNA, mRNA encoding the dSaCas9-2xVP64 effector fusion protein, and the SaCas9 IL-2-targeting gRNA IL-2_X. For the results shown in FIG.34D, the SpCas9 MED12-targeting gRNA MED12_7 was used. As a control, an SaCas9 non-target guide (SaNT, CCGAAUAAUUGGUGUAGUGAG, SEQ ID NO:439) and/or the SpNT guide (SEQ ID NO:441) was used. IL-2 expression was assessed on days 8, 12, and 19. [0830] As shown in FIG.34C and FIG.34D, the simultaneous presence of the SaCas9 IL- 2-targeting gRNA and the SpCas9 MED12-targeting gRNA (alongside their respective dCas9 effector fusion proteins) resulted in increased IL-2 expression at at least one timepoint for multiple SpCas9 MED12-targeting gRNAs, compared to in the presence of the SpCas9 MED12- targeting gRNA alone, the SaCas9 IL-2-targeting gRNA alone, or in the presence of non-target guides only. For multiple MED12-targeting gRNAs, increased IL-2 expression was observed at multiple timepoints. [0831] Further, IL-2 expression and fold IL-2+ Her2 CAR T cell expansion after multiple rounds of co-culture with target cells was evaluated for CAR T cells produced from two healthy donors. For the results shown in FIG.34E and FIG.34F, Her2 CAR T cells were delivered various combinations of the following: mRNA encoding the dSpCas9-KRAB-DNMT3A/L effector fusion protein, the SpCas9 MED12-targeting gRNA MED12_3, mRNA encoding the dSaCas9-2xVP64 effector fusion protein, and one or both of the SaCas9 IL-2-targeting gRNA IL-2_U and the SaCas9 IL-2-targeting gRNA IL-2_X. The SaNT (SEQ ID NO:439) and/or SpNT guides (SEQ ID NO:441) were added to maintain molar equivalency of guide:effector ratio across samples. Three days following delivery of the DNA-targeting systems, CAR T cells were co-cultured with target cells for four days, after which the CAR T cells were rechallenged against target cells. As shown in FIG.34E and FIG.34F, which show IL-2 expression 24 hours after the first and second stimulations, respectively, MED12 repression combined with multiplexed IL-2 activation (both IL-2-targeting gRNAs present) showed increased performance as compared to the CAR-only, single-edited, or non-IL-2-multiplexed groups. As shown in FIG. 34F, MED12 repression combined with multiplexed IL-2 activation led to a >30-fold average increase in absolute viable CD3+CAR+IL-2+ producing cells after re-challenge, compared to the CAR-only control. [0832] Together, these results indicate that combined activation and repression of different genes for increasing T cell effector function can be achieved when using dCas9 effector fusion sf-6059407 22474-20028.40 proteins from different Staphylococcus species. Without wishing to be bound by theory, the use of dCas9 effector fusion proteins from different Staphylococcus species may promote increased T cell effector function by preventing cross-binding of dCas9 effector fusion proteins and gRNAs that results in activation of the gene intended to be repressed, and vice versa. Example 18: Testing IL-2 Activator Guides with Fusion Protein Containing NFN Domain [0833] Exemplary fusion protein dSaCas9-NFN-VP64 (set forth in SEQ ID NO: 529, encoding SEQ ID NO: 528), which is schematically represented in the left panel of FIG.35 and contains dSaCas9 covalently linked to two exemplary transcriptional activator effector domains VP64 (SEQ ID NO: 142) and NCOA3-FOXO3-NCOA3 (NFN; SEQ ID NO: 528), was tested for the ability to transcriptionally activate IL-2, modulate T cell effector function, and maintain cell viability, as compared to previously tested exemplary dSaCas9 or dSpCas9 fusion proteins dSaCas9-2xVP64 (set forth in SEQ ID NO:385, encoding SEQ ID NO:386), as schematically represented in the right panel of FIG.35, and dSpCas9-2xVP64 (set forth in SEQ ID NO:76, encoding SEQ ID NO:77). For the dSpCas9-2xVP64 fusion protein, the exemplary IL-2 targeting gRNA IL-2_H (spacer sequence set forth in SEQ ID NO: 420) was used to target IL-2. For the exemplary dSaCas9 fusion protein, two multiplexed guide RNAs were used: IL-2_U (spacer set forth in SEQ ID NO: 433) paired with IL-2_X (spacer set forth in SEQ ID NO: 436). A. Modulation of T cell IL-2 Activation with Fusion Protein Containing NFN Domain [0834] For assessing T cell effector function using the exemplary fusion protein dSaCas9- NFN-VP64, experiments were performed using serial stimulation experiments performed substantially as described in Example 14, using a readout of intracellular expression of IL-2 by flow cytometry following intracellular cytokine staining (ICS) with IL-2 antibodies. [0835] FIG.36 depicts the intracellular expression of IL-2 after first stimulation using an average of two donors as %IL-2 positive cells in the left panel and mean fluorescent levels (MFI; corresponding to average expression levels) in the right panel, in cells that were delivered mRNA encoding dSaCas9-2xVP64, dSaCas9-VP64-NFN, or dSpCas9-2xVP64 mRNA with corresponding IL-2 targeting gRNA(s). No delivery of exemplary fusion protein (CAR only) was used as a negative control. As shown in FIG.36, all exemplary fusion proteins led to increased transcriptional activation of IL-2 as compared to the negative control. Each fusion sf-6059407 22474-20028.40 protein led to a different level of IL-2 expression, indicating that different combinations of effector domains and species of Cas9 proteins can be combined in various ways and can be used to tune or modulate transcriptional activation of targeted genes, e.g., IL-2. B. Modulation of T cell phenotypes following IL-2 activation and Med12 repression with further repression of TGFBR2 and/or CISH [0836] Following validation of dSaCas9-NFN-VP64 in combination with repression of different genes to modulate T cell effector function, further experiments were carried out to test T cell effector function in which multiplex DNA-targeting systems were used to activate IL-2 expression using dSaCas9-NFN-VP64 and repress Med12, TGFBR2 and/or CISH expression. [0837] IL-2 expression, proliferation, and tumor cell killing using Her2 CAR T cells during multiple rounds of co-culture with target cells substantially as described in Example 14 were evaluated after a first, second, or third stimulation with target cells using dCas9 effector fusion proteins from different Staphylococcus species for IL-2 activation and TGFBR2, MED12 and/or CISH repression. [0838] For the results shown in FIGs.37-39, Her2 CAR T cells were delivered various combinations of DNA-targeting modules for the following: (a) mRNA encoding the dSpCas9- KRAB-DNMT3A/L effector fusion protein, and two or more of (i) the SpCas9 MED12- targeting gRNA (MED12_3, targeting sequence set forth in SEQ ID NO:82), (ii) the SpCas9 CISH-targeting gRNA (CISH_1, targeting sequence set forth in SEQ ID NO: 28), and (iii) the SpCas9 TGFBR2-targeting gRNA (TGFBR2_2, targeting sequence set forth in SEQ ID NO: 301; and (b) the mRNA encoding the dSaCas9-NFN-VP64 effector fusion protein, and the corresponding two SaCas9 IL-2-targeting gRNAs. For comparison, DNA-targeting modules containing the mRNA encoding the dSpCas9-KRAB-DNMT3A/L effector fusion protein and respective gRNA for targeted repression of MED12 with a non-targeting gRNA SaNT, whose spacer sequence is set forth in SEQ ID NO:439; TGFBR2 and MED12; and TGFBR2, MED12 and CISH were tested. For some experiments, a DNA-targeting module containing the mRNA encoding the dSpCas9-KRAB-DNMT3A/L effector fusion protein and respective gRNA for targeted repression of TFGBR2 and a non-targeting gRNA was also used. In addition, for comparison, a DNA-targeting module containing mRNA encoding the dSaCas9-NFN-VP64 effector fusion protein and the IL-2 targeting gRNAs also was tested. As negative controls, cells were also delivered only the non-targeting gRNA SpNT (CAR alone) or no delivery (mock). sf-6059407 22474-20028.40 [0839] Three days following delivery of the DNA-targeting systems, CAR T cells were co- cultured with target cells for four days, after which the CAR T cells were rechallenged against target cells twice, resulting in three total stimulations. As shown in FIG.37, which shows the fold change of IL-2 expressing cells after a second stimulation (left) or third stmulation (right) in the presence of TGF-beta (TGFb), multiplexed repression of TGFBR2, MED12, and CISH combined with IL-2 activation showed increased performance as compared to the CAR-only, or as compared to controls repression only controls or activation only controls. As shown in FIG. 37, the repression of TGFBR2, MED12, and CISH combined with multiplexed IL-2 activation led to a >7-fold average increase in absolute viable CD3+CAR+IL-2+ producing cells after the third stimulation, compared to the CAR-only control. [0840] Proliferation was measured in the presence of 2.5 ng/µL TGF-beta (TGFb) and measured every four hours following first (top) or second (bottom) stimulation in T cells derived from a first donor (FIG.38A) and a second donor (FIG.38B). As shown in FIG.38A and 38B, the presence of TGFb, commonly found in microtumor environments and that can inhibit T cell growth, resulted in little observed proliferation for negative control cells (CAR alone and mock), as expected. Further, despite the presence of TGFb, multiplexed repression of TGFBR2, MED12, and CISH combined with IL-2 activation resulted in the greatest proliferation as compared to the negative control cells, or as compared to repression only controls or activation only controls. [0841] Tumor cell killing (i.e., cytotoxic activity) was measured substantially as previously described in Example 5 following a first, second, and/or third stimulation (described as stim 1, 2, and/or 3) in two separate donors. As shown in FIG.39, any of the tested conditions generally resulted in a decrease in tumor cells, or cytotoxic activity, as compared to a mock delivery (mock) or delivery of a non-targeting guide (CAR alone). Further, multiplexed repression of TGFBR2, MED12, and/or CISH combined with IL-2 activation resulted in more durable or sustained cytotoxic killing after a second or third stimulation as compared to repression alone or IL-2 activation alone. These results support that multiplexed repression may promote persistence of T cell activity. [0842] Together, these results indicate that combined activation and repression of different genes for increasing T cell effector function can be achieved when using dSaCas9-NFN-VP64 for transcriptional activation. Further, the functional effects are tunable or can be modulated by targeting a different combination of genes for activation or repression. sf-6059407 22474-20028.40 C. T cell Viability with Alternative Fusion Proteins [0843] Following validation that multiplexed activation and repression using dSaCas-NFN- VP64 leads to increased T cell effector function, further experiments were performed to assess how such multiplexed activation and repression affected cell health. To assess viability of T cells following electroporation of different DNA-targeting modules, cells were diluted with AO/PI dye and then read viability and live cell counts using a Nexcelom cell counter. Viability was assessed in T cells derived a first donor (FIG.40; left) and a second donor (FIG.40; right) 72 hours post-electroporation of cells with various combinations of DNA-targeting modules. DNA-targeting modules included all the conditions previously described in Example 18B. As controls, delivery of no DNA-targeting modules (CAR only) or DNA-targeting modules where dSpCas2-2xVP64 was used in place of dSaCas9-NFN-VP64 for transcriptional activation were also performed. These conditions included delivering: (a) mRNA encoding dSpCas9-2xVP64, and gRNA IL-2_H or (b) mRNA encoding dSaCas9-2xVP64, and gRNAs IL-2_U and IL-2_X, and mRNA encoding dSpCas9-KRAB-DNMT3A/L, and gRNA MED12_3 (IL-2 dSaCas9- 2xVP64 + MED12). [0844] As shown in FIG.40, cells that had been delivered dSaCas9-NFN-VP64 had comparable or slightly lower viability as compared to controls that did not contain dSaCas9- NFN-VP64 in T cells derived from a first donor (left) and second donor (right). These results indicate that cells remain viable even after delivery of DNA-targeting modules that contained the exemplary fusion protein dSaCas-NFN-VP64 for transcriptional activation. [0845] Together, these results indicate that activation alone as well as combined activation and repression of different genes for increasing T cell effector function can be achieved when using alternative fusion proteins, especially dSpCas9-NFN-VP64. Further, activation alone and combined activation and repression of different genes can be achieved with different fusion proteins, including dSpCas9-NFN-VP64, while maintaining tolerable cell health and viability. Example 19: Design and Validation of ZFPs for Modulation of IL-2, TGFBR2, and MED-12 [0846] Engineered zinc finger protein (ZFP) fusion proteins containing either transcriptional activator or transcriptional repressor effector domains were designed then validated in primary human T-cells to identify ZFPs that upregulated or downregulated cytokines upon T-cell stimulation. Libraries of ZFPs targeting the IL-2 gene, TGFBR2 gene, or Med12 gene were sf-6059407 22474-20028.40 designed. Per given region of interest in a target gene, ten or twelve ZFPs were prioritized for testing based on proximity to a top-performing gRNA and binding score. For the IL-2 gene, ten ZFPs targeting the transcriptional start site (TSS) were selected partially based on proximity to gRNA IL-2_1 (target site set forth in SEQ ID NO: 78), ten ZFPs targeting region 4 were selected partially based on proximity to gRNA IL-2_H (target site set forth in SEQ ID NO: 395), and ten ZFPs targeting region 5 of the IL-2 gene were selected partially based on proximity to gRNA IL-2_J (target site set forth in SEQ ID NO: 397). For the MED12 gene, twelve ZFPs targeting the TSS were selected partially based on proximity to gRNAs MED12_3 (target site set forth in SEQ ID NO: 82) and MED12_4 (target site set forth in SEQ ID NO: 83). For the TGFBR2 gene, twelve ZFPs targeting the TSS were selected partially based on proximity to gRNA TGFBR2_2 (target site set forth in SEQ ID NO: 301. [0847] FIGS. 41-45 show alignments for a subset of designed ZFPs target sites (indicated by rectangles) for a given target region that were used to help select which ZFPs to test: IL-2 gene region 4, IL-2 region 5, IL-2 TSS, MED-12 TSS, and TGFBR2 TSS, respectively. Top- performing ZFPs (see Table E17) and guide RNAs (gRNAs) are annotated. [0848] Fusion proteins containing one of the priotized ZFPs were then designed and screened for cell health and ability to modulate target genes, such as IL-2, as measured by flow cytometry in CAR T cells that were prepared substantially as described previously in Example 14. CAR T cells were electroporated with different concentrations of mRNA encoding ZFP fusion protein. Fusion proteins containing an IL-2 targeting ZFP were designed to contain the exemplary transcriptional activator effector domain NCOA3-FOXO3-NCOA3 (NFN) (SEQ ID NO: 549) and fusion proteins containing a MED12- or TGFBR2-targeting ZFP were designed to contain the exemplary transcriptional repression effector domains KRAB (SEQ ID NO: 70) and DNMT3A/L (SEQ ID NO: 135). As controls, there was either no delivery (CAR alone) or delivery of mRNA encoding a corresponding dCas9 fusion proteins (dSpCas9-2xVP64 for transcriptional activation or dSpCas9-KRAB-DNMT3A/L for transcriptional repression) and gRNA targeting IL-2 (IL-2_H, whose spacer sequence is set forth in SEQ ID NO: 420), MED- 12 (MED-12_3, whose spacer sequence is set forth in SEQ ID NO: 93), or TGFBR2 (TGFBR2_2, whose spacer sequence is set forth in SEQ ID NO: 304), corresponding to the target gene of the ZFP. Among the panel of ZFP fusion proteins tested, ZFP fusion proteins with the strongest transcriptional activity and viability were selected. Representative, top-performing ZFPs and their corresponding ZFP fusion proteins are set forth in Table E17. sf-6059407 22474-20028.40 Table E17: Representative ZFP fusion proteins for targeting IL-2, MED12, and TGFBR2
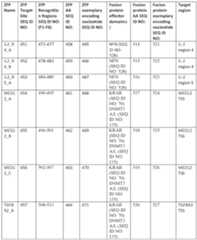
sf-6059407 22474-20028.40 [0849] FIG.46 depicts the expression of IL-2 following delivery of the representative top- performing IL-2-targeting ZFP fusion proteins set forth in Table E17. IL-2 expression was represented as % IL-2 positivity following a first stimulation 72 hours post-delivery (top) or cell counts of IL-2-positive CAR T-cells following a second stimulation (bottom). The results indicate that the three representative top-performing IL-2-targeting ZFP fusion proteins were able to transcriptionally activate IL-2 as compared to CAR alone. At certain concentrations, the level of IL-2 activation achieved by the IL-2 fusion proteins was comparable to the level of expression achieved using dSpCas9-2xVP64 paired with an IL-2 targeting gRNA (IL2_H). [0850] FIGS. 47A and 47B depict the expression of IL-2 and TGFBR2 following delivery of the representative top-performing TGFBR2 ZFP fusion protein set forth in Table E17. In addition to the fusion protein sequence listed in Table E17, each TGFBR2 fusion protein also contained a 3x FLAG tag (SEQ ID NO: 287) on its N-terminus. IL-2 expression was represented as % IL-2 positive CAR T cells following a second stimulation (FIG.47A) and expression of TGFBR2 as % knockdown (KD) of TGFBR2 after either 72 hours post-delivery (FIG.47B; left) or 15 days post-delivery (FIG.47B; right), as assessed by qRT-PCR. The results indicate that the representative TGFBR2_A ZFP-KRAB-DNMT3A/L was able to modulate IL-2 and TGFBR2 expression at levels comparable or better than dSpCas9-KRAB-DNMT3A/L paired with a TGFBR2-targeting gRNA. [0851] FIGS. 48A and 48B depict the expression of IL-2 and CD25 following delivery of the representative top-performing MED12 ZFPs set forth in Table E17. In addition to the fusion protein sequence listed in Table E17, each MED12 fusion protein also contained a 3x FLAG tag (SEQ ID NO: 287) on its N-terminus. IL-2 expression is represented as % IL-2 positive CAR T cells following a second stimulation (FIG.48A) and expression of CD25 as % positivity after either 7 days post-delivery (FIG.48B; top) or 15 days post-delivery (FIG.48B; bottom), as assessed by flow cytometery. The results indicate that the representative top-performing MED12-targeting ZFP fusion proteins were able to modulate IL-2 and CD25 expression at levels comparable or better than dSpCas9-KRAB-DNMT3A/L paired with a MED12-targeting gRNA. [0852] Cell health was assessed as overall cell count 72 hours post-delivery. As shown in FIG.49A-C, which depicts cell count from the representative top-performing IL-2 (FIG.49A), TGFBR2 (FIG.49B), and MED12 (FIG.49C)-targeting ZFP fusion proteins set forth in Table sf-6059407 22474-20028.40 E17, delivery of several of the ZFP fusion proteins led to cell health comparable or better than delivery of a dSpCas9 fusion protein paired with a gRNA targeting the same gene. [0853] All together, these results indicate that ZFP fusion proteins can transcriptionally activate and repress target genes that lead to T cell effector function while maintaining cell viability. Without wishing to be bound by theory, the delivery of ZFPs may be preferable or easier than Cas9 proteins in specific situations due to the more compact size of ZFPs. Example 20: Testing ZFP and dCas9 Fusion Proteins for Multiplexed Repression and Activation [0854] Following validation of ZFP fusion proteins, further experiments were carried out to test T cell effector function in which multiplex DNA-targeting systems were used to activate IL- 2 expression using exemplary dSaCas9 fusion proteins dSaCas9-NFN-VP64 (set forth in SEQ ID NO: 530, encoding SEQ ID NO: 529) or dSaCas9-NFN (set forth in SEQ ID NO: 540, encoding SEQ ID NO: 541) or exemplary IL-2 ZFP fusion proteins (IL2_R4_B ZFP fusion protein set forth in SEQ ID NO: 515 or IL2_R5_A ZFP fusion protein set forth in SEQ ID NO: 516) and to repress MED12, TGFBR2 and/or CISH expression using dSpCas9-KRAB- DNMT3A/L paired with a MED12-, TGFBR2-, and/or CISH-targeting gRNA or, alternatively, an exemplary TGFBR2_A_ZFP fusion protein (set forth in SEQ ID NO: 520) to repress TGFBR2. To modulate a target gene using a dSaCas9 fusion protein, the following gRNAs were used unless otherwise noted: (a) gRNA IL-2_U to target IL-2, (b) gRNA MED12_3 to target MED12, (c) gRNA CISH_1 to target CISH, and (d) TGFBR2_2 to target TGFBR2. [0855] IL-2 expression, percentage of polyfunctional cells, and CAR T cell frequency was evaluated following a first, second, or third stimulation (stim 1, stim 2, or stim 3, respectively) in T cells derived from two donors prepared substantially as previously described in Example 15. Tested conditions included: (a) repression of TGFBR2, MED12, and CISH using dSpCas9- KRAB-DNMT3A/L and corresponding gRNAs, (b) repression of TGFBR2 and MED12 using dSpCas9-KRAB-DNMT3A/L and corresponding gRNAs, (c) repression of MED12 and CISH using dSpCas9-KRAB-DNMT3A/L and corresponding gRNAs, (d) multiplexed repression of TGFBR2, MED12, and CISH using dSpCas9-KRAB-DNMT3A/L paired with activation of IL-2 using dSaCas9-NFN-VP64 with corresponding gRNAs (TGFBR2+MED12+CISH+IL2), (e) multiplexed repression of TGFBR2 and MED12 using dSpCas9-KRAB-DNMT3A/L paired with activation of IL-2 using dSaCas9-NFN-VP64 with corresponding gRNAs, (f) multiplexed sf-6059407 22474-20028.40 repression of TGFBR2 using dSpCas9-KRAB-DNMT3A/L paired with activation of IL-2 using dSaCas9-NFN-VP64 with corresponding gRNAs, (g) multiplexed repression of MED12 using dSpCas9-KRAB-DNMT3A/L paired with activation of IL-2 using dSaCas9-NFN-VP64 with corresponding gRNAs, (h) multiplexed repression of TFGBR2, MED12, and CISH using dSpCas9-KRAB-DNMT3A/L and corresponding gRNAs paired with activation of IL-2 using either IL2_R4_B ZFP fusion protein at a concentration of 50 µg/mL or IL2_R5_A_ZFP fusion protein at a concentration of 12.5 µg/mL, (i) TGFBR2_A ZFP fusion protein at a concentration of 25 µg/mL paired with activation of IL-2 using dSpCas9-2xVP64 and gRNA IL-2_H (spacer sequence set forth in SEQ ID NO: 420), or (j) multiplexed repression of TGFBR2, MED12, and CISH using dSpCas9-KRAB-DNMT3A/L paired with activation of IL-2 using dSaCas9-NFN with corresponding gRNAs (TGFBR2+MED12+CISH+IL2-NFN). As controls, cells were also delivered: (a) dSaCas9-NFN with a non-targeting gRNA SaNT (SEQ ID NO: 439), (b) dSaCas9-NFN-VP64 with a non-targeting gRNA SaNT (SEQ ID NO: 439), (c) no delivery of fusion protein (CAR only), or (d) mock delivery (mock). [0856] FIG.50A depicts the expression of IL-2, shown by % IL-2 positive cells, after the first stimulation using fresh cells. FIG.50B and FIG.50C depict the number of IL-2 positive cells for each condition, normalized to CAR alone, after a second or third stimulation, respectively, in the presence of TGF-beta (TGFb). As shown in FIGs.50A-C, every tested condition resulted in increased IL-2 expression as compared to any of the controls, indicating that targeting the given target genes with any tested combination of representative ZFP or dCas9 fusion proteins leads to modulation of IL-2 expression. Further, each condition led to a different amount of increased IL-2 expression, suggesting that IL-2 expression can be tuned depending on the appropriate combination of target gene and fusion protein type. [0857] FIG.51A-C depict level of polyfunctional T cells after a first, second, or third simulation, respectively. Every tested condition resulted in increased numbers of polyfunctional T cells as compared to any of the controls, indicating that targeting the given target genes with any tested combination of representative ZFP or dCas9-based fusion proteins also leads to modulation of T cell effector function. Further, each condition led to a different amount of increased polyfunctional cells, suggesting that intensity of T cell function can be tuned depending on the appropriate combination of target gene and fusion protein type. [0858] FIG.52 depicts frequency of CAR T cells after each stimulation in cells derived from the first donor (left panel) or second donor (right panel). Cells were previously frozen and sf-6059407 22474-20028.40 then thawed prior to performing the experiment. Every tested condition resulted in increased numbers of CAR T cells as compared to any of the controls. Together, these results indicate that multiplexed activation and repression of exemplary target genes using both Cas9- and ZFP- based epigenetic editors can be tuned to effect T cell effector function. Example 21: Design and Validation of Dual ZFP Fusion Protein Constructs [0859] Constructs encoding both (a) an ZFP fusion protein to transcriptionally activate IL-2 and (b) an ZFP fusion protein to transcriptionally repress MED12, also referred to as a dual ZFP fusion protein construct, were designed and tested to determine if delivery of both ZFP fusion proteins on the same transcript led to transcriptional activation of the IL-2 gene, transcriptional repression of the MED12 gene, and IL-2 cytokine upregulation upon T-cell stimulation. Two constructs were designed, both of which used the exemplary IL2_R5_A ZFP fusion protein (amino acid sequence set forth in SEQ ID NO: 516; exemplary nucleotide sequence encoding fusion protein set forth in SEQ ID NO: 545) as previously described in Example 19 as the ZFP fusion protein to transcriptionally activate IL-2 and the exemplary MED12_A ZFP fusion protein (amino acid set forth in SEQ ID NO: 517; exemplary nucleotide sequence encoding fusion protein set forth in SEQ ID NO: 546) as previously described in Example 19 as the ZFP fusion protein to transcriptionally repress MED12. Both constructs separated each fusion protein with a P2A sequence as set forth in SEQ ID NO: 352, although other 2A peptide sequences or an IRES sequence could also be used. The construct was operably linked to a promoter, specifically a CMV promoter (as set forth in SEQ ID NO: 544), although other promoters suitable for expression of mRNA could also be used. As schematicized in FIG.53, one of the constructs (Construct 1; exemplary nucleic acid sequence set forth in SEQ ID NO: 542) contained, from N- terminus to C-terminus: (a) a sequence encoding the ZFP fusion protein to transcriptionally repress MED12 (SEQ ID NO: 546), (b) P2A (SEQ ID NO: 352), and (c) a sequence encoding the ZFP fusion protein to transcriptionally activate IL-2 (SEQ ID NO: 545). The other construct (Construct 2; exemplary nucleic acid sequence set forth in SEQ ID NO: 543) flipped the position of the two ZFP fusion proteins, so that it contained, from N-terminus to C-terminus: (a) a sequence encoding the ZFP fusion protein to transcriptionally activate IL-2 (SEQ ID NO: 545), (b) P2A (SEQ ID NO: 352), and (c) a sequence encoding the ZFP fusion protein to transcriptionally repress MED12 (SEQ ID NO: 546). sf-6059407 22474-20028.40 [0860] CD8 T cells from at least two healthy donors were thawed and activated on day 0 with the anti-CD3/anti-CD28 T cell activation reagents as previously described and transduced with a polynucleotide encoding a chimeric antigen receptor (CAR) 24 hours after CD3/CD28 activation. CAR T cells were derived from various donors, and a first, second, or third donor in any given experiment or corresponding figure does not necessarily correspond to the same first, second, or third donor from another experiment or figure. In these experiments, the polynucleotide encoded an exemplary Her2-targeted chimeric antigen receptor (Her2 CAR), although any other suitable CARs could be used. Further, in this experiment, the polynucleotide encoding the CAR also included a truncated EGFR (EGFRt) marker as a surrogate marker for T cells expressing the CAR (i.e. Her2 CAR T cells). Her2 CAR T cells were electroporated, generally on day 4, with constructs encoding either of the dual ZFP fusion proteins. As controls, cells were either mock electroporated without delivery of any constructs or electoporated with constructs encoding dSpCas9-2xVP64 (SEQ ID NO: 76, encoding SEQ ID NO: 77) paired with the IL-2 targeting gRNA Sp IL-2_J (target site set forth in SEQ ID NO: 397), or constructs encoding DNMT3A/L-XTEN80-dSpCas9-KRAB (SEQ ID NO:337) paired with the MED12- targeting gRNA MED12_3 (target site set forth in SEQ ID NO: 82). In addition, cells were also electroporated with constructs encoding either the exemplary IL2_R5_A fusion protein as set forth in SEQ ID NO: 516 or the exemplary MED12_A ZFP fusion protein as set forth in SEQ ID NO: 517. For some conditions, only one type of construct was delivered to the cells. For other conditions, a combination of both constructs was delivered together at a concentration of either 75 ug/mL per construct or 37.5 ug/mL per construct. [0861] To evaluate how the dual ZFP constructs affected IL-2 expression, qPCR was performed on RNA harvested from cells derived from either Donor 1 (FIG.54A) or Donor 2 (FIG.54B) one day post-electroporation and levels of IL-2 for each condition were quantified relative to housekeeping gene TATA-binding protein (TBP) and normalized to cells that had been mock electoporated. As shown in FIG.54A and FIG.54B, all constructs targeting IL-2 had IL-2 expression levels higher than that of the mock electroporation, whose expression is indicated by a dotted line, indicating that all these fusion proteins increase IL-2 expression. The increased expression of IL-2 when using the dual ZFP fusion protein constructs, as compared to a mock electroporation, further indicates that the exemplary IL-2-targeting ZFP fusion protein is functional in the dual ZFP fusion protein construct. sf-6059407 22474-20028.40 [0862] To evaluate how the dual ZFP constructs affected MED12 expression, qPCR was performed on RNA harvested from cells derived from any one of the three indicated donors two days post-electroporation and levels of MED12 for each condition were quantified relative to housekeeping gene TBP and normalized to cells that had been mock electoporated. As shown in FIG.55, all constructs targeting MED12 had MED12 expression levels lower than that of the mock electroporation, indicating that all fusion proteins targeting MED12 decreased MED12 expression. The decreased expression of MED12 when using the dual ZFP fusion protein constructs, as compared to a mock electroporation, further indicates that the exemplary MED12- targeting ZFP fusion protein is functional in the dual ZFP fusion protein construct. [0863] To evaluate IL-2+ CAR T cell frequency, flow cytometry was used to measure the amount of cells positive for both IL-2 and truncated EGFR, where truncated EGFR was used as a surrogate marker for T cells expressing the CAR, following intracellular cytokine staining after stimulating the cells three times. As shown in FIG.56, which measures the amount of EGFR and IL-2 postive cells per microliter, all constructs, including the dual ZFP fusion protein constructs, led to greater number of EGFR+/IL-2+ cells than the mock electroporation across cells derived from three different donors, indicating that the use of dual ZFP fusion protein constructs can increase the number of CAR T cells positive for the cytokine IL-2. Taken together, these results indicate that ZFP-based epigenetic editors expressed from the same construct can be used for multiplexed activation and repression of exemplary target genes, such as to effect T cell effector function. [0864] The present invention is not intended to be limited in scope to the particular disclosed embodiments, which are provided, for example, to illustrate various aspects of the invention. Various modifications to the compositions and methods described will become apparent from the description and teachings herein. Such variations may be practiced without departing from the true scope and spirit of the disclosure and are intended to fall within the scope of the present disclosure. SEQUENCES [0865] All sequences of U.S. provisional application No.63/530,049 filed July 31, 2023 (SEQ ID NOs: 1-452), U.S. provisional application No.63/581,952 filed September 11, 2023 (SEQ ID NOs: 1-452), and U.S. provisional application No.63/570,751 filed March 27, 2024 (SEQ ID NOs: 1-541) are incorporated by reference in their entireties. sf-6059407