WO2024234006A1 - Systems, compositions, and methods for targeting liver sinusodial endothelial cells (lsecs) - Google Patents
Systems, compositions, and methods for targeting liver sinusodial endothelial cells (lsecs)Download PDFInfo
- Publication number
- WO2024234006A1 WO2024234006A1PCT/US2024/029153US2024029153WWO2024234006A1WO 2024234006 A1WO2024234006 A1WO 2024234006A1US 2024029153 WUS2024029153 WUS 2024029153WWO 2024234006 A1WO2024234006 A1WO 2024234006A1
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- lnp
- composition
- atgrna
- lipid
- polynucleotide
- Prior art date
- Legal status (The legal status is an assumption and is not a legal conclusion. Google has not performed a legal analysis and makes no representation as to the accuracy of the status listed.)
- Pending
Links
Classifications
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/87—Introduction of foreign genetic material using processes not otherwise provided for, e.g. co-transformation
- C12N15/88—Introduction of foreign genetic material using processes not otherwise provided for, e.g. co-transformation using microencapsulation, e.g. using amphiphile liposome vesicle
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K48/00—Medicinal preparations containing genetic material which is inserted into cells of the living body to treat genetic diseases; Gene therapy
- A61K48/0008—Medicinal preparations containing genetic material which is inserted into cells of the living body to treat genetic diseases; Gene therapy characterised by an aspect of the 'non-active' part of the composition delivered, e.g. wherein such 'non-active' part is not delivered simultaneously with the 'active' part of the composition
- A61K48/0025—Medicinal preparations containing genetic material which is inserted into cells of the living body to treat genetic diseases; Gene therapy characterised by an aspect of the 'non-active' part of the composition delivered, e.g. wherein such 'non-active' part is not delivered simultaneously with the 'active' part of the composition wherein the non-active part clearly interacts with the delivered nucleic acid
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K48/00—Medicinal preparations containing genetic material which is inserted into cells of the living body to treat genetic diseases; Gene therapy
- A61K48/005—Medicinal preparations containing genetic material which is inserted into cells of the living body to treat genetic diseases; Gene therapy characterised by an aspect of the 'active' part of the composition delivered, i.e. the nucleic acid delivered
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K14/00—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof
- C07K14/435—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof from animals; from humans
- C07K14/745—Blood coagulation or fibrinolysis factors
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K14/00—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof
- C07K14/435—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof from animals; from humans
- C07K14/745—Blood coagulation or fibrinolysis factors
- C07K14/755—Factors VIII, e.g. factor VIII C (AHF), factor VIII Ag (VWF)
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/63—Introduction of foreign genetic material using vectors; Vectors; Use of hosts therefor; Regulation of expression
- C12N15/79—Vectors or expression systems specially adapted for eukaryotic hosts
- C12N15/85—Vectors or expression systems specially adapted for eukaryotic hosts for animal cells
- C12N15/86—Viral vectors
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/87—Introduction of foreign genetic material using processes not otherwise provided for, e.g. co-transformation
- C12N15/90—Stable introduction of foreign DNA into chromosome
- C12N15/902—Stable introduction of foreign DNA into chromosome using homologous recombination
- C12N15/907—Stable introduction of foreign DNA into chromosome using homologous recombination in mammalian cells
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K48/00—Medicinal preparations containing genetic material which is inserted into cells of the living body to treat genetic diseases; Gene therapy
- A61K48/0008—Medicinal preparations containing genetic material which is inserted into cells of the living body to treat genetic diseases; Gene therapy characterised by an aspect of the 'non-active' part of the composition delivered, e.g. wherein such 'non-active' part is not delivered simultaneously with the 'active' part of the composition
- A61K48/0025—Medicinal preparations containing genetic material which is inserted into cells of the living body to treat genetic diseases; Gene therapy characterised by an aspect of the 'non-active' part of the composition delivered, e.g. wherein such 'non-active' part is not delivered simultaneously with the 'active' part of the composition wherein the non-active part clearly interacts with the delivered nucleic acid
- A61K48/0041—Medicinal preparations containing genetic material which is inserted into cells of the living body to treat genetic diseases; Gene therapy characterised by an aspect of the 'non-active' part of the composition delivered, e.g. wherein such 'non-active' part is not delivered simultaneously with the 'active' part of the composition wherein the non-active part clearly interacts with the delivered nucleic acid the non-active part being polymeric
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/10—Type of nucleic acid
- C12N2310/20—Type of nucleic acid involving clustered regularly interspaced short palindromic repeats [CRISPR]
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2320/00—Applications; Uses
- C12N2320/30—Special therapeutic applications
- C12N2320/32—Special delivery means, e.g. tissue-specific
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2710/00—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA dsDNA viruses
- C12N2710/00011—Details
- C12N2710/10011—Adenoviridae
- C12N2710/10311—Mastadenovirus, e.g. human or simian adenoviruses
- C12N2710/10341—Use of virus, viral particle or viral elements as a vector
- C12N2710/10343—Use of virus, viral particle or viral elements as a vector viral genome or elements thereof as genetic vector
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2750/00—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA ssDNA viruses
- C12N2750/00011—Details
- C12N2750/14011—Parvoviridae
- C12N2750/14111—Dependovirus, e.g. adenoassociated viruses
- C12N2750/14141—Use of virus, viral particle or viral elements as a vector
- C12N2750/14143—Use of virus, viral particle or viral elements as a vector viral genome or elements thereof as genetic vector
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2800/00—Nucleic acids vectors
- C12N2800/30—Vector systems comprising sequences for excision in presence of a recombinase, e.g. loxP or FRT
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N9/00—Enzymes; Proenzymes; Compositions thereof; Processes for preparing, activating, inhibiting, separating or purifying enzymes
- C12N9/14—Hydrolases (3)
- C12N9/16—Hydrolases (3) acting on ester bonds (3.1)
- C12N9/22—Ribonucleases [RNase]; Deoxyribonucleases [DNase]
Definitions
- LSECsLiver sinusoidal endothelial cells
- FVIIIHuman factor VIII
- Current gene therapiestarget hepatocytes in order to treat hemophilia A.
- the hepatocyteis not the natural site for FVIII synthesis and current gene therapy protocols are eliciting immune responses that require immune suppression (El-Maarri et al., Hamostaseologie Nov;40(S 01):S26- S31. doi: 10.1055/a- 1282-2286 (2020)). Therefore, there is a need in the art that addresses and overcome these shortcomings and enables the delivery of cargo into LSECs for therapeutic purposes.
- compositions and LNP formulationscapable of delivering a cargo into a liver sinusoidal endothelial cell (LSEC).
- the LNP formulations provided hereinare capable of delivering the components needed for genetic engineering (e.g., components needed for Programmable Addition via Site-Specific Targeting) into an LSEC.
- the liveris made up of parenchymal and nonparenchymal cells, like hepatocytes, LSECs, Kupffer, and stellate cells.
- LNPsneed to be precisely engineered due to cellular physiological differences.
- LSECsthe most permeable liver cells, facilitate macromolecule transport between the sinusoidal lumen and hepatocytes due to their high endocytic capacity and numerous fenestrae. Small nanoparticles can pass through these fenestrae, given their diameter (-100 nm in humans, -140 nm in mice). Therefore, LNP size and composition can influence endothelium- specific delivery. This disclosure provides compositions and formulations aimed at facilitating delivery to LSECs.
- compositionincludes: a lipid nanoparticle (LNP) formulation comprising a plurality of LNPs, wherein each LNP comprises: (i) a PEG-modified lipid; (ii) a neutral lipid, or (iii) both (i) and (ii); wherein each LNP encapsulates: (i) the gene editor polynucleotide; (ii) the at least a first attachment- site containing guide RNA (atgRNA); or (iii) both (i) and (ii), wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (LSEC) the (i) a gene editor polynucleotide; (ii) at least a first attachment- site containing guide RNA (atgRNA); (iii) or both (i) and (ii).
- LNPlipid nanoparticle
- the LNP compositionincludes: (a) a gene editor polynucleotide; (b) at least a first attachment- site containing guide RNA (atgRNA); (c) an ionizable lipid; (d) a cholesterol; (e) a PEG-modified lipid; and (f) a neutral lipid, wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (a), (b), (c), (d), (e), (f), or a combination thereof.
- replacement of helper lipids with charged alternatives in lipid nanoparticlesfacilitates targeted mRNA delivery to the LSECs.
- the present disclosureprovides a compositions and formulations for site-specific genetic engineering of LSECs using Programmable Addition via Site-Specific Targeting Elements (PASTE) (see lonnidi et al. doi: 10.1101/2021.11.01.466786; the entirety of lonnidi et al. is incorporated by reference), transposon-mediated gene editing, or other suitable gene editing or gene incorporation technology.
- PASTESite-Specific Targeting Elements
- the present disclosurealso provides compositions and formulations for site-specific genetic engineering of LSECs using Programmable Addition via Site-Specific Targeting Elements (PASTE) as described in U.S. Patent No. 11,572,556 and PCT Publication No. WO 2022/087235A; each of which is hereby incorporated by reference in its entirety.
- PASTESite-Specific Targeting Elements
- this disclosurefeatures a composition
- a lipid nanoparticle (LNP) formulationcomprising a plurality of LNPs, wherein each LNP comprises: (i) an ionizable lipid; (ii) a cholesterol; (iii) a polyethylene glycol (PEG) -modified lipid; (iv) optionally a neutral lipid, or (v) a combination of (i) - (iv); each LNP encapsulates a cargo; wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (LSEC) the encapsulated cargo.
- LSECliver sinusoidal endothelial cell
- the present disclosureprovides a composition comprising: a lipid nanoparticle (LNP) formulation comprising a plurality of LNPs, wherein each LNP comprises: (i) an ionizable lipid; (ii) a cholesterol; and (iii) a polyethyleneglycol (PEG)-modified lipid; and (iv) optionally a neutral lipid, each LNP encapsulates a cargo; wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (LSEC) the encapsulated cargo.
- LNPlipid nanoparticle
- the cargois selected from: an mRNA, an siRNA, an antisense oligonucleotide, a DNA, a guide RNA (gRNA), a protein, a peptide, a chemotherapeutic agent, a small molecule drug, or a combination thereof.
- the cargois an mRNA and a gRNA.
- the mRNAis a gene editor polynucleotide encoding a gene editor polypeptide and the gRNA is an attachment site-containing guide RNA (atgRNA).
- this disclosurefeatures a composition
- a lipid nanoparticle (LNP) formulationcomprising a plurality of LNPs, wherein each LNP comprises: (i) a polyethylene glycol (PEG) -modified lipid; (ii) a neutral lipid, or (iii) both (i) and (ii); wherein each LNP encapsulates: (i) a gene editor polynucleotide; (ii) at least a first attachment-site containing guide RNA (atgRNA); (iii) or both (i) and (ii), wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (LSEC) (i) the gene editor polynucleotide; (ii) the at least a first attachment- site containing guide RNA (atgRNA); or (iii) both (i) and (ii).
- LSECliver sinusoidal endothelial cell
- this disclosurefeatures a lipid nanoparticle (LNP) composition
- LNPlipid nanoparticle composition
- a gene editor polynucleotidecomprising: (a) a gene editor polynucleotide; (b) at least a first attachment- site containing guide RNA (atgRNA); (c) an ionizable lipid; (d) a cholesterol; (e) a polyethylene glycol (PEG)-modified lipid; and (f) optionally a neutral lipid
- LNP formulationis capable of delivering into a liver sinusoidal endothelial cell (a), (b), (c), (d), (e), (f), or a combination thereof.
- the present disclosureprovides a lipid nanoparticle (LNP) composition
- a lipid nanoparticle (LNP) compositioncomprising: (a) a gene editor polynucleotide; (b) at least a first attachment- site containing guide RNA (atgRNA); (c) an ionizable lipid; (d) a cholesterol; and (e) a polyethylene glycol (PEG)- modified lipid; and (f) optionally a neutral lipid, wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (LSEC) the gene editor polynucleotide and the at least first attachment- site containing guide RNA (atgRNA).
- LSECliver sinusoidal endothelial cell
- the ionizable lipidis an ionizable cationic lipid.
- the ionizable cationic lipidis selected from N- (2,3dioleyloxy)propyl)-N,N,Ntrimethylammonium chloride (DOTMA); N- (2,3dioleoyloxy)propyl)-N,N,N-trimethylamntonium chloride (DODAP); N,N-dioleyl-N,N- dimethylammonium chloride (DODAC); 1,2 bis (oleoyloxy)-3-(trimethylammonio) propane (DOTAP); N,NdistearylN,N-dimethylammonium bromide (DDAB); 3-(N — (N,N- dimethylaminoethane)-carbam-oyl)cholesterol (DC-Choi); diheptadecylamidoglycylspermidine (DHGS) and N-(l,2-dimyristyloxyprop-3-yl)-N,N-di
- DOTMAN- (2
- composition or LNP compositionfurther comprising a anionic lipid.
- the ionizable lipidis replaced with an anionic lipid.
- the anionic lipidis selected from: a phospholipid, phospholipid derivative, a sterol, a fatty acid, or a combination thereof.
- the PEG-modified lipidis select from 1,2-Dimyristoyl-sn-glycerol methoxypolyethylene glycol (PEG-DMG), PEG-disteryl glycerol (PEG-DSG), PEG- dipalmetoleyl, PEG-dioleyl, PEG-distearyl, PEG-diacylglycamide (PEG-DAG), PEG- dipalmitoyl phosphatidylethanolamine (PEG-DPPE), and PEG-l,2-dimyristyloxlpropyl-3-amine (PEG-c-DMA), or a combination thereof.
- PEG-DMG1,2-Dimyristoyl-sn-glycerol methoxypolyethylene glycol
- PEG-DSGPEG-disteryl glycerol
- PEG- dipalmetoleylPEG-dioleyl
- PEG-distearylPEG-diacylg
- the PEG-modified lipidcomprises a moiety capable binding to a receptor on a liver sinusoidal endothelial cell.
- the moietyis capable of binding to a mannose receptor, a stabilin receptor, an Fc receptor, or a combination thereof.
- the moietyis a mannose.
- the PEG-modified lipidis a mannose-PEG lipid.
- the neutral lipidis a phospholipid.
- the phospholipidis a natural phospholipid selected from: phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylglycerol (PG), phosphatidylserine (PS), phosphatidylinositol (PI), Phosphatidic acid (phosphatidate) (PA), dipalmitoylphosphatidylcholine, monoacyl-phosphatidylcholine (lyso PC), l-palmitoyl-2-oleoyl- sn-glycero-3-phosphocholine (POPC), N-Acyl-PE, phosphoinositides, and phospho sphingolipids, or a combination thereof.
- PCphosphatidylcholine
- PEphosphatidylethanolamine
- PGphosphatidylglycerol
- PSphosphatidylserine
- PIphosphatidylinositol
- PAP
- the phospholipidis a phospholipid derivative selected from: phosphatidic acid (DMPA, DPPA, DSPA), phosphatidylcholine (DDPC, DLPC, DMPC, DPPC, DSPC, DOPC, POPC, DEPC), phosphatidylglycerol (DMPG, DPPG, DSPG, DOPG), phosphatidylethanolamine (DMPE, DPPE, DSPE DOPE), and phosphatidylserine (DOPS), or a combination thereof.
- DMPAphosphatidic acid
- DDPCphosphatidylcholine
- DDPCDLPC
- DMPCDPPC
- DSPCDOPC
- POPCPOPC
- DEPCphosphatidylglycerol
- DMPEDPPG, DSPG, DOPG
- DOPGphosphatidylethanolamine
- DOPSphosphatidylserine
- the phospholipidis selected from: l,2-dioleoyl-sn-glycero-3- phospho-rac-(l-glycerol) sodium salt (DOPG); l,2-dilinoleoyl-sn-glycero-3-phosphocholine (DLPC); 1,2-dimyristoyl-sn-glycero-phosphocholine (DMPC); 1 ,2-dioleoyl-sn-glycero-3- phosphocholine (DOPC); l,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC); 1,2-distearoyl- sn-glycero-3-phosphocholine (DSPC); 1,2-diundecanoyl-sn-glycero-phosphocholine (DUPC); 1- palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC); l,2-di-
- the phospholipidis DOPG.
- each LNP in the LNP formulationcomprises: (i) modified PEGylated lipids; and (ii) neutral lipids.
- each LNP in the LNP formulationcomprises (i) PEG-modified lipids comprising a moiety capable binding to a receptor on a liver sinusoidal endothelial cell (LSEC); and (ii) phospholipids.
- LSECliver sinusoidal endothelial cell
- each LNP in the LNP formulationcomprises mannose-PEG lipids and DOPGs.
- the LNPfurther comprises: cholesterol and an ionizable lipid.
- each LNP in the LNP formulationranges from about 100 nm to about 200 nm. In some embodiments, each LNP in the LNP formulation ranges from about 85 nm to about 120 nm. In some embodiments, each LNP in the LNP formulation ranges from about 45 nm to about 65 nm. In some embodiments, each LNP in the LNP formulation ranges from about 40 nm to about 50 nm.
- the encapsulation efficiencyis about 80% to about 100%. In some embodiments, the encapsulation efficiency is no less than 84%. In some embodiments, the encapsulation efficiency is no less than 90%. [0033] In some embodiments, the polydispersity index of the composition ranges from about 0.005 to 0.5. In some embodiments, the polydispersity index of the composition ranges from about 0.025 to 0.4.
- the gene editor polynucleotidecomprises a nucleotide sequence encoding a nickase and a nucleotide sequence encoding a reverse transcriptase.
- the nickaseis linked to the reverse transcriptase by a linker.
- the first atgRNAcomprises: (i) a domain that is capable of guiding the nickase to a target sequence; and (ii) a reverse transcriptase (RT) template that comprises at least a portion of an at least first integration recognition site, whereby the at least portion of the at least first integration recognition site is integrated into the genome of the cell at the target sequence.
- RTreverse transcriptase
- the LNPsfurther comprises a nicking gRNA.
- the LNPsfurther comprise a second atgRNA.
- the first atgRNA and the second atgRNAare an at least first pair of atgRNAs, wherein the at least first pair of atgRNAs have domains that are capable of guiding the prime editor system to a target sequence; the first atgRNA further includes a first RT template that comprises at least a portion of the first integration recognition site; the second atgRNA further includes a second RT template that comprises at least a portion of the first integration recognition site, and the first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site, whereby the first integration recognition site is integrated into the genome of the cell at the target sequence.
- the LNP or LNP compositionfurther comprising an integration enzyme or a polynucleotide encoding an integration enzyme.
- the integration enzyme or the polynucleotide encoding the integration enzymeis encapsulated in an LNP along with (i) a gene editor polynucleotide; (ii) at least a first attachment- site containing guide RNA (atgRNA); or (iii) both (i) and (ii),
- the LNP or LNP compositionfurther comprising a donor polynucleotide template.
- this disclosurefeatures a liver sinusoidal endothelial cell (LSEC) comprising any of the compositions or LNP compositions described herein.
- this disclosurefeatures a genetically engineered liver sinusoidal endothelia cell (LSEC) made with any of the compositions or LNP compositions described herein.
- LSECliver sinusoidal endothelia cell
- this disclosurefeatures a method for site-specifically incorporating an integration recognition site into a liver sinusoidal endothelial cell (LSEC) genome at a target DNA sequence, comprising: incorporating an integration target recognition site into the genome by delivering into the cell: any of the compositions or LNP compositions described herein.
- LSECliver sinusoidal endothelial cell
- this disclosurefeatures a method for site-specifically integrating a donor polynucleotide template into a liver sinusoidal endothelial cell (LSEC) genome at a target DNA sequence, comprising: incorporating an integration recognition site into the genome by delivering into the cell: any of the compositions or LNP compositions described herein; and integrating the donor polynucleotide template into the genome by delivering into the cell: an integration enzyme; and a donor polynucleotide template, wherein the donor polynucleotide template is linked to a sequence that is an integration cognate of the integration recognition site present in the atgRNA, and wherein the donor polynucleotide template is integrated into the genome at the incorporated genomic integration recognition site by the integration enzyme.
- LSECliver sinusoidal endothelial cell
- this disclosurefeatures a genetically engineered liver sinusoidal endothelia cell (LSEC) made by using any of the composition or LNP compositions described herein.
- LSECliver sinusoidal endothelia cell
- FIG. lshows a non-limiting illustration of a gene editor construct packaged within a lipid nanoparticle (LNP).
- LNPlipid nanoparticle
- FIG. 2illustrates the donor template (i.e., “cargo” or “payload” or “template polynucleotide”)) packaged within a vector.
- FIG. 3illustrates integrase-mediated self-circularization of the donor template (template polynucleotide) within viral genome.
- the circularized donor templateis capable of being genomically incorporated into an orthogonal integrase target recognition site (i.e., “beacon”).
- FIG. 4shows non-limiting illustrations of a gene editor construct packaged within a lipid nanoparticle and an atgRNA, ngRNA, and donor template (i.e., template polynucleotide encoding a gene of interest) packaged within a vector.
- GOIgene of interest.
- PGIprogrammable gene insertion.
- U6U6 promoter.
- atgRNAattachment site-containing guide RNA (atgRNA).
- FIG. 5shows non-limiting illustrations of a gene editor construct (e.g., mRNA encoding PE2-BxBl) and a nicking guide RNA (ngRNA) packaged within a lipid nanoparticle (LNP) and an atgRNA and donor template (i.e., template polynucleotide encoding a gene of interest) packaged within a vector.
- a gene editor constructe.g., mRNA encoding PE2-BxBl
- ngRNAnicking guide RNA
- LNPlipid nanoparticle
- an atgRNA and donor templatei.e., template polynucleotide encoding a gene of interest
- FIGs. 6A-6Bshow non-limiting illustrations of three self-complementary AAV (scAAV) genomes capable of recombinase/integrase-mediated self-circularization.
- FIG. 6Ashows the structure of the three self-complementary AAV (scAAV) genomes capable of recombinase/integrase-mediated self-circularization.
- FIG. 6Ashows the structure of the three self-complementary AAV (scAAV) genomes capable of recombinase/integrase-mediated self-circularization.
- FIG. 6Bshows non-limiting examples of sequences that enable self-circularization (e.g., LoxP AttP GT (SEQ ID NO: 568 and SEQ ID NO: 569); FRT AttP GT (SEQ ID NO: 570 and SEQ ID NO: 571); and AttB CC AttP GT (SEQ ID NO: 572 and SEQ ID NO: 573)).
- GTindicates an AttP site with a GT dinucleotide.
- AttB CCindicates an AttB site with a CC dinucleotide.
- LoxPa LoxP recombinase recognition site.
- FRTa FRT recombinase recognition site.
- FIG. 7shows a non-limiting illustration of recombinase/integrase-mediated intramolecular circularization products.
- FIGs. 8A-8Bshow non-limiting illustrations of a ddPCR assay and intramolecular circularization ddPCR detection probes.
- FIG. 8Ashows a non-limiting illustration of the ddPCR strategy.
- FIG. 8Bshows non-limiting examples of the universal probe (SEQ ID NO: 574 and SEQ ID NO: 575) and an AttR probe (SEQ ID NO: 576 and SEQ ID NO: 577) that can be used in the assay shown in FIG. 8A.
- FIG. 9shows a non-limiting illustration of a pDNA genome and AAV transfection and screening protocol.
- FIG. 10shows data for circularization of AAV pDNA and packaged AAV genomic DNA with Bxbl.
- FIG. 11shows data for Cre-, FLPe-, and Bxbl-mediated circularization of AAV pDNA confirmed by ddPCR.
- FIG. 12shows Cre-, FLPe-, and Bxbl-mediated circularization of packaged AAV confirmed by ddPCR
- FIG. 13shows percent circularization between the Bxbl-mediated attR scar ddPCR probe (“attR probe” described in FIG. 8B) and the universal ddPCR probe (“universal probe” described in FIG. 8B).
- FIGs. 14A-14Eshows analysis of AttP variants.
- FIG. 14Ashows a non-limiting schematic of AttP mutations tested for improving integration efficiency (SEQ ID NOS: 394 and 540-542, respectively, in order of appearance).
- FIG. 14Bshows integration efficiencies of wildtype and mutant AttP sites across a panel of AttB lengths.
- FIG. 14Cshows a non-limiting schematic of multiplexed integration of different cargo sets at specific genomic loci. Three fluorescent cargos (GFP, mCherry, and YFP) are inserted orthogonally at three different loci (ACTB, LMNB1, NOLC1) for in-frame gene tagging.
- FIG. 14Ashows a non-limiting schematic of AttP mutations tested for improving integration efficiency (SEQ ID NOS: 394 and 540-542, respectively, in order of appearance).
- FIG. 14Bshows integration efficiencies of wildtype and mutant AttP sites across a panel of AttB lengths.
- FIG. 14Cshows
- FIG. 14Dshows orthogonality of top 4 AttB/AttP dinucleotide pairs evaluated for GFP integration with PASTE at the ACTB locus.
- FIG. 15illustrates a schematic of single atgRNA and dual atgRNA approaches for beacon placement (“integration recognition site”).
- FIG. 16shows percent beacon placement in primary mouse hepatocytes (PMH) following delivery of mRNA to deliver a polynucleotide encoding a gene editor polynucleotide construct and an AAV to deliver the first and second atgRNA according to the following conditions: (i) concurrent delivery (“co-dose”), (ii) AAV delivery followed by a “1-day delay” before delivery of the mRNA, or (iii) AAV delivery followed by a “2-day delay” before delivery of the mRNA.
- FIG. 17shows percent beacon placement in primary human hepatocytes (PHH) following delivering of mRNA to deliver a polynucleotide encoding a gene editor polynucleotide construct and an AAV to deliver the first and second atgRNA. The mRNA and AAV were delivered concurrently.
- FIG. 18shows percent in vivo beacon placement in the Nolcl locus of mice following delivery of a polynucleotide encoding a gene editor polynucleotide construct using a lipid nanoparticle (LNP) and a first atgRNA and second atgRNA using an AAV.
- %BP% beacon placement.
- LNPLNP formulation #1.
- LNP #F2LNP formulation #F2.
- LNP #F3LNP formulation #F3.
- FIG. 19show percent in vivo integration of a template polynucleotide in AttP mice following delivering of the Bxbl using adenovirus (AdV) and the template polynucleotide using an AAV (“AAV Cargo”).
- Bxbl Advwas administered to the mice at a dose of either 3E10 or 1E11 vector genomes (vg) per animal.
- AAV Cargowas administered to the mice at a dose of 1E12.
- FIG. 20Ashows ddPCR data for percent in vivo beacon placement in the Nolcl locus of neonatal mice at eight days post-delivery of a single dose of a mixture of two LNPs.
- First LNPcontained mRNA encoding a prime editing system and a first synthetic atgRNA (atgRNA 1) at a 1:1 ratio.
- Second LNPcontained mRNA encoding a prime editing system and a second synthetic atgRNA (atgRNA2) at a 1:1 ratio.
- Each of the first and second atgRNAstargeted the mouse Nolcl locus, encoded a portion of an integration recognition site (“beacon”), and together included a 6bp overlap.
- the first and second LNPswere combined 1:1 as mixture and administered at either 1 mg/kg or 3 mg/kg.
- LNP #F2LNP formulation #F2.
- FIG. 20Bshow NGS data for percent in vivo beacon placement in the Nolcl locus of the same neonatal mice and treatment conditions as described in FIG. 20A.
- NGS datashows beacon placement eight days after administration of the LNP mixture.
- LNP #F2LNP formulation #F2.
- FIG. 20Cshows NGS data for percentage of in vivo beacons placed in the Nolcl NGS data is for the same mice with the same treatment conditions as described in FIG. 20A.
- NGS datashows data for eight days after administration of the LNP mixture.
- LNP #F2LNP formulation #F2.
- FIG. 21Ashows ddPCR data for percent in vivo beacon placement in the Nolcl locus of neonatal mice at 6 weeks post-delivery of a single dose of a mixture of two LNPs.
- First LNPcontained mRNA encoding a prime editing system and a first synthetic atgRNA (atgRNA 1) at a 1:1 ratio.
- Second LNPcontained mRNA encoding a prime editing system and a second synthetic atgRNA (atgRNA2) at a 1:1 ratio.
- Each of the first and second atgRNAstargeted the mouse Nolcl locus, encoded a portion of an integration recognition site (“beacon”), and together included a 6bp overlap.
- the first and second LNPswere combined 1:1 as mixture and administered at either 1 mg/kg or 3 mg/kg.
- LNP #F2LNP formulation #F2.
- FIG. 21Bshows NGS data for percent in vivo beacon placement in the Nolcl locus of the same neonatal mice and treatment conditions as described in FIG. 21A.
- NGS datashows beacon placement 6 weeks after administration of the LNP mixture.
- LNP #F2LNP formulation #F2.
- FIG. 22Ashows ddPCR data for percent in vivo beacon placement in the Factor IX (“mF9”) locus of 6-8 week old mice at day 8 post-delivery of a single dose of a mixture of two LNPs.
- First LNPcontained mRNA encoding a prime editing system and a first synthetic atgRNA (atgRNAl) at a ratio of 1:0.5, 1:1, or 1:2.
- Second LNPcontained mRNA encoding a prime editing system and a second synthetic atgRNA (atgRNA2) at a ratio of 1:1, 1:0.5, or 1:2.
- Each of the first and second atgRNAstargeted the mouse Factor IX locus, encoded a portion of an integration recognition site (“beacon”), and together included a 6bp overlap.
- the first and second LNPswere combined 1:1 as mixture with the final ratio of mRNA:atgRNAl:atgRNA2 at 1:0.25:0.25; 1:0.5:0.5, or 1:1:1.
- LNP #F2LNP formulation #F2.
- FIG. 22Bshows NGS data for percent in vivo beacon placement in the mF9 locus of the same neonatal mice and treatment conditions as described in FIG. 22A.
- NGS datashows beacon placement 8 days after administration of the LNP mixture.
- LNP #F2LNP formulation #F2.
- FIGs. 23A-23Bshow the experimental conditions tested in Example 11.
- FIG. 23Ashows LNP formulations tested in Example 11.
- FIG. 23Bshows control conditions for Example 11.
- FIGs. 24A-24Bshow representative images of liver sinusoidal endothelial cells (LSECs) contacted with the indicated LNP formulations: cKK (500 ng GFP) or cKK-DOPG (500 ng GFP). Top panels are GFP images. Bottom panels are brightfield.
- FIG. 24Ashows 10X magnification of LSECs contacted with cKK (500 ng GFP) or cKK-DOPG (500 ng GFP).
- FIG. 24Bshows 20X magnification of LSEC contacted with cKK (500 ng GFP) or cKK-DOPG (500 ng GFP).
- FIG. 25shows representative images of LSECs contacted with the indicated LNP formulations: cKK-Mannose (500 ng GFP) or cKK-DOPG/Mannose (500 ng GFP).
- FIG. 26shows representative images of control LSECs: transfected with GFP mRNA at 1000 ng per well or untreated LSECs.
- FIG. 27shows the experimental conditions tested in Example 12.
- FIG. 28shows a plot of percent (%) Indels in genomic DNA harvested from LSECs following introduction of the indicated guide RNAs (gRNAs).
- F8refers to Factor VIII.
- FIG. 29shows a plot of percent (%) Indels in genomic DNA harvested from LSECs following introduction of guide RNA 2 (G2) using the indicated LNP formulations at the indicated amounts.
- FIG. 30shows a plot of percent (%) Indels in genomic DNA harvested from LSECs following introduction of guide RNA 4 (G4) using the indicated LNP formulations at the indicated amounts.
- FIG. 31shows a plot of percent (%) Indels in genomic DNA harvested from LSEC and hepatocytes.
- LSECare treated with LNPs with and without modifications e.g., a PEG-modified lipid or a phospholipid (e.g., one of the indicated phospholipids)).
- Hepatocytesare treated with LNPs with and without modifications.
- FIG. 32shows a plot of ratio of LSEC targeting to hepatocyte targeting (“LSEC:Hepatocyte targeting”) comparing LNPs with and without modifications e.g., a PEG- modified lipid or a phospholipid (e.g., one of the indicated phospholipids)).
- Gene editoris a protein that that can be used to perform gene editing, gene modification, gene insertion, gene deletion, or gene inversion.
- gene editor polynucleotiderefers to polynucleotide sequence encoding the gene editor protein.
- Such an enzyme or enzyme fusionmay contain DNA or RNA targetable nuclease protein (i.e., Cas protein, ADAR, or AD AT), wherein target specificity is mediated by a complexed nucleic acid (i.e., guide RNA).
- RNA targetable nuclease proteini.e., Cas protein, ADAR, or AD AT
- target specificityis mediated by a complexed nucleic acid (i.e., guide RNA).
- Such an enzyme or enzyme fusionmay be a DNA/RNA targetable protein, wherein target specificity is mediated by internal, conjugated, fused, or linked amino acids, such as within TALENs, ZFNs, or meganucleases.
- a gene editor comprising a targetable proteinmay be fused, linked, complexed, operate in cis or trans to one or more proteins or protein fragment motifs.
- Gene editorsmay be fused or linked to one or more integrase, recombinase, polymerase, telomerase, reverse transcriptase, or invertase.
- a gene editorcan be a prime editor fusion protein or a gene writer fusion protein.
- Prime editinguses CRISPR enzyme that nicks or cuts only single strand of double stranded DNA, i.e., a nickase; the nickase can occur either naturally or by mutation or modification of a nuclease that makes double stranded cuts.
- the nickaseis programmed (directed) with a prime-editing guide RNA (pegRNA).
- pegRNAprime-editing guide RNA
- attachment site containing guide RNAthat both specifies the target and encodes for the desired integrase target recognition site.
- the nickasemay be programmed (directed) with an atgRNA.
- the nickaseis a catalytically impaired Cas9 endonuclease, a Cas9 nickase, that is fused to the reverse transcriptase.
- the Cas9 nickase part of the proteinis guided to the DNA target site by the atgRNA (or pegRNA), whereby a nick or single stranded cut occurs.
- the reverse transcriptase domainthen uses the atgRNA (or pegRNA) to template reverse transcription of the desired edit, directly polymerizing DNA onto the nicked target DNA strand.
- the edited DNA strandreplaces the original DNA strand, creating a heteroduplex containing one edited strand and one unedited strand.
- the prime editor (PE)guides resolution of the heteroduplex to favor copying the edit onto the unedited strand, completing the process (typically achieved with a nickase gRNA).
- Other enzymes that can be used to nick or cut only a single strand of double stranded DNAincludes a cleavase (e.g., cleavase I enzyme).
- an additional agent or agentsmay be added that improve the efficiency and outcome purity of the prime edit.
- the agentmay be chemical or biological and disrupt DNA mismatch repair (MMR) processes at or near the edit site (i.e., PE4 and PE5 and PEmax architecture by Chen et al. Cell, 184, 1-18, October 28, 2021; Chen et al. is incorporated herein by reference).
- MMRDNA mismatch repair
- the agentis a MMR-inhibiting protein.
- the MMR-inhibiting proteinis dominant negative MMR protein.
- the dominant negative MMR proteinis MLHldn.
- the MMR-inhibiting agentis incorporated into the co-delivery method described herein.
- the MMR-inhibiting agentis linked or fused to the prime editor protein fusion, which may or may not have a linked or fused integrase. In some embodiments, the MMR-inhibiting agent is linked or fused to the Gene WriterTM protein, which may or may not have a linked or fused integrase.
- the prime editor or gene editor systemcan be used to achieve DNA deletion and replacement.
- the DNA deletion replacementis induced using a pair of atgRNAs or pegRNA that target opposite DNA strands, programming not only the sites that are nicked but also the outcome of the repair (i.e., PrimeDel by Choi et al. Nat. Biotechnology, October 14, 2021; Choi et al. is incorporated herein by reference and TwinPE by Anzalone et al. BioRxiv, November 2, 2021; Anzalone et al. is incorporated herein by reference).
- the DNA deletionis induced using a single atgRNA.
- the DNA deletion and replacementis induced using a wild type Cas9 prime editor (PE-Cas9) system (i.e., PED AR by Jiang et al. Nat. Biotechnology, October 14, 2021; Jiang et al. is incorporated herein by reference in its entirety).
- the DNA replacementis an integrase target recognition site or recombinase target recognition site.
- the constructs and methods described hereinmay be utilized to incorporate the pair of pegRNAs (or atgRNAs) used in PrimeDel, TwinPE (WO2021226558 incorporated by reference herein in its entirety), or PED AR, the prime editor fusion protein or Gene Writer protein, optionally a nickase guide RNA (ngRNA), an integrase, a nucleic acid cargo, and optionally a recombinase into a LNP delivery system or vector delivery system (e.g., AAV or Adenovirus).
- the integrasemay be directly linked, for example by a peptide linker, to the prime editor fusion or gene writer protein.
- the prime editorscan refer to a retrovirus or lentivirus reverse transcriptase such as a Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase (RT) fused to a CRISPR enzyme nickase such as a Cas9 H840A nickase, a Cas9nickase.
- the prime editorscan refer to a retrovirus or lentivirus reverse transcriptase such as a Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase (RT) fused to a cleavase.
- the RTcan be fused at, near or to the C-terminus of a Cas9nickase, e.g., Cas9 H840A. Fusing the RT to the C-terminus region, e.g., to the C-terminus, of the Cas9 nickase may result in higher editing efficiency.
- a complexis called PEI.
- the CRISPR enzyme nickasee.g., Cas9(H840A), i.e., a Cas9nickase
- the CRISPR enzyme nickaseinstead of being a Cas9 (H840A), i.e., instead of being a Cas9 nickase, the CRISPR enzyme nickase instead can be a CRISPR enzyme that naturally is a nickase or cuts a single strand of double stranded DNA; for instance, the CRISPR enzyme nickase can be Casl2a/b. Alternatively, the CRISPR enzyme nickase can be another mutation of Cas9, such as Cas9(D10A).
- a CRISPR enzymesuch as a CRISPR enzyme nickase, such as Cas9 (wild type), Cas9(H840A), Cas9(D10A) or Cas 12a/b nickase can be fused in some embodiments to a pentamutant of M-MLV RT (D200N/ L603W/ T330P/ T306K/ W313F), whereby there can be up to about 45-fold higher efficiency, and this is called PE2.
- a CRISPR enzyme nickasesuch as Cas9 (wild type), Cas9(H840A), Cas9(D10A) or Cas 12a/b nickase
- a pentamutant of M-MLV RTD200N/ L603W/ T330P/ T306K/ W313F
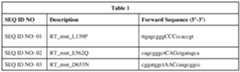
- the M-MLV RTcomprise one or more of the mutations Y8H, P51L, S56A, S67R, E69K, V129P, L139P, T197A, H204R, V223H, T246E, N249D, E286R, Q2911, E302K, E302R, F309N, M320L, P330E, L435G, L435R, N454K, D524A, D524G, D524N, E562Q, D583N, H594Q, E607K, D653N, and L671P. Specific M-MLV RT mutations are shown in Table 1.
- the reverse transcriptasecan also be a wild-type or modified transcription xenopolymerase (RTX), avian myeloblastosis virus reverse transcriptase (AMV RT), Feline Immunodeficiency Virus reverse transcriptase (FIV-RT), FeLV-RT (Feline leukemia virus reverse transcriptase), HIV-RT (Human Immunodeficiency Virus reverse transcriptase).
- RTXtranscription xenopolymerase
- AMV RTavian myeloblastosis virus reverse transcriptase
- FV-RTFeline Immunodeficiency Virus reverse transcriptase
- FeLV-RTFeLV-RT
- Feline leukemia virus reverse transcriptaseFeLV-RT
- HIV-RTHuman Immunodeficiency Virus reverse transcriptase
- the reverse transcriptasecan be a fusion of MMuLV to the Sto7d DNA binding domain (see lonnidi et al.
- PE3, PE3b, PE4, PE5, and/or PEmaxwhich a skilled person can incorporate into the co-delivery system described herein, involves nicking the non-edited strand, potentially causing the cell to remake that strand using the edited strand as the template to induce HR.
- the nicking of the non-edited strandcan involve the use of a nicking guide RNA (ngRNA).
- ngRNAnicking guide RNA
- Prime editing or CRISPR systemExamples of prime editors can be found in the following: W02020/191153, W02020/191171, WO2020/191233,
- the prime editor proteinPrior to RT-mediated edit incorporation, the prime editor protein (or system) (1) site-specifically targets a genomic locus and (2) performs a catalytic cut or nick. These steps are typically performed by a CRISPR-Cas.
- the Cas proteinmay be substituted by other nucleic acid programmable DNA binding proteins (napDNAbp) such as zinc finger nucleases (ZFNs), transcription activator- like effector nucleases (TAEENs), or meganucleases.
- ZFNszinc finger nucleases
- TAEENstranscription activator- like effector nucleases
- meganucleasesmeganucleases
- a Gene Writer proteincomprises: (A) a polypeptide or a nucleic acid encoding a polypeptide, wherein the polypeptide comprises (i) a reverse transcriptase domain, and either (x) an endonuclease domain that contains DNA binding functionality or (y) an endonuclease domain and separate DNA binding domain; and (B) a template RNA comprising (i) a sequence that binds the polypeptide and (ii) a heterologous insert sequence. Examples of such Gene WriterTM proteins and related systems can be found in US20200109398, which is incorporated by reference herein in its entirety.
- the prime editor or Gene Writer protein fusion or prime editor protein linked or fused to an integraseis expressed as a split construct.
- the split constructin reconstituted in a cell.
- the split constructcan be fused or ligated via intein protein splicing.
- the split constructcan be reconstituted via protein-protein inter- molecular bonding and/or interactions.
- the split constructcan be reconstituted via chemical, biological, or environmental induced oligomerization.
- the split constructcan be adapted into one or more delivery vectors described herein.
- an integrase or recombinaseis directly linked or fused, for example by a peptide linker, which may be cleavable or non-cleavable, to the prime editor fusion protein (i.e., fused Cas9 nickase-reverse transcriptase) or Gene Writer protein.
- a peptide linkerwhich may be cleavable or non-cleavable, to the prime editor fusion protein (i.e., fused Cas9 nickase-reverse transcriptase) or Gene Writer protein.
- Suitable linkersfor example between the Cas9, RT, and integrase, may be selected from Table 3:
- the prime editor or Gene Writer protein fusion or prime editor protein linked or fused to an integraseis expressed as a split construct.
- the split constructin reconstituted in a cell.
- the split constructcan be fused or ligated via intein protein splicing.
- the split constructcan be reconstituted via protein-protein inter- molecular bonding and/or interactions.
- the split constructcan be reconstituted via chemical, biological, or environmental induced oligomerization.
- the split constructcan be adapted into one or more nucleic acid constructs described herein.
- SpCas9Streptococcus pyogenes Cas9
- RECrecognition
- NUCnuclease
- the REC lobecan be divided into three regions, a long a helix referred to as the bridge helix (residues 60-93), the RECI (residues 94-179 and 308-713) domain, and the REC2 (residues 180-307) domain.
- the NUC lobeconsists of the RuvC (residues 1-59, 718-769, and 909-1098), HNH (residues 775-908), and PAM-interacting (PI) (residues 1099- 1368) domains.
- the negatively charged sgRNA:target DNA heteroduplexis accommodated in a positively charged groove at the interface between the REC and NUC lobes.
- the RuvC domainis assembled from the three split RuvC motifs (RuvC I— III) and interfaces with the PI domain to form a positively charged surface that interacts with the 30 tail of the sgRNA.
- the HNH domainlies between the RuvC II— III motifs and forms only a few contacts with the rest of the protein. Structural aspects of SpCas9 are described by Nishimasu et al., Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA, Cell 156, 935-949, February 27, 2014.
- the REC lobeincludes the RECI and REC2 domains.
- the REC2 domaindoes not contact the bound guide:target heteroduplex, indicating that truncation of REC lobe may be tolerated by SpCas9.
- SpCas9 mutant lacking the REC2 domain(D175-307) retained -50% of the wild-type Cas9 activity, indicating that the REC2 domain is not critical for DNA cleavage.
- PAM-Interacting domain'The NUC lobe contains the PAM-interacting (PI) domain that is positioned to recognize the PAM sequence on the noncomplementary DNA strand.
- the PI domain of SpCas9is required for the recognition of 5’-NGG-3’ PAM, and deletion of the PI domain (A1099-1368) abolished the cleavage activity, indicating that the PI domain is critical for SpCas9 function and a major determinant for the PAM specificity.
- RuvC domain'The RuvC nucleases of SpCas9 have an RNase H fold and four catalytic residues, AsplO (Ala), Glu762, His983, and Asp986, that are critical for the two-metal cleavage of the noncomplementary strand of the target DNA.
- AsplOAsplO
- Glu762, His983, and Asp986,that are critical for the two-metal cleavage of the noncomplementary strand of the target DNA.
- the Cas9 RuvC domainhas other structural elements involved in interactions with the guide:target heteroduplex (an end-capping loop between a42 and a43) and the PI domain/stem loop 3 (P hairpin formed by P3 and [34).
- SpCas9 HNH nucleaseshave three catalytic residues, Asp839, His840, and Asn863 and cleave the complementary strand of the target DNA through a single-metal mechanism.
- sgRNADNA recognition'.
- the sgRNA guide regionis primarily recognized by the REC lobe.
- the backbone phosphate groups of the guide regioninteract with the RECI domain (Argl65, Glyl66, Arg403, Asn407, Lys510, Tyr515, and Arg661) and the bridge helix (Arg63, Arg66, Arg70, Arg71, Arg74, and Arg78).
- the 20-hydroxyl groups of Gl, C15, U16, and G19hydrogen bond with Vall009, Tyr450, Arg447/Ile448, and Thr404, respectively.
- SpCas9recognizes the guide:target heteroduplex in a sequence-independent manner.
- the backbone phosphate groups of the target DNA(nucleotides 1, 9-11, 13, and 20) interact with the RECI (Asn497, Trp659, Arg661, and Gln695), RuvC (Gln926), and PI (Glul 108) domains.
- the C2’ atoms of the target DNAform van der Waals interactions with the RECI domain (Leul69, Tyr450, Met495, Met694, and His698) and the RuvC domain (Ala728).
- the terminal base pair of the guide:target heteroduplex(GEC20’) is recognized by the RuvC domain via end-capping interactions; the sgRNA G1 and target DNA C20’ nucleobases interact with the Tyrl013 and Vall015 side chains, respectively, whereas the 20-hydroxyl and phosphate groups of sgRNA G1 interact with Vall009 and Gln926, respectively.
- the nucleobases of G21 and U50 in the G2EU50wobble pair stack with the terminal C20:G10 pair in the guide:target heteroduplex and Tyr72 on the bridge helix, respectively, with the U5004 atom hydrogen bonded with Arg75.
- A51adopts the syn conformation and is oriented in the direction opposite to U50.
- the nucleobase of A51is sandwiched between Phel 105 and U63, with its Nl, N6, and N7 atoms hydrogen bonded with G62, Glyl l03, and Phel 105, respectively.
- Stem-loop recognition'Stem loop 1 is primarily recognized by the REC lobe, together with the PI domain.
- the backbone phosphate groups of stem loop 1interact with the RECI domain (Leu455, Ser460, Arg467, Thr472, and Ile473), the PI domain (Lysl l23 and Lysl l24), and the bridge helix (Arg70 and Arg74), with the 20-hydroxyl group of G58 hydrogen bonded with Leu455.
- A52interacts with Phel l05 through a face-to-edge p-p stacking interaction, and the flipped U59 nucleobase hydrogen bonds with Asn77.
- the single-stranded linker and stem loops 2 and 3are primarily recognized by the NUC lobe.
- the backbone phosphate groups of the linker(nucleotides 63-65 and 67) interact with the RuvC domain (Glu57, Lys742, and Lysl097), the PI domain (Thrl l02), and the bridge helix (Arg69), with the 20-hydroxyl groups of U64 and A65 hydrogen bonded with Glu57 and His721, respectively.
- the C67 nucleobaseforms two hydrogen bonds with Vail 100.
- Stem loop 2is recognized by Cas9 via the interactions between the NUC lobe and the non-Watson-Crick A68:G81 pair, which is formed by direct (between the A68 N6 and G81 06 atoms) and water-mediated (between the A68 N1 and G81 N1 atoms) hydrogen-bonding interactions.
- the A68 and G81 nucleobasescontact Serl351 and Tyrl356, respectively, whereas the A68:G81 pair interacts with Thrl358 via a water-mediated hydrogen bond.
- the 20-hydroxyl group of A68hydrogen bonds with His 1349, whereas the G81 nucleobase hydrogen bonds with Lys33.
- Stem loop 3interacts with the NUC lobe more extensively, as compared to stem loop 2.
- the backbone phosphate group of G92interacts with the RuvC domain (Arg40 and Lys44), whereas the G89 and U90 nucleobases hydrogen bond with Glnl272 and Glul225/Alal227, respectively.
- the A88 and C91 nucleobasesare recognized by Asn46 via multiple hydrogenbonding interactions.
- Cas9 proteins smaller than SpCas9allow more efficient packaging of nucleic acids encoding CRISPR systems, e.g., Cas9 and sgRNA into one rAAV (“all-in-one-AAV”) particle.
- efficient packaging of CRISPR systemscan be achieved in other viral vector systems (i.e., lentiviral, integration deficient lentiviral, hd-AAV, etc.) and non-viral vector systems (i.e., lipid nanoparticle).
- Small Cas9 proteinscan be advantageous for multidomain-Cas-nuclease-based systems for prime editing.
- Cas9 proteinsinclude Staphylococcus aureus (SauCas9, 1053 amino acid residues) and Campylobacter jejuni (CjCas9, 984 amino residues).
- SerCas9Staphylococcus aureus
- CjCas9Campylobacter jejuni
- Staphylococcus lugdunensis (Siu) Cas9as having genome-editing activity and provided homology mapping to SpCas9 and SauCas9 to facilitate generation of nickases and inactive (“dead”) enzymes (Schmidt et al., 2021, Improved CRISPR genome editing using small highly active and specific engineered RNA-guided nucleases. Nat Commun 12, 4219. doi.org/10.1038/s41467-021-24454-5) and engineered nucleases with higher cleavage activity by fragmenting and shuffling Cas9 DNAs.
- the small Cas9s and nickasesare useful in the instant disclosure.
- the Cas9 proteins used hereinmay also include other “Cas9 variants” having at least about 70% identical, at least about 80% identical, at least about 90% identical, at least about 95% identical, at least about 96% identical, at least about 97% identical, at least about 98% identical, at least about 99% identical, at least about 99.5% identical, or at least about 99.9% identical to any reference Cas9 protein, including any wild type Cas9, or mutant Cas9 (e.g., a dead Cas9 or Cas9 nickase), or fragment Cas9, or circular permutant Cas9, or other variant of Cas9 disclosed herein or known in the art.
- Cas9 variantshaving at least about 70% identical, at least about 80% identical, at least about 90% identical, at least about 95% identical, at least about 96% identical, at least about 97% identical, at least about 98% identical, at least about 99% identical, at least about 99.5% identical, or at least about 99.9% identical to any reference Cas9 protein, including any wild
- a Cas9 variantmay have 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 21, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50 or more amino acid changes compared to a reference Cas9.
- the Cas9 variantcomprises a fragment of a reference Cas9 (e.g., a gRNA binding domain or a DNA-cleavage domain), such that the fragment is at least about 70% identical, at least about 80% identical, at least about 90% identical, at least about 95% identical, at least about 96% identical, at least about 97% identical, at least about 98% identical, at least about 99% identical, at least about 99.5% identical, or at least about 99.9% identical to the corresponding fragment of wild type Cas9.
- a reference Cas9e.g., a gRNA binding domain or a DNA-cleavage domain
- the fragmentis at least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 95% identical, at least 96%, at least 97%, at least 98%, at least 99%, or at least 99.5% of the amino acid length of a corresponding wild type Cas9 (e.g., SEQ ID NO: 18).
- a corresponding wild type Cas9e.g., SEQ ID NO: 18
- the disclosurealso may utilize Cas9 fragments that retain their functionality and that are fragments of any herein disclosed Cas9 protein.
- the Cas9 fragmentis at least 100 amino acids in length.
- the fragmentis at least 100, 150, 200, 250, 300, 350, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, 1100, 1150, 1200, 1250, or at least 1300 amino acids in length.
- the prime editors disclosed hereinmay comprise one of the Cas9 variants described as follows, or a Cas9 variant thereof having at least about 70% identical, at least about 80% identical, at least about 90% identical, at least about 95% identical, at least about 96% identical, at least about 97% identical, at least about 98% identical, at least about 99% identical, at least about 99.5% identical, or at least about 99.9% identical to any reference Cas9 variants.
- prime editors utilized hereincomprise CRISPR-Cas system enzymes other than type II enzymes.
- prime editorscomprise type V or type VI CRISPR-Cas system enzymes. It will be appreciated that certain CRISPR enzymes exhibit promiscuous ssDNA cleavage activity and appropriate precautions should be considered.
- prime editorscomprise a nickase or a dead CRISPR with nuclease function comprised in a different component.
- the nucleic acid programmable DNA binding proteins utilized hereininclude, without limitation, Cas9 (e.g., dCas9 and nCas9), Casl2a (Cpfl), Casl2bl (C2cl), Casl2b2, Casl2c (C2c3), Casl2d (CasY), Casl2e (CasX), C2c4, C2c5, C2c8, C2c9, C2cl0, Casl3a (C2c2), Casl3b (C2c6), Casl3c (C2c7), Casl3d, and Argonaute.
- Cas9e.g., dCas9 and nCas9
- Casl2aCasl2a
- Casl2aCasl2a
- Casl2blCasl2b2
- Casl2c3Casl2d
- CasYCasl2
- Cas-equivalentsfurther include those described in Makarova et al., “C2c2 is a single-component programmable RNA- guided RNA-targeting CRISPR effector,” Science 2016; 353(6299) and Makarova et al., “Classification and Nomenclature of CRISPR-Cas Systems: Where from Here?,” The CRISPR Journal, Vol.l. No.5, 2018, the contents of which are incorporated herein by reference.
- One example of a nucleic acid programmable DNA-binding protein that has different PAM specificity than Cas9is Clustered Regularly Interspaced Short Palindromic Repeats from Prevotella and Francisella 1 (i.e, Casl2a (Cpfl)).
- Casl2a(Cpfl) is also a Class 2 CRISPR effector, but it is a member of type V subgroup of enzymes, rather than the type II subgroup. It has been shown that Casl2a (Cpfl) mediates robust DNA interference with features distinct from Cas9.
- Casl2a (Cpfl)is a single RNA-guided endonuclease lacking tracrRNA, and it utilizes a T- rich protospacer-adjacent motif (TTN, TTTN, or YTN). Moreover, Cpfl cleaves DNA via a staggered DNA double- stranded break.
- Cpfl proteinsare known in the art and have been described previously, for example Yamano et al., “Crystal structure of Cpfl in complex with guide RNA and target DNA.” Cell (165) 2016, p.949-962; the entire contents of which is hereby incorporated by reference.
- prime editors used hereincomprise the type V CRISPR family includes Francisella novicida U112 Cpfl (FnCpfl) also known as FnCasl2a.
- FnCpfladopts a bilobed architecture with the two lobes connected by the wedge (WED) domain.
- the N-terminal REC lobeconsists of two a-helical domains (RECI and REC2) that have been shown to coordinate the crRNA-target DNA heteroduplex.
- the C-terminal NUC lobeconsists of the C-terminal RuvC and Nuc domains involved in target cleavage, the arginine-rich bridge helix (BH), and the PAM- interacting (PI) domain.
- the repeat-derived segment of the crRNAforms a pseudoknot stabilized by intra-molecular base-pairing and hydrogen-bonding interactions.
- the pseudoknotis coordinated by residues from the WED, RuvC, and REC2 domains, as well as by two hydrated magnesium cations.
- nucleotides 1-5 of the crRNAare ordered in the central cavity of FnCasl2a and adopt an A-form-like helical conformation. Conformational ordering of the seed sequence is facilitated by multiple interactions between the ribose and phosphate moieties of the crRNA backbone and FnCpfl residues in the WED and RECI domains.
- FnCasl2a-crRNA complexfurther reveals that the bases of the seed sequence are solvent exposed and poised for hybridization with target DNA.
- Structural aspects of FnCpflare described by Swarts et al., Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRTS PR-Cas 12a, Molecular Cell 66, 221-233, April 20, 2017.
- the crRNA-target DNA strand heteroduplexis enclosed in the central cavity formed by the REC and NUC lobes and interacts extensively with the RECI and REC2 domains.
- the PAM-containing DNA duplexcomprises target strand nucleotides dT0-dT8 and non-target strand nucleotides dA(8)*-dA0* and is contacted by the PI, WED, and RECI domains.
- the 5’-TTN-3’ PAMis recognized in FnCasl2a by a mechanism combining the shapespecific recognition of a narrowed minor groove, with base-specific recognition of the PAM bases by two invariant residues, Lys671 and Lys613.
- the duplex of the target DNAis disrupted by the side chain of residue Lys667, which is inserted between the DNA strands and forms a cation-7t stacking interaction with the dAO-dTO* base pair.
- the phosphate group linking target strand residues dT(- 1) and dTOis coordinated by hydrogen-bonding interactions with the side chain of Lys823 and the backbone amide of Gly826.
- Target strand residue dT(-l)bends away from residue TO, allowing the target strand to interact with the seed sequence of the crRNA.
- the non-target strand nucleotides dTl*-dT5*interact with the Arg692- Ser702 loop in FnCasl2a through hydrogen-bonding and ionic interactions between backbone phosphate groups and side chains of Arg692, Asn700, Ser702, and Gln704, as well as main-chain amide groups of Lys699, Asn700, and Ser702.
- Alanine substitution of Q704 or replacement of residues Thr698-Ser702 in FnCasl2a with the sequence Ala-Gly3 (SEQ ID NO: 115)substantially reduced DNA cleavage activity, suggesting that these residues contribute to R-loop formation by stabilizing the displaced conformation of the nontarget DNA strand.
- the crRNA-target strand heteroduplexis terminated by a stacking interaction with a conserved aromatic residue (Tyr410). This prevents base pairing between the crRNA and the target strand beyond nucleotides U20 and dA(-20), respectively. Beyond this point, the target DNA strand nucleotides re-engage the non-target DNA strand, forming a PAM-distal DNA duplex comprising nucleotides dC(-21)-dA(-27) and dG21*-dT27*, respectively. The duplex is confined between the REC2 and Nuc domains at the end of the central channel formed by the REC and NUC lobes.
- Target DNA cleavageFnCpfl can independently accommodate both the target and non-target DNA strands in the catalytic pocket of the RuvC domain.
- the RuvC active sitecontains three catalytic residues (D917, E1006, and D1255). Structural observations suggest that both the target and non-target DNA strands are cleaved by the same catalytic mechanism in a single active site in Cpfl/Casl2a enzymes.
- Another type V CRISPRis AsCpfl from Acidaminococcus sp BV3L6 (Yamano et al., Crystal structure of Cpfl in complex with guide RNA and target DNA, Cell 165, 949-962, May 5, 2016)
- the nucleasecomprises a Casl2f effector.
- Small CRISPR- associated effector proteins belonging to the type V-F subtypehave been identified through the mining of sequence databases and members classified into Casl2fl (Casl4a and type V-U3), Casl2f2 (Casl4b) and Casl2f3 (Casl4c, type V-U2 and U4).
- Casl2flCasl2fl
- Casl4bCasl2f2
- Casl4ctype V-U2 and U4
- Exemplary CRISPR-Cas proteins and enzymes used in the prime editors hereininclude the following without limitation.
- Table 6Casl2b (C2cl) orthologs
- protospacer adjacent sequenceor “protospacer adjacent motif’ or “PAM” refers to an approximately 2-6 base pair DNA sequence (or a 2-, 3-, 4-, 5-, 6-, 7-, 8-, 9-, 10-, 11-, 12-long nucleotide sequence) that is an important targeting component of a Cas9 nuclease.
- PAM sequenceis on either strand, and is downstream in the 5' to 3' direction of Cas9 cut site.
- the canonical PAM sequence(i.e., the PAM sequence that is associated with the Cas9 nuclease of Streptococcus pyogenes or SpCas9) is 5'-NGG-3' wherein “N” is any nucleobase followed by two guanine (“G”) nucleobases.
- Nis any nucleobase followed by two guanine (“G”) nucleobases.
- Gguanine
- Different PAM sequencescan be associated with different Cas9 nucleases or equivalent proteins from different organisms.
- any given Cas9 nucleasemay be modified to alter the PAM specificity of the nuclease such that the nuclease recognizes alternative PAM sequence.
- the PAM specificitycan be modified by introducing one or more mutations, including (a) DI 135V, R1335Q, and T1337R “the VQR variant”, which alters the PAM specificity to NGAN or NGNG, (b) D1135E, R1335Q, and T1337R “the EQR variant”, which alters the PAM specificity to NGAG, and (c) DI 135V, G1218R, R1335E, and T1337R “the VRER variant”, which alters the PAM specificity to NGCG.
- the D1135E variant of canonical SpCas9still recognizes NGG, but it is more selective compared to the wild type SpCas9 protein.
- Cas9 enzymes from different bacterial speciescan have varying PAM specificities and some embodiments are therefore chosen based on the desired PAM recognition.
- Cas9 from Staphylococcus aureus(SaCas9) recognizes NGRRT or NGRRN.
- Cas9 from Neisseria meningitis (NmCas)recognizes NNNNGATT.
- Cas9 from Streptococcus thermophilis(StCas9) recognizes NNAGAAW.
- Cas9 from Treponema denticola (TdCas)recognizes NAAAAC. These examples are not meant to be limiting.
- non- SpCas9sbind a variety of PAM sequences, which makes them useful to expand the range of sequences that can be targeted according to the disclosure.
- non-SpCas9smay have other characteristics that make them more useful than SpCas9.
- Cas9 from Staphylococcus aureus(SaCas9) is about 1 kilobase smaller than SpCas9, so it can be packaged into adeno-associated virus (AAV).
- AAVadeno-associated virus
- Prime editor fusion proteindescribes a protein that is used in prime editing.
- Prime editinguses CRISPR enzyme that nicks or cuts only single strand of double stranded DNA, i.e., a
- I l l nickase; and a nickasecan occur either naturally or by mutation or modification of a nuclease that makes double stranded cuts.
- Such an enzymecan be a catalytically-impaired Cas9 endonuclease (a nickase).
- a nickasecan be a Casl2a/b, MAD7, or variant thereof.
- the nickaseis fused to an engineered reverse transcriptase (RT).
- RTengineered reverse transcriptase
- the nickaseis programmed (directed) with a primeediting guide RNA (pegRNA).
- pegRNAprimeediting guide RNA
- the nickaseis a catalytically-impaired Cas9 endonuclease, a Cas9 nickase, that is fused to the reverse transcriptase.
- the Cas9 nickase part of the proteinis guided to the DNA target site by the pegRNA, whereby a nick or single stranded cut occurs.
- the reverse transcriptase domainthen uses the pegRNA to template reverse transcription of the desired edit, directly polymerizing DNA onto the nicked target DNA strand.
- the edited DNA strandreplaces the original DNA strand, creating a heteroduplex containing one edited strand and one unedited strand.
- the prime editorguides resolution of the heteroduplex to favor copying the edit onto the unedited strand, completing the process (typically achieved with a nickase gRNA).
- PEIrefers to a PE complex comprising a fusion protein comprising Cas9(H840A) and a wild type MMLV RT having the following N-terminus to C-terminus structure: [NLS]-[Cas9(H840A)]- [linker] -[MMLV_RT(wt)] + a desired atgRNA (or PEgRNA).
- the prime editors disclosed hereinis comprised of PEI.
- PE2refers to a PE complex comprising a fusion protein comprising Cas9(H840A) and a variant MMLV RT having the following N-terminus to C-terminus structure:
- the prime editors disclosed hereinare comprised of PE2.
- the prime editors disclosed hereinis comprised of PE2 and co-expression of MMR protein MLHldn, that is PE4.
- PE3refers to PE2 plus a second-strand nicking guide RNA that complexes with the PE2 and introduces a nick in the non-edited DNA strand. The induction of the second nick increases the chances of the unedited strand, rather than the edited strand, to be repaired.
- the prime editors disclosed hereinare comprised of PE3.
- the prime editors disclosed hereinare comprised of PE3 and co-expression of MMR protein MLHldn, that is PE5.
- PE3brefers to PE3 but wherein the second-strand nicking guide RNA is designed for temporal control such that the second strand nick is not introduced until after the installation of the desired edit. This is achieved by designing a gRNA with a spacer sequence with mismatches to the unedited original allele that matches only the edited strand. Using this strategy, mismatches between the protospacer and the unedited allele should disfavor nicking by the sgRNA until after the editing event on the PAM strand takes place.
- a prime editing complexconsists of a type II CRISPR PE protein containing an RNA-guided DNA-nicking domain fused to a reverse transcriptase (RT) domain and complexed with a pegRNA.
- the pegRNAcomprises (5’ to 3’) a spacer that is complementary to the target sequence of a genomic DNA, a nickase (e.g. Cas9) binding site, a reverse transcriptase template including editing positions, and primer binding site (PBS).
- the PE-pegRNA complexbinds the target DNA and the CRISPR protein nicks the PAM- containing strand.
- the resulting 3' end of the nicked targethybridizes to the primer-binding site (PBS) of the pegRNA, then primes reverse transcription of new DNA containing the desired edit using the RT template of the pegRNA.
- PBSprimer-binding site
- the overall structure of the pegRNAis like that of a typical type II sgRNA with a reverse transcriptase template/primer binding site appended to the 3’ end. The structure leaves the PBS at the 3’ end of the pegRNA free to bind to the nicked strand complementary to the target which forms the primer for reverse transcription.
- Guide RNAs of CRISPRsdiffer in overall structure. For example, while the spacer of a type II gRNA is located at the 5’ end, the spacer of a type V gRNA is located towards the 3’ end, with the CRISPR protein (e.g. Casl2a) binding region located toward the 5’ end. Accordingly, the regions of a type V pegRNA are rearranged compared to a type II pegRNA.
- the overall structure of the pegRNAis like that of a typical type II sgRNA with a reverse transcriptase template/primer binding site appended to the 3’ end.
- the pegRNAcomprises (5’ to 3’) a CRISPR protein-binding region, a spacer which is complementary to the target sequence of a genomic DNA, a reverse transcriptase template including editing positions, and primer binding site (PBS).
- an atgRNAcomprises a reverse transcriptase template that encodes, partially or in its entirety, an integration recognition site (also referred to as an integration target recognition site) or a recombinase recognition site (also referred to as a recombinase target recognition site).
- the integration target recognition sitewhich is to be placed at a desired location in the genome or intracellular nucleic acid, is referred to as a “beacon” site or an “attachment site” or a “landing pad” or “landing site.”
- An integration target recognition site or recombinase target recognition site incorporated into the pegRNAis referred to as an attachment site containing guide RNA (atgRNA).
- the term “attachment site-containing guide RNA”refers to an extended single guide RNA (sgRNA) comprising a primer binding site (PBS), a reverse transcriptase (RT) template sequence, and wherein the RT template encodes for an integration recognition site or a recombinase recognition site that can be recognized by a recombinase, integrase, or transposase.
- the RT templatecomprises a clamp sequence and an integration recognition site.
- an atgRNAmay be referred to as a guide RNA.
- An integration recognition site or recombinase target recognition site incorporated into the pegRNAis referred to as an attachment site containing guide RNA (atgRNA).
- cognate integration recognition siteor “integration cognate” or “cognate pair” refers to a first integration recognition site (e.g., any of the integration recognition sites described herein) and a second integration recognition site (e.g., any of the integration recognition sites described herein) that can be recombined. Recombination between a first integration recognition site (e.g., any of the integration recognition sites described herein) and a second recognition site (e.g., any of the integration recognition sites described herein) is mediated by functional symmetry between the two integration recognition sites and the central dinucleotide of each of the two integration recognition sites.
- a first integration recognition sitee.g., any of the integration recognition sites described herein
- a second integration recognition sitee.g., any of the integration recognition sites described herein
- a non-limiting example of a cognate pairinclude an attB site and an attP site, whereby a serine integrase mediates recombination between the attB site and the attP site.
- an atgRNAcomprises a reverse transcriptase template that encodes, partially or in its entirety, an integration recognition site (also referred to as an integration target recognition site) or a recombinase recognition site (also referred to as a recombinase target recognition site).
- the integration target recognition sitewhich is to be placed at a desired location in the genome or intracellular nucleic acid, is referred to as a “beacon,” a “beacon” site or an “attachment site” or a “landing pad” or “landing site.”
- An integration target recognition site or recombinase target recognition site incorporated into the pegRNAis referred to as an attachment site containing guide RNA (atgRNA).
- the primer binding siteallows the 3’ end of the nicked DNA strand to hybridize to the atgRNA, while the RT template serves as a template for the synthesis of edited genetic information.
- the atgRNAis capable for instance, without limitation, of (i) identifying the target nucleotide sequence to be edited and (ii) encoding new genetic information that replaces (or in some cases adds) the targeted sequence.
- the atgRNAis capable of (i) identifying the target nucleotide sequence to be edited and (ii) encoding an integration site that replaces (or inserts/deletes within) the targeted sequences.
- the co-delivery system described hereinincludes a polynucleotide sequence encoding an attachment site-containing guide RNA (atgRNA) packaged in an LNP.
- the co-delivery system described hereinincludes a vector comprising a polynucleotide sequence encoding an atgRNA.
- the atgRNAcomprises a domain that is capable of guiding the prime editor fusion protein to a target sequence, thereby identifying the target nucleotide sequence to be edited; and a reverse transcriptase (RT) template that comprises a first integration recognition site.
- the atgRNAcomprises a domain that is capable of guiding the prime editor fusion protein (or prime editor system) to a target sequence, thereby identifying the target nucleotide sequence to be edited; and a reverse transcriptase (RT) template that comprises at least a portion first integration recognition site.
- a domainthat is capable of guiding the prime editor fusion protein (or prime editor system) to a target sequence, thereby identifying the target nucleotide sequence to be edited
- RTreverse transcriptase
- the co-delivery system described hereinincludes a polynucleotide sequence encoding a first attachment site-containing guide RNA (atgRNA) and a polynucleotide nucleotide sequence encoding a second attachment site-containing guide RNA (atgRNA) packaged into the same LNP.
- the co-delivery system described hereinincludes a polynucleotide sequence encoding a first attachment site-containing guide RNA (atgRNA) packaged into a first LNP and a polynucleotide nucleotide sequence encoding a second attachment site-containing guide RNA (atgRNA) packaged into a second LNP.
- the co-delivery system described hereinincludes a vector comprising a polynucleotide sequence encoding a first attachment site-containing guide RNA (atgRNA), a polynucleotide sequence encoding a second atgRNA, or both.
- the co-delivery system described hereinincludes a polynucleotide sequence encoding a first attachment site-containing guide RNA (atgRNA) packaged into a first LNP and a vector comprising a polynucleotide sequence encoding a second atgRNA.
- the co-delivery systemcontains a first atgRNA and a second atgRNA
- the first atgRNA and the second atgRNAare an at least first pair of atgRNAs, where the at least first pair of atgRNAs have domains that are capable of guiding the gene editor protein or prime editor fusion protein to a target sequence
- the first atgRNAfurther includes a first RT template that comprises at least a portion of the first integration recognition site
- the second atgRNAfurther includes a second RT template that comprises at least a portion of the first integration recognition site
- the first atgRNA and the second atgRNAscollectively encode the entirety of the first integration recognition site.
- the first atgRNA’ s reverse transcriptase templateencodes for a first single-stranded DNA sequence (i.e., a first DNA flap) that contains a complementary region to a second single-stranded DNA sequence (i.e., a second DNA flap) encoded by a second atgRNA comprising a second reverse transcriptase template.
- the complementary region between the first and second single- stranded DNA sequencesis comprised of more than 5 consecutive bases of an integrase target recognition site.
- the complementary region between the first and second single- stranded DNA sequencesis comprised of more than 10 consecutive bases of an integrase target recognition site.
- the complementary region between the first and second single-stranded DNA sequencesis comprised of more than 20 consecutive bases of an integrase target recognition site. In certain embodiments, the complementary region between the first and second single-stranded DNA sequences is comprised of more than 30 consecutive bases of an integrase target recognition site.
- Use of two guide RNAs that are (or encode DNA that is) partially complementarity to each other and comprised of consecutive bases of an integrase target recognition siteare referred to as dual, paired, annealing, complementary, or twin attachment site-containing guide RNAs (atgRNAs).
- RNAsthat are (or encode DNA that is) full complementarity to each other and comprised of consecutive bases of an integrase target recognition site are referred to as dual, paired, annealing, complementary, or twin attachment sitecontaining guide RNAs (atgRNAs).
- atgRNAsguide RNAs
- Table 9includes atgRNAs, sgRNAs and nicking guides that can be used herein. Spacers are labeled in capital font (SPACER), RT regions in bold capital (RT REGION), AttB sites in bold lower case (attB site), and PBS in capital italics (PBS). Unless otherwise denoted, the AttB is for Bxbl.
- the co-delivery system described hereincontains an integrase and/or a recombinase.
- the co-delivery systemincludes an integrase and/or a recombinase packaged in a LNP.
- the co-delivery systemincludes a polynucleotide encoding an integrase and/or a recombinase.
- the co-delivery systemincludes an integrase or a recombinase packaged in a vector (e.g., a viral vector).
- the co-delivery systemincludes at least a first integrase (e.g., a first integrase and a second integrase) and/or at least a first recombinase (e.g., a first recombinase and a second recombinase).
- a first integrasee.g., a first integrase and a second integrase
- a first recombinasee.g., a first recombinase and a second recombinase
- the integration enzymee.g., the integrase or recombinase
- the integration enzymeis selected from the group consisting of Dre, Vika, Bxbl, cpC31, RDF, cpBTl, Rl, R2, R3, R4, R5, TP901-1, Al 18, (pFCl, (pCl, MR11, TGI, cp370.1, W , BL3, SPBc, K38, Peaches, Veracruz, Rebeuca, Theia, Benedict, KSSJEB, PattyP, Doom, Scowl, Lockley, Switzer, Bob3, Troube, Abrogate, Anglerfish, Sarfire, SkiPole, Concept!, Museum, Severus, Airmid, Benedict, Hinder, ICleared, Sheen, Mundrea, BxZ2, cpRV, retrotransposases encoded by a Tcl/mariner family member including but not limited to retrotransposases encoded by LI,
- Xu et aldescribes methods for evaluating integrase activity in E. coli and mammalian cells and confirmed at least R4, (pC31, (pBTl, Bxbl, SPBc, TP901-1 and WP integrases to be active on substrates integrated into the genome of HT1080 cells (Xu et al., 2013, Accuracy and efficiency define Bxbl integrase as the best of fifteen candidate serine recombinases for the integration of DNA into the human genome. BMC Biotechnol. 2013 Oct 20;13:87. doi: 10.1186/1472-6750-13-87).
- Durrantdescribes new large serine recombinases (LSRs) divided into three classes distinguished from one another by efficiency and specificity, including landing pad LSRs which outperform wild-type Bxbl in episomal and chromosomal integration efficiency, LSRs that achieve both efficient and sitespecific integration without a landing pad, and multi-targeting LSRs with minimal site- specificity. Additionally, embodiments can include any serine recombinase such as BceINT, SSCINT, SACINT, and INT10 (see lonnidi et al., 2021; Drag-and-drop genome insertion without DNA cleavage with CRISPR directed integrases. bioRxiv 2021. ! 1.0 !
- the integration sitecan be selected from an attB site, an attP site, an attL site, an attR site, a lox71 site a Vox site, or a ERT site.
- integrases, transposases and the likecan depend on nuclear localization.
- prokaryotic enzymesare adapted to modulate nuclear localization.
- eukaryotic or vertebrate enzymesare adapted to modulate nuclear localization.
- the disclosureprovides fusion or hybrid proteins. Such modulation can comprise addition or removal of one or more nuclear localization signal (NLS) and/or addition or removal of one or more nuclear export signal (NES).
- NLSnuclear localization signal
- NESnuclear export signal
- nuclear export signal (NES) of transposasesaffects the transposition activity of mariner-like elements Ppmarl and. Ppmar2 of moso bamboo. Mob DNA. 2019 Aug 19; 10:35. doi:10.1186/sl3100-019-0179-y).
- the methods and constructsare used to modulate nuclear localization of system components of the disclosure.
- the integrase used hereinis selected from below (Table 10).
- FIGs. 14A-14Eshows analysis of effect of variant AttP sites on integration efficiency.
- kits for deliveringe.g., co-delivery or dual delivery
- a systemcapable of site- specifically integrating a template polynucleotide into the genome of a cell
- the methodsincludes delivering to a (i) gene editor construct and a (ii) template polynucleotide, and (iii) at least a first attachment site-containing guide (atgRNA).
- atgRNAfirst attachment site-containing guide
- LNPlipid nanoparticle
- atgRNAfirst attachment site-containing guide RNA
- the first atgRNAcomprises (i) a domain that is capable of guiding the prime editor system to a target sequence; and (ii) a reverse transcriptase (RT) template that comprises at least a portion of a first integration recognition site.
- the RT templatecomprises the entirety of the first integration recognition site.
- the vectoralso includes a sequence encoding a nicking guide RNA (ngRNA).
- LNPlipid nanoparticle
- atgRNAfirst attachment site-containing guide RNA
- atgRNAsecond attachment site-containing guide RNA
- the first atgRNA and the second atgRNAare an at least first pair of atgRNAs, wherein the at least first pair of atgRNAs have domains that are capable of guiding the prime editor system to a target sequence, the first atgRNA further includes a first RT template that comprises at least a portion of the at least first integration recognition site; and the second atgRNA further includes a second RT template that comprises at least a portion of the first integration recognition site, and the first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site.
- the first atgRNA and second atgRNAinclude at least a 6bp overlap (e.g., 6bp of complementarity).
- lipid nanoparticlecomprising: (i) a gene editor polynucleotide (e.g., a gene editor polynucleotide construct), and (ii) a first attachment sitecontaining guide RNA (atgRNA); and a vector comprising: (i) a template polynucleotide, and (ii) a second atgRNA.
- LNPlipid nanoparticle
- the first atgRNA and the second atgRNAare an at least first pair of atgRNAs, wherein the at least first pair of atgRNAs have domains that are capable of guiding the prime editor system to a target sequence, the first atgRNA further includes a first RT template that comprises at least a portion of the a first integration recognition site; and the second atgRNA further includes a second RT template that comprises at least a portion of the first integration recognition site, and the first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site.
- the first atgRNA and second atgRNAinclude at least a 6bp overlap (e.g., 6bp of complementarity).
- lipid nanoparticlecomprising: (i) a gene editor polynucleotide (e.g., a gene editor polynucleotide construct), (ii) a first attachment site-containing guide RNA (atgRNA), and (iii) a second atgRNA; and a vector comprising (i) a template polynucleotide.
- the first atgRNA and the second atgRNAare an at least first pair of atgRNAs, wherein the at least first pair of atgRNAs have domains that are capable of guiding the prime editor system to a target sequence, the first atgRNA further includes a first RT template that comprises at least a portion of the at least first integration recognition site; and the second atgRNA further includes a second RT template that comprises at least a portion of the at least first integration recognition site, and the first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site.
- the first atgRNA and second atgRNAinclude at least a 6bp overlap (e.g., 6bp of complementarity).
- lipid nanoparticlecomprising: (i) a gene editor polynucleotide (e.g., a gene editor polynucleotide construct) and (ii) a first attachment site-containing guide RNA (atgRNA); and a vector comprising: (i) a template polynucleotide, and (ii) a nicking atgRNA.
- LNPlipid nanoparticle
- atgRNAfirst attachment site-containing guide RNA
- the first atgRNAcomprises (i) a domain that is capable of guiding the prime editor system to a target sequence; and (ii) a reverse transcriptase (RT) template that comprises at least a portion of a first integration recognition site.
- the RT templatecomprises the entirety of the first integration recognition site.
- the LNP and the first vectorare delivered at least 1 day, 2 days, 3 days, 4 days, 5 days, 6 days, 7 days, 2 weeks, 3 weeks, 4 weeks, 5 weeks, 6 weeks, 7 weeks, or 8 weeks apart.
- the methodincludes delivering an LNP and a second vector
- the LNP and the second vectorare delivered at least 1 day, 2 days, 3 days, 4 days, 5 days, 6 days, 7 days, 2 weeks, 3 weeks, 4 weeks, 5 weeks, 6 weeks, 7 weeks, or 8 weeks apart.
- lipid nanoparticlecomprising a gene editor polynucleotide (e.g., a gene editor polynucleotide construct) and a vector comprising a template polynucleotide and at least a first attachment site-containing guide RNA (atgRNA).
- atgRNAattachment site-containing guide RNA
- the first atgRNAcomprises (i) a domain that is capable of guiding the prime editor system to a target sequence; and (ii) a reverse transcriptase (RT) template that comprises at least a portion of a first integration recognition site.
- the RT templatecomprises the entirety of the first integration recognition site. In some embodiments, where the vector comprises a polynucleotide encoding a first atgRNA, the vector also includes a sequence encoding a nicking guide RNA (ngRNA).
- ngRNAnicking guide RNA
- lipid nanoparticlecomprising a gene editor polynucleotide (e.g., a gene editor polynucleotide construct) and a vector comprising a template polynucleotide and a first attachment site-containing guide RNA (atgRNA) and a second attachment site-containing guide RNA (atgRNA).
- LNPlipid nanoparticle
- atgRNAfirst attachment site-containing guide RNA
- atgRNAsecond attachment site-containing guide RNA
- the first atgRNA and the second atgRNAare an at least first pair of atgRNAs, wherein the at least first pair of atgRNAs have domains that are capable of guiding the prime editor system to a target sequence, the first atgRNA further includes a first RT template that comprises at least a portion of the a first integration recognition site; and the second atgRNA further includes a second RT template that comprises at least a portion of the first integration recognition site, and the first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site.
- the first atgRNA and second atgRNAinclude at least a 6bp overlap.
- lipid nanoparticlecomprising: (i) a gene editor polynucleotide (e.g., a gene editor polynucleotide construct), and (ii) a first attachment site-containing guide RNA (atgRNA); and a vector comprising: (i) a template polynucleotide, and (ii) a second atgRNA.
- LNPlipid nanoparticle
- atgRNAfirst attachment site-containing guide RNA
- the first atgRNA and the second atgRNAare an at least first pair of atgRNAs, wherein the at least first pair of atgRNAs have domains that are capable of guiding the prime editor system to a target sequence, the first atgRNA further includes a first RT template that comprises at least a portion of the a first integration recognition site; and the second atgRNA further includes a second RT template that comprises at least a portion of the first integration recognition site, and the first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site.
- the first atgRNA and second atgRNAinclude at least a 6bp overlap.
- lipid nanoparticlecomprising: (i) a gene editor polynucleotide (e.g., a gene editor polynucleotide construct), (ii) a first attachment site-containing guide RNA (atgRNA), and (iii) a second atgRNA; and a vector comprising (i) a template polynucleotide.
- the first atgRNA and the second atgRNAare an at least first pair of atgRNAs, wherein the at least first pair of atgRNAs have domains that are capable of guiding the prime editor system to a target sequence, the first atgRNA further includes a first RT template that comprises at least a portion of the a first integration recognition site; and the second atgRNA further includes a second RT template that comprises at least a portion of the first integration recognition site, and the first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site.
- the first atgRNA and second atgRNAinclude at least a 6bp overlap.
- lipid nanoparticlecomprising: (i) a gene editor polynucleotide (e.g., a gene editor polynucleotide construct) and (ii) a first attachment site-containing guide RNA (atgRNA); and a vector comprising: (i) a template polynucleotide, and (ii) a nicking atgRNA.
- LNPlipid nanoparticle
- atgRNAfirst attachment site-containing guide RNA
- the first atgRNAcomprises (i) a domain that is capable of guiding the prime editor system to a target sequence; and (ii) a reverse transcriptase (RT) template that comprises at least a portion of a first integration recognition site.
- the RT templatecomprises the entirety of the first integration recognition site.
- the LNP comprising a gene editor polynucleotide constructis capable delivering to a cell cytoplasm the gene editor polynucleotide construct. In some embodiments, the LNP comprising a gene editor polynucleotide construct is capable delivering to a cell nucleus the gene editor polynucleotide construct. In some embodiments, the LNP comprises a gene editor protein and associated guide nucleic acids. In some embodiments, the LNP comprises a gene editor protein and associated guide nucleic acids that are capable of localizing to cell nucleus.
- a gene editor polynucleotide constructis delivered to a cell by a fusosome.
- a gene editor polynucleotide constructis delivered to a cell cytoplasm by a fusosome.
- the fusosomecomprises a gene editor protein and associated guide nucleic acids.
- a gene editor polynucleotide constructis delivered to a cell by an exosome.
- a gene editor polynucleotide constructis delivered to a cell cytoplasm by an exosome.
- the exosomecomprises a gene editor protein and associated guide nucleic acids.
- the prime editor or Gene Writer protein fusioneither of which may have a fused/linked integrase, is incorporated (i.e., packaged) into LNP as protein. Further, associated atgRNA and optional ngRNAs may be co-packaged with gene editor proteins in LNP.
- the gene editor polynucleotide constructcomprises (a) a polynucleotide sequence encoding a prime editor fusion protein or a Gene WriterTM protein, (b) a polynucleotide sequence encoding an attachment site-containing guide RNA (atgRNA), (c) optionally, a polynucleotide sequence encoding a nickase guide RNA (ngRNA), (d) a polynucleotide sequence encoding an integrase, (e) and optionally, a polynucleotide sequence encoding a recombinase.
- atgRNAattachment site-containing guide RNA
- ngRNAnickase guide RNA
- a polynucleotide sequence encoding an integrasee
- a polynucleotide sequence encoding a recombinaseoptionally, a polynucleotide sequence encoding a recombina
- the prime editor or Gene Writer protein fusionis expressed as a split construct.
- the split constructin reconstituted in a cell.
- the split constructcan be fused or ligated via intein protein splicing.
- the split constructcan be reconstituted via protein-protein inter-molecular bonding and/or interactions.
- the split constructcan be reconstituted via chemical, biological, or environmental induced oligomerization.
- the split constructcan be adapted into one or more nucleic acid constructs described herein.
- the systems describedinclude a gene editor polynucleotide that is delivered to a cell using the methods described herein.
- the gene editor polynucleotideis delivered as a polynucleotide (e.g., an mRNA).
- the gene editor polynucleotideis delivered as a protein.
- the gene editor polynucleotide or proteinis packaged, and thereby vectorized, within a lipid nanoparticle (LNP).
- LNPlipid nanoparticle
- the gene editor polynucleotide or proteinis packaged in a LNP and is codelivered with a template polynucleotide (i.e., nucleic acid “cargo” or nucleic acid “payload”) packaged into a separate vector (e.g., a viral vector (e.g., an AAV or adenovirus)) or a second lipid nanoparticle (LNP).
- a template polynucleotidei.e., nucleic acid “cargo” or nucleic acid “payload” packaged into a separate vector (e.g., a viral vector (e.g., an AAV or adenovirus)) or a second lipid nanoparticle (LNP).
- a separate vectore.g., a viral vector (e.g., an AAV or adenovirus)
- LNPsecond lipid nanoparticle
- the gene editor polynucleotideis delivered to the cells as a polynucleotide.
- the gene editor polynucleotideis delivered to the cells as an mRNA encoding the gene editor polynucleotide (e.g., the gene editor protein or the prime editor system).
- the mRNAcomprises one or more modified uridines.
- the mRNAcomprises a sequence where each of the uridines is a modified uridine.
- the mRNAis uridine depleted.
- the mRNA encoding the nickasecomprises one or more modified uridines.
- the mRNA encoding the reverse transcriptasecomprises one or more modified uridines. In some embodiments, the mRNA encoding the nickase comprises one or more modified uridines, and the mRNA encoding the reverse transcriptase comprises one or more modified uridines. In some embodiments, where the integrase is encoded in an mRNA, the mRNA comprises modified uridines. In some embodiments, a modified uridine is a Nl-Methylpseudouridine-5’ -Triphosphate. In some embodiments, a modified uridine is a pseudouridine. In some embodiments, the mRNA comprises a 5’ cap. In some embodiments, the 5’ cap comprises a molecular formula of C32H43N15O24P4 (free acid).
- the gene editor polynucleotide(e.g., a gene editor polynucleotide construct) comprises a polynucleotide sequence encoding a primer editor system (e.g., any of the prime editor systems described herein).
- the prime editor systemcomprises a nucleotide sequence encoding a nickase (e.g., any of the Cas proteins or variants thereof (e.g., nickases) and nickases described herein, see Tables 4-8) and a nucleotide sequence encoding a reverse transcriptase (e.g., any of the reverse transcriptases described herein).
- the nucleotide sequence encoding the nickase and the nucleotide sequence encoding the reverse transcriptaseare positioned in the construct such that when expressed the nickase is linked to the reverse transcriptase.
- the nickaseis linked to the reverse transcriptase by in-frame fusion.
- the nickaseis linked to the reverse transcriptase by a linker.
- the linkeris a peptide fused in-frame between the nickase and reverse transcriptase.
- the gene editor polynucleotide(e.g., a gene editor polynucleotide construct) further comprises a polynucleotide sequence encoding at least a first integrase (e.g., any of the integrases described herein, e.g., as described in Table 10 and also in Yamall et al., Nat. Biotechnol., 2022, doi.org/10.1038/s41587-022-01527-4 and Durrant et al., Nat. Biotechnol., 2022, doi.org/10.1038/s41587-022-01494-w, each of which are herein incorporated by reference in their entireties).
- the linked nickase-reverse transcriptaseare further linked to the first integrase.
- the gene editor polynucleotide constructfurther comprises a polynucleotide sequence encoding at least a first recombinase (e.g., any of the recombinases described herein). 5.9.2. Split Nickase and Reverse Transcriptase
- nucleotide sequence encoding the nickase and the nucleotide sequence encoding the reverse transcriptaseare positioned in a construct such that when expressed the nickase is not linked to the reverse transcriptase.
- a sequence encoding a self-cleaving peptidee.g., a P2A
- a sequence encoding a self-cleaving peptideis positioned between the nucleotide sequence encoding the nickase and the nucleotide sequence encoding the reverse transcriptase such that when expressed the nickase is not linked to the reverse transcriptase.
- nucleotide sequence encoding the nickase and the nucleotide sequence encoding the reverse transcriptaseare on separate polynucleotides.
- the nickase and/or the reverse transcriptasecan be engineered such that when expressed they are linked to a binding domain, where the binding domain is part of a binding pair that enables recruitment of the reverse transcriptase to the genomic location of the nickase (e.g., nCas9).
- a binding paircan be any pair of molecules wherein upon forming the binding pair the binding domains enable recruitment of the reverse transcriptase to the genomic location of the nickase. Once in sufficient proximity with each other the binding domains can dimerize. This dimerization (or other type of interaction) brings the nickase and RT into proximity with each other, thereby enhancing the reverse transcription of the RT template in the atgRNA.
- Non-limiting examples of binding pairs that can aid recruitment of the RT to the genomic location of the nickaseinclude without limitation: dimerizing leucine zippers, an antigen and a corresponding antigen binding domains (e.g., antibody), RNA aptamers and corresponding RNA aptamer binding proteins, affinity tags and corresponding affinity tag binding proteins.
- dimerizing leucine zippers that can serve as a binding pairincludes two binding domains: a Leucine Zip LZ1 domain having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 568 and a Leucine Zip LZ2 domain having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 569.
- the Leucine Zip LZ1 domainis positioned either N- terminally or C-terminally to the nickase and the Leucine Zip LZ2 is positioned either N-terminally or C-terminally to the RT.
- the Leucine Zip LZ1 domainis positioned either N-terminally or C-terminally to the RT and the Leucine Zip LZ2 domain is positioned either N- terminally or C-terminally to the Cas9.
- the system and methods described hereininclude a construct that encodes a polypeptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 576 (a Leucine Zip LZ1 domain positioned C-terminally to the nickase), and a construct that encodes a polypeptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 577 (a Leucine Zip LZ2 domain positioned N-terminally to the RT).
- the system and methods described hereininclude a construct that encodes a polypeptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 578 (a Leucine Zip LZ1 domain positioned N-terminally to the nickase), and a construct that encodes a polypeptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 577 (a Leucine Zip LZ2 domain positioned N-terminally to the RT).
- dimerizing leucine zippers that can serve as a binding pairincludes two binding domains: a EE1234L peptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 570 and a RR1234L peptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 571.
- the EE1234L peptideis positioned either N-terminally or C-terminally to the nickase and the RR1234L peptide is positioned either N-terminally or C-terminally to the RT.
- the EE1234L peptideis positioned either N-terminally or C-terminally to the RT and the RR1234L peptide is positioned either N-terminally or C-terminally to the Cas9.
- the system and methods described hereininclude a construct that encodes a polypeptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 579 (a EE1234L peptide positioned N-terminally to the nickase), and a construct that encodes a polypeptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 580 (a RR1234L peptide positioned N-terminally to the RT).
- an affinity tagis a SpyTag and the corresponding affinity tag binding protein is SpyCatcher, where upon binding of the SpyTag with the SpyCatcher the fusion protein is a covalently stabilized multi-protein complex.
- the affinity tage.g., SpyTag
- the affinity tag binding proteine.g.,. SpyCatcher
- the affinity tag binding proteine.g., SpyCatcher
- the affinity tag binding proteinis positioned either N-terminally or C-terminally to the nickase.
- an antigen and corresponding antigen binding domainincludes an antigen such as a peptide present within a tag or a tag itself and an antigen binding domain that binds specifically to the antigen.
- an antigenis a GCN4 peptide within a SunTag and the antigen binding domain is an anti-GCN4 scFv.
- the SunTag (GCN4 peptide)is positioned either N-terminally or C-terminally to the nickase and the anti-GCN4 antigen binding domain is positioned either N-terminally or C-terminally to the RT.
- the SunTag(GCN4 peptide) is positioned either N-terminally or C-terminally to the RT and the anti-GCN4 antigen binding domain is positioned either N-terminally or C-terminally to the nickase.
- the system and methods described hereininclude a construct that encodes a polypeptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 574 (SunTag (GCN4 peptide) positioned C-terminally to the nickase), and a construct that encodes a polypeptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 575 (anti-GCN4 antigen binding domain positioned N-terminally to the RT).
- a constructincludes a nucleotide sequence encoding a self-cleaving peptide (e.g., a P2A) positioned between the nucleotide sequence encoding the nickase and the nucleotide sequence encoding the RT.
- a nucleotide sequence encoding a self-cleaving peptidee.g., a P2A
- the nucleotide sequence encoding the RTis such that when expressed the nickase is not linked to the reverse transcriptase.
- the constructincludes the nucleotide sequences positioned 5 ’-3’: nucleotide sequence encoding the nickase, nucleotide sequence encoding the self-cleaving peptide (e.g., a P2A), and nucleotide sequence encoding the RT. In one embodiment, the construct includes the nucleotide sequences positioned 5 ’-3’: nucleotide sequence encoding the RT, nucleotide sequence encoding the selfcleaving peptide (e.g., a P2A), and nucleotide sequence encoding the nickase.
- the construct that includes a nucleotide sequence encoding a selfcleaving peptide (e.g., a P2A) positioned between the nucleotide sequence encoding the nickase and the nucleotide sequence encoding the RThas an amino acid sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 581 or 582.
- a selfcleaving peptidee.g., a P2A
- the systems and methods described hereininclude a vector that is capable of co-delivering a template polynucleotide, one or more attachment site-containing gRNA, one or more integrases, one or more recombinases, a gene editor polynucleotide, one or more integration recognition sites, one or more recombinase recognition sites, or a combination thereof.
- Non-limiting examples of vectors that can be used in the methods or systems described hereininclude the vectors described in FIGs. 3-6.
- the vectorincludes a polynucleotide sequence encoding an attachment site-containing guide RNA (atgRNA).
- the polynucleotide sequence encoding the attachment site-containing guide RNA (atgRNA)is operably linked to a regulatory element (e.g., a U6 promoter) that is capable of driving expression of the atgRNA.
- the atgRNAcomprises (i) a domain that is capable of guiding the prime editor system to a target sequence; and (ii) a reverse transcriptase (RT) template that comprises at least a portion of a first integration recognition site.
- the RT templatecomprises the entirety of the first integration recognition site.
- the vector or the LNPincludes a polynucleotide sequence encoding a nicking gRNA.
- the vectorincludes a polynucleotide sequence encoding a first attachment site-containing guide RNA (atgRNA) and a polynucleotide sequence encoding a second attachment site-containing guide RNA (atgRNA).
- atgRNAfirst attachment site-containing guide RNA
- atgRNAsecond attachment site-containing guide RNA
- the first atgRNA and the second atgRNAare an at least first pair of atgRNAs, wherein the at least first pair of atgRNAs have domains that are capable of guiding the prime editor system to a target sequence, the first atgRNA further includes a first RT template that comprises at least a portion of the a first integration recognition site; the second atgRNA further includes a second RT template that comprises at least a portion of the first integration recognition site, and the first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site.
- the first atgRNA and second atgRNAinclude at least a 6bp overlap.
- the vectorincludes a template polynucleotide and a sequence that is an integration cognate of an integration recognition site site-specifically incorporated into the genome of a cell.
- the vectorincludes a template polynucleotide and a second integration recognition site that is a cognate pair with the first integration recognition site site- specifically incorporated into the genome of the cell.
- the sequence that is an integration cognatee.g., a second integration recognition site
- the vector comprising a template polynucleotideis a recombinant adenovirus, a helper dependent adenovirus, an AAV, a lentivirus, an HSV, an annelovirus, a retrovirus, a DoggyboneTM DNA (dbDNA), a minicircle, a plasmid, a miniDNA, an exosome, a fusosome, or an nanoplasmid.
- the vectoris capable of localizing to the nucleus.
- the template polynucleotideis delivered to the cytoplasm and localizes to the nucleus. In certain embodiments, the template polynucleotide is delivered to the cytoplasm by LNP. In certain embodiments, the donor template polynucleotide construct comprises a recognition sequence that is recognized by a DNA binding protein (DNA binding domain) or a transcription factor binding domain. In certain embodiments, the donor template polynucleotide construct is delivered to the nucleus by an integrase or recombinase.
- the template polynucleotideis delivered to the mitochondria.
- the donor template polynucleotide constructcomprises a mitochondria targeting sequence.
- the vector comprising a template polynucleotideis AAV.
- the AAVcontains a 5’ inverted terminal repeat (ITR).
- the AAVcontains a 3’ inverted terminal repeat (ITR).
- the AAVcontains a 5’ and a 3’ ITR.
- the 5’ and 3’ ITRare not derived from the same serotype of virus.
- the ITRsare derived from adenovirus, AAV2, and/or AAV5.
- the vector comprising a template polynucleotideis single stranded AAV (ssAAV).
- the vector comprising a donor template polynucleotide constructis self-complementary AAV (sc AAV).
- a vectorcomprises an attachment site-containing guideRNA (atgRNA), a nicking-guideRNA (ngRNA), and template polynucleotide.
- the vector comprising an attachment site-containing guideRNA (atgRNA), a nicking-guideRNA (ngRNA), and template polynucleotideis recombinant adenovirus, helper dependent adenovirus, AAV, lentivirus, HSV, annelovirus, retrovirus, DoggyboneTM DNA (dbDNA), minicircle, plasmid, miniDNA, exosome, fusosome, or nanoplasmid.
- the vectoris capable of localizing to the nucleus.
- the attachment site-containing guideRNA (atgRNA) sequence and the nicking-guideRNA (ngRNA) sequencecontain a terminal poly dT.
- a vectorcomprises an attachment site-containing guideRNA (atgRNA), and donor template.
- the vector comprising an attachment sitecontaining guideRNA (atgRNA) and donor templateis recombinant adenovirus, helper dependent adenovirus, AAV, lentivirus, HSV, annelovirus, retrovirus, DoggyboneTM DNA (dbDNA), minicircle, plasmid, miniDNA, exosome, fusosome, or nanoplasmid.
- the vectoris capable of localizing to the nucleus.
- the attachment sitecontaining guideRNA (atgRNA) sequencecontain a terminal poly dT.
- the template polynucleotideis capable of being integrated into a genomic locus that contains an integrase target recognition site or a recombinase target recognition site.
- the template polynucleotidecomprises at least one of the following: a gene, a gene fragment, an expression cassette, a logic gate system, or any combination thereof. In some embodiments, the template polynucleotide comprises at least one intron or exon.
- the template polynucleotidefurther comprises at least one integrase target recognition site or a recombinase target integrase site.
- at least one integrase target recognition site or a recombinase target integrase siteis placed within the donor template vector inverted terminal repeat.
- the delivery system(e.g., co-delivery system) includes a vector having a sub-sequence that is capable of self-circularizing to form a self-circular nucleic acid.
- the vectorcomprises a physical portion or region of the vector that is capable of self-circularizing to form a circular construct.
- sub-sequencerefers to a portion of the vector that is capable of self-circularizing, where the sub-sequence is flanked by integration recognition sites or recombinase recognition sites positioned to enable selfcircularization.
- self-circular nucleic acidrefers to a double- stranded, circular nucleic acid construct produced as a result of recombination of a cognate pair of integrase or recombinase recognition sites present on the vector. Recombination occurs when the vector is contacted with an integrase or a recombinase under conditions that allow for recombination of the cognate pair of integrase or recombinase recognition sites.
- the sub-sequence of the vectorincludes a first recombinase recognition site and a second recombinase recognition site, wherein the first and second recombinase recognition sites are capable of being recombined by a recombinase.
- the sub-sequence of the vectorincludes a first recombinase recognition site, a second recombinase recognition site, and a second integration recognition site (e.g., the second integration recognition site is a cognate pair of the first integration recognition site), where the first and second recombinase recognition sites flank the integration recognition site.
- the first recombinase recognition site, the second recombinase recognition, and a recombinaseenable the self-circularizing and formation of the circular construct.
- the sub-sequence of the vectorincludes a third integration recognition site and a fourth integration recognition site, wherein the third and fourth integration recognition sites are a cognate pair.
- the subsequence of the vectorincludes the second integration recognition site, the third integration recognition site, the fourth integration recognition site, where the third and fourth integration recognition sites flank the second integration recognition site (where the second integration recognition site is a cognate pair of the first integration recognition site).
- the third integration recognition site, the fourth integration recognition site, and an integraseenable self -circularization and formation of the circular construct.
- the third integration recognition site and/or the fourth integration recognition sitescannot recombine with the first integration recognition site and/or the second integration recognition site due, in part, to having different central dinucleotides than the first and second integration recognition sites.
- each integration recognition site or each pair of integration recognitionis capable of being recognized by a different integrase. In some embodiments where the subsequence includes three or more integration recognition sites, each integration recognition site or each pair of integration recognition comprises a different central dinucleotide.
- self-circularizingis mediated at the integration recognition sites or recombinase recognition sites. In some embodiments, the self-circularizing is mediated by an integrase or a recombinase.
- the self-circular nucleic acid comprising the second integration recognition siteis capable of being integrated into the cell’s genome at the target sequence that contains the first integration recognition site.
- the self-circular nucleic acidcomprises one or more additional integration recognition sites that enable integration of an additional nucleic acid cargo.
- the additional nucleic acid cargoincludes a sequence that is a cognate pair with one or more of the additional integration recognition sites in the selfcircular nucleic acid.
- integration of the self-circular nucleic acid into the genome of a cellresults in integration of the one or more additional integration recognition sites into the genome along with the nucleic acid cargo.
- the integrated one or more additional integration recognition sitesserve as an integration recognition site (beacon) for placing the additional nucleic acid cargo.
- the additional nucleic acid cargois integrated into the cell’s genome.
- the self-circularized nucleic acidcomprises a DNA cargo
- the DNA cargois a gene or gene fragment.
- the DNA cargois an expression cassette.
- the DNA cargois a logic gate or logic gate system.
- the logic gate or logic gate systemmay be DNA based, RNA based, protein based, or a mix of DNA, RNA, and protein.
- the nucleic acid cargois a genetic, protein, or peptide tag and/or barcode.
- the system or methods described hereininclude a second vector.
- the gene editor polynucleotideencodes a prime editor system comprising a nickase (e.g., any of the Cas proteins or variants thereof (e.g., nickases) and nickases described herein, see Tables 4-8) and a reverse transcriptase (e.g., any of the reverse transcriptase described herein)
- the second vectorcomprises a polynucleotide sequence encoding an integrase (e.g., any of the integrases described herein, e.g., as described in Table 10 and also in Yamall et al., Nat.
- the second vectorcomprises a polynucleotide sequence encoding at least a first recombinase.
- the gene editor polynucleotide encodes a prime editor system comprising a nickase, a reverse transcriptase, and an integrasethe second vector comprises a polynucleotide sequence encoding at least a first recombinase.
- the second vectorcomprises a polynucleotide sequence encoding at least a second integrase.
- the second vectorincludes a template polynucleotide and a sequence that is an integration cognate of an integration recognition site site-specifically incorporated into the genome of a cell.
- the second vectorincludes a template polynucleotide and a second integration recognition site that is a cognate pair with the first integration recognition site site-specifically incorporated into the genome of the cell.
- the sequence that is an integration cognatee.g., a second integration recognition site
- a second integration recognition siteenables integration of the template polynucleotide or portion thereof when contacted with an integrase and the site-specifically incorporated first integration recognition site.
- the second vectoris a vector selected from: adenovirus, AAV, lentivirus, HSV, annelovirus, retrovirus, DoggyboneTM DNA (dbDNA), minicircle, plasmid, miniDNA, exosome, fusosome, or nanoplasmid.
- the polynucleotide sequence encoding the prime editor systemis encoded on at least two different vectors.
- a first vectorcomprises a polynucleotide sequence encoding a nickase and a second vector comprises a polynucleotide sequence encoding a reverse transcriptase. In such cases, the first vector and second are delivered concurrently.
- the polynucleotide sequence(s) encoding the prime editor systemis encoded on at least two (non-contiguous) polynucleotide sequences.
- a first polynucleotide sequenceencodes a nickase and a second polynucleotide sequence encodes a reverse transcriptase.
- the first vector and secondare delivered concurrently (e.g., in a first LNP).
- the methodincludes co-delivering to a cell a first gene editor polynucleotide construct and a first attachment site-containing guide RNA (atgRNA) are packaged, and thereby vectorized, within the first LNP, and a second gene editor polynucleotide construct and a second attachment site containing guide RNR (atgRNA) are packaged, and thereby vectorized, within the second LNP, where the first atgRNA and the second atgRNA are an at least first pair of atgRNA.
- the at least first pair of atgRNAscomprise domains that are capable of guiding the prime editor system to a target sequence.
- the first atgRNAfurther includes a first RT template that comprises at least a portion of a first integration recognition site.
- the second atgRNAfurther includes a second RT template that comprises at least a portion of the first integration recognition site.
- the first atgRNA and the second atgRNAscollectively encode the entirety of the first integration recognition site.
- the first atgRNA and second atgRNAinclude at least a 6bp overlap.
- the methodincludes delivering a first LNP (e.g., a first LNP comprising a first gene editor polynucleotide construct and a first atgRNA) and a second LNP (e.g., a second LNP comprising a second gene editor polynucleotide construct and a second atgRNA), the first LNP and the second LNP are mixed prior to delivering to a cell.
- a first LNPe.g., a first LNP comprising a first gene editor polynucleotide construct and a first atgRNA
- a second LNPe.g., a second LNP comprising a second gene editor polynucleotide construct and a second atgRNA
- the first LNP and the second LNPare mixed at a ratio of first LNP to second LNP of 1:10, 1:9, 1:8, 1:7, 1:6, 1:5, 1:4, 1:3, 1:2, 1:1, 1:0.75, 0.75:1, 2:1, 3:1, 4:1, 5:1, 6:1, 7:1, 8:1, 9:1, or 10:1.
- the first LNP and the second LNPare mixed at a ratio of 1:1.
- a first LNP comprising a first gene editor polynucleotide construct and a first attachment site-containing guide RNAcomprises a ratio of ratio of gene editor polynucleotide construct (e.g., mRNA) to atgRNAl of 1:10, 1:9, 1:8, 1:7, 1:6, 1:5, 1:4, 1:3, 1:2, 1:1, 1:0.75, 0.75:1, 2:1, 3:1, 4:1, 5:1, 6:1, 7:1, 8:1, 9:1, or 10:1.
- the first LNPcomprises a ratio of mRNA to atgRNAl of 2:1.
- a second LNPcomprising a second gene editor polynucleotide construct and a second attachment site-containing guide RNA (atgRNA2) comprises a ratio of gene editor polynucleotide construct (e.g., mRNA) to atgRNA2 of 1:10, 1:9, 1:8, 1:7, 1:6, 1:5, 1:4, 1:3, 1:2, 1:1, 1:0.75, 0.75:1, 2:1, 3:1, 4:1, 5:1, 6:1, 7:1, 8:1, 9:1, or 10:1.
- the second LNPcomprises a ratio of mRNA to atgRNA2 of 2:1.
- the methodincludes delivering a first LNP (e.g., a first LNP comprising a first gene editor polynucleotide construct and a first atgRNA) and a second LNP (e.g., a second LNP comprising a second gene editor polynucleotide construct and a second atgRNA)
- the first LNP and the second LNPare mixed such that the ratio of gene editor polynucleotide construct (e.g., mRNA) to first atgRNA (atgRNAl) to second atgRNA (atgRNA2) is 1:0.25:0.25, l:0.5:0.5, 1:0.75:0.75, or 1:1:1.
- the method of co-delivering to a cell a mixture of LNPsincludes co-delivering three or more LNPs, four or more LNPs, five or more LNPs, six or more LNPs, seven or more LNPs, eight or more LNPs, nine or more LNPs, or ten or more LNPs.
- a systemcapable of site-specifically integrating at least a first integration recognition site into the genome of a cell, the system comprising: a first gene editor polynucleotide construct and a first attachment site-containing guide RNA (atgRNA) are packaged, and thereby vectorized, within the first LNP, and a second gene editor polynucleotide construct and a second attachment site containing guide RNR (atgRNA) are packaged, and thereby vectorized, within the second LNP, where the first atgRNA and the second atgRNA are an at least first pair of atgRNA.
- the at least first pair of atgRNAscomprise domains that are capable of guiding the prime editor system to a target sequence.
- the first atgRNAfurther includes a first RT template that comprises at least a portion of a first integration recognition site.
- the second atgRNAfurther includes a second RT template that comprises at least a portion of the first integration recognition site.
- the first atgRNA and the second atgRNAscollectively encode the entirety of the first integration recognition site.
- the first atgRNA and second atgRNAinclude at least a 6bp overlap.
- the systemcomprises a first LNP (e.g., any of the first LNPs described herein) and a second LNP (e.g., any of the second LNPs described herein) at a ratio of first LNP to second LNP of 1:10, 1:9, 1:8, 1:7, 1:6, 1:5, 1:4, 1:3, 1:2, 1:1, 1:0.75, 0.75:1, 2:1, 3:1, 4:1, 5:1, 6:1, 7:1, 8:1, 9:1, or 10:1.
- the systemcomprise the first LNP and the second LNP at a ratio of 1 : 1.
- the systemcomprises a first LNP having a ratio of a first gene editor polynucleotide construct to a first attachment site-containing guide RNA (atgRNAl) of 1:10, 1:9, 1:8, 1:7, 1:6, 1:5, 1:4, 1:3, 1:2, 1:1, 1:0.75, 0.75:1, 2:1, 3:1, 4:1, 5:1, 6:1, 7:1, 8:1, 9:1, or 10:1.
- the systemincludes a first LNP having a ratio of mRNA (i.e., mRNA encoding the gene editor protein) to atgRNAl of 2:1.
- the systemcomprise a second LNP having a ratio of a second gene editor polynucleotide construct to a second attachment site-containing guide RNA (atgRNA2) of 1:10, 1:9, 1:8, 1:7, 1:6, 1:5, 1:4, 1:3, 1:2, 1:1, 1:0.75, 0.75:1, 2:1, 3:1, 4:1, 5:1, 6:1, 7:1, 8:1, 9:1, or 10:1.
- the systemincludes a second LNP having a ratio of mRNA (i.e., mRNA encoding the gene editor protein) to atgRNA2 of 2:1.
- the systemcomprises a ratio of gene editor polynucleotide construct (e.g., mRNA encoding the gene editor protein) to first atgRNA (atgRNAl) to second atgRNA (atgRNA2) of 1:0.25:0.25, l:0.5:0.5, 1:0.75:0.75, or 1:1:1.
- the systemcomprises a mixture of LNPs comprising three or more LNPs, four or more LNPs, five or more LNPs, six or more LNPs, seven or more LNPs, eight or more LNPs, nine or more LNPs, or ten or more LNPs.
- a vectorcomprising a template polynucleotide and a sequence that is an integration cognate (i.e., cognate to an integration recognition site site-specifically incorporated into the genome of a cell) can be delivered to the cell concurrently with the split LNPs or after delivery of the split LNPs.
- a vector that includes a template polynucleotide and a second integration recognition site that is a cognate pair with the first integration recognition siteis delivered to the cell.
- the sequence that is an integration cognatee.g., a second integration recognition site
- a second integration recognition siteenables integration of the template polynucleotide or portion thereof when contacted with an integrase and the site- specifically incorporated first integration recognition site.
- the disclosureinvolves vectors, e.g. for delivering or introducing in a cell, but also for propagating these components (e.g. in prokaryotic cells).
- a "vector”is a tool that allows or facilitates the transfer of an entity from one environment to another. It is a replicon, such as a plasmid, phage, or cosmid, into which another DNA segment may be inserted so as to bring about the replication of the inserted segment.
- a vectoris capable of replication when associated with the proper control elements.
- the term “vector”refers to a nucleic acid molecule capable of transporting another nucleic acid to which it has been linked.
- Vectorsinclude, but are not limited to, nucleic acid molecules that are single- stranded, doublestranded, or partially double-stranded; nucleic acid molecules that comprise one or more free ends, no free ends (e.g. circular); nucleic acid molecules that comprise DNA, RNA, or both; and other varieties of polynucleotides known in the art.
- a ’’plasmidrefers to a circular double stranded DNA loop into which additional DNA segments can be inserted, such as by standard molecular cloning techniques.
- viral vectorwherein virally- derived DNA or RNA sequences are present in the vector for packaging into a virus (e.g.
- Viral vectorsalso include polynucleotides carried by a virus for transfection into a host cell.
- Certain vectorsare capable of autonomous replication in a host cell into which they are introduced (e.g. bacterial vectors having a bacterial origin of replication and episomal mammalian vectors).
- Other vectorse.g., non-episomal mammalian vectors are integrated into the genome of a host cell upon introduction into the host cell, and thereby are replicated along with the host genome.
- vectorsare capable of directing the expression of genes to which they are operatively-linked. Such vectors are referred to herein as "expression vectors.” Vectors for and that result in expression in a eukaryotic cell can be referred to herein as “eukaryotic expression vectors.” Common expression vectors of utility in recombinant DNA techniques are often in the form of plasmids.
- Recombinant expression vectorscan comprise a nucleic acid of the disclosure in a form suitable for expression of the nucleic acid in a host cell, which means that the recombinant expression vectors include one or more regulatory elements, which may be selected on the basis of the host cells to be used for expression, that is operatively-linked to the nucleic acid sequence to be expressed.
- "operably linked"is intended to mean that the nucleotide sequence of interest is linked to the regulatory element(s) in a manner that allows for expression of the nucleotide sequence (e.g. in an in vitro transcription/translation system or in a host cell when the vector is introduced into the host cell).
- Vector deliverye.g., plasmid, viral delivery:
- the CRISPR enzymefor instance a Type V protein such as C2cl or C2c3, and/or any of the present RNAs, for instance a guide RNA
- Effector proteins and one or more guide RNAscan be packaged into one or more vectors, e.g., plasmid or viral vectors.
- the vectore.g., plasmid or viral vector is delivered to the tissue of interest by, for example, an intramuscular injection, while other times the delivery is via intravenous, transdermal, intranasal, oral, mucosal, or other delivery methods. Such delivery may be either via a single dose, or multiple doses.
- the actual dosage to be delivered hereinmay vary greatly depending upon a variety of factors, such as the vector choice, the target cell, organism, or tissue, the general condition of the subject to be treated, the degree of transformation/modification sought, the administration route, the administration mode, the type of transformation/modification sought, etc.
- Such a dosagemay further contain, for example, a carrier (water, saline, ethanol, glycerol, lactose, sucrose, calcium phosphate, gelatin, dextran, agar, pectin, peanut oil, sesame oil, etc.), a diluent, a pharmaceutically-acceptable carrier (e.g., phosphate -buffered saline), a pharmaceutically-acceptable excipient, and/or other compounds known in the art.
- a carrierwater, saline, ethanol, glycerol, lactose, sucrose, calcium phosphate, gelatin, dextran, agar, pectin, peanut oil, sesame oil, etc.
- a pharmaceutically-acceptable carriere.g., phosphate -buffered saline
- a pharmaceutically-acceptable excipiente.g., phosphate -buffered saline
- the dosagemay further contain one or more pharmaceutically acceptable salts such as, for example, a mineral acid salt such as a hydrochloride, a hydrobromide, a phosphate, a sulfate, etc.; and the salts of organic acids such as acetates, propionates, malonates, benzoates, etc.
- auxiliary substancessuch as wetting or emulsifying agents, pH buffering substances, gels or gelling materials, flavorings, colorants, microspheres, polymers, suspension agents, etc. may also be present herein.
- Suitable exemplary ingredientsinclude microcrystalline cellulose, carboxymethylcellulose sodium, polysorbate 80, phenylethyl alcohol, chlorobutanol, potassium sorbate, sorbic acid, sulfur dioxide, propyl gallate, the parabens, ethyl vanillin, glycerin, phenol, parachlorophenol, gelatin, albumin and a combination thereof.
- the deliveryis via an adenovirus, which may be at a single booster dose containing at least 1 x 10 5 particles (also referred to as particle units, pu) of adenoviral vector.
- the dosepreferably is at least about 1 x 10 6 particles (for example, about 1 x 10 6 - 1 x 10 11 particles), more preferably at least about 1 x 10 7 particles, more preferably at least about 1 x 10 8 particles (e.g., about 1 x 10 8 - 1 x 10 11 particles or about 1 x 10 9 - 1 x 10 12 particles), and most preferably at least about 1 x IO 10 particles (e.g., about 1 x 10 9 - 1 x IO 10 particles or about 1 x 10 9 -l x 10 12 particles), or even at least about 1 x IO 10 particles (e.g., about 1 x 10 10 -l x 10 12 particles) of the adenoviral vector.
- the dosecomprises no more than about 1 x 10 14 particles, preferably no more than about 1 x 10 13 particles, even more preferably no more than about 1 x 10 12 particles, even more preferably no more than about 1 x 10 11 particles, and most preferably no more than about 1 x IO 10 particles (e.g., no more than about 1 x 10 9 particles).
- the dosemay contain a single dose of adenoviral vector with, for example, about 1 x 10 6 particle units (pu), about 2 x 10 6 pu, about 4 x 10 6 pu, about 1 x 10 7 pu, about 2 x 10 7 pu, about 4 x 10 7 pu, about 1 x 10 8 pu, about 2 x 10 8 pu, about 4 x 10 8 pu, about 1 x 10 9 pu, about 2 x 10 9 pu, about 4 x 10 9 pu, about 1 x IO 10 pu, about 2 x IO 10 pu, about 4 x IO 10 pu, about 1 x 10 11 pu, about 2 x 10 11 pu, about 4 x 10 11 pu, about 1 x 10 12 pu, about 2 x 10 12 pu, or about 4 x 10 12 pu of adenoviral vector.
- adenoviral vectorwith, for example, about 1 x 10 6 particle units (pu), about 2 x 10 6 pu, about 4 x 10 6 pu, about 1 x 10 7 pu, about 2 x
- the adenoviral vectorsin U.S. Pat. No. 8,454,972 B2 to Nabel, et. al., granted on Jun. 4, 2013; incorporated by reference herein, and the dosages at col 29, lines 36-58 thereof.
- the adenovirusis delivered via multiple doses.
- the deliveryis via an AAV.
- a therapeutically effective dosage for in vivo delivery of the AAV to a humanis believed to be in the range of from about 20 to about 50 ml of saline solution containing from about 1 x 10 10 to about 1 x IO 50 functional AAV/ml solution. The dosage may be adjusted to balance the therapeutic benefit against any side effects.
- the AAV doseis generally in the range of concentrations of from about 1 x 10 5 to 1 x IO 50 genomes AAV (sometimes referred to herein as “vector genomes” or “vg”), from about 1 x 10 8 to 1 x IO 20 genomes AAV, from about 1 x 10 10 to about 1 x 10 16 genomes, or about 1 x 10 11 to about 1 x 10 16 genomes AAV.
- a human dosagemay be about 1 x 10 13 genomes AAV.
- concentrationsmay be delivered in from about 0.001 ml to about 100 ml, about 0.05 to about 50 ml, or about 10 to about 25 ml of a carrier solution.
- the promoter used to drive nucleic acid-targeting effector protein coding nucleic acid molecule expressioncan include: AAV ITR can serve as a promoter: this is advantageous for eliminating the need for an additional promoter element (which can take up space in the vector). The additional space freed up can be used to drive the expression of additional elements (gRNA, etc.). Also, ITR activity is relatively weaker, so can be used to reduce potential toxicity due to over expression of nucleic acid-targeting effector protein. For ubiquitous expression, can use promoters: CMV, CAG, CBh, PGK, SV40, Ferritin heavy or light chains, etc.
- promotersFor brain or other CNS expression, can use promoters: SynapsinI for all neurons, CaMKIIalpha for excitatory neurons, GAD67 or GAD65 or VGAT for GABAergic neurons, etc.
- For liver expressioncan use Albumin promoter.
- For lung expressioncan use SP-B.
- For endothelial cellscan use ICAM.
- For hematopoietic cellscan use IFNbeta or CD45.
- Osteoblastscan use OG-2.
- the promoter used to drive guide RNAcan include: Pol III promoters such as U6 or Hl Use of Pol II promoter and intronic cassettes to express guide RNA Adeno Associated Virus (AAV).
- Pol III promoterssuch as U6 or Hl Use of Pol II promoter and intronic cassettes to express guide RNA Adeno Associated Virus (AAV).
- AAVAddeno Associated Virus
- Nucleic acid-targeting effector protein and one or more guide RNAcan be delivered using adeno associated virus (AAV), lentivirus, adenovirus or other plasmid or viral vector types, in particular, using formulations and doses from, for example, U.S. Pat. No. 8,454,972 (formulations, doses for adenovirus), U.S. Pat. No. 8,404,658 (formulations, doses for AAV) and U.S. Pat. No. 5,846,946 (formulations, doses for DNA plasmids) and from clinical trials and publications regarding the clinical trials involving lentivirus, AAV and adenovirus.
- AAVadeno associated virus
- lentiviruslentivirus
- adenovirus or other plasmid or viral vector typesin particular, using formulations and doses from, for example, U.S. Pat. No. 8,454,972 (formulations, doses for adenovirus), U.S.
- the route of administration, formulation and dosecan be as in U.S. Pat. No. 8,454,972 and as in clinical trials involving AAV.
- the route of administration, formulation and dosecan be as in U.S. Pat. No. 8,404,658 and as in clinical trials involving adenovirus.
- the route of administration, formulation and dosecan be as in U.S. Pat. No. 5,846,946 and as in clinical studies involving plasmids.
- Dosesmay be based on or extrapolated to an average 70 kg individual (e.g., a male adult human), and can be adjusted for patients, subjects, mammals of different weight and species.
- the viral vectorscan be injected into the tissue of interest.
- the expression of nucleic acid-targeting effectorcan be driven by a cell-type specific promoter.
- liver- specific expressionmight use the Albumin promoter and neuron- specific expression (e.g., for targeting CNS disorders) might use the Synapsin I promoter.
- AAVIn terms of in vivo delivery, AAV is advantageous over other viral vectors for a couple of reasons: Low toxicity (this may be due to the purification method not requiring ultra centrifugation of cell particles that can activate the immune response) and Low probability of causing insertional mutagenesis because it doesn't integrate into the host genome.
- AAVhas a packaging limit of 4.5 or 4.75 Kb.
- nucleic acid-targeting effector proteinsuch as a Type V protein such as C2cl or C2c3
- a promoter and transcription terminatorhave to be all fit into the same viral vector. Therefore embodiments of the disclosure include utilizing homologs of nucleic acid-targeting effector protein (such as a Type V protein such as C2cl or C2c3) that are shorter.
- the AAVcan be AAV1, AAV2, AAV5 or any combination thereof.
- AAV8is useful for delivery to the liver. The herein promoters and vectors are preferred individually.
- Packaging cellsare typically used to form virus particles that are capable of infecting a host cell. Such cells include 293 cells, which package adenovirus, and psi2 cells or PA317 cells, which package retrovirus.
- Viral vectors used in gene therapyare usually generated by producing a cell line that packages a nucleic acid vector into a viral particle.
- the vectorstypically contain the minimal viral sequences required for packaging and subsequent integration into a host, other viral sequences being replaced by an expression cassette for the polynucleotide(s) to be expressed.
- the missing viral functionsare typically supplied in trans by the packaging cell line.
- AAV vectors used in gene therapytypically only possess ITR sequences from the AAV genome which are required for packaging and integration into the host genome.
- Viral DNAis packaged in a cell line, which contains a helper plasmid encoding the other AAV genes, namely rep and cap, but lacking ITR sequences.
- the cell linemay also be infected with adenovirus as a helper.
- the helper viruspromotes replication of the AAV vector and expression of AAV genes from the helper plasmid.
- the helper plasmidis not packaged in significant amounts due to a lack of ITR sequences. Contamination with adenovirus can be reduced by, e.g., heat treatment to which adenovirus is more sensitive than AAV. Additional methods for the delivery of nucleic acids to cells are known to those skilled in the art. See, for example, US20030087817, incorporated herein by reference.
- Millington- Ward et al.(Molecular Therapy, vol. 19 no. 4, 642-649 April 2011) describes adeno-associated virus (AAV) vectors to deliver an RNA interference (RNAi)-based rhodopsin suppressor and a codon-modified rhodopsin replacement gene resistant to suppression due to nucleotide alterations at degenerate positions over the RNAi target site.
- RNAiRNA interference
- An injection of either 6.0 x 10 8 vp or 1.8 x IO 10 vp AAVwere subretinally injected into the eyes by Millington- Ward et al.
- the AAV vectors of Millington- Ward et al.may be applied to the system of the present disclosure, contemplating a dose of about 2 x 10 11 to about 6 x 10 11 vp administered to a human.
- Dalkara et al.also relates to in vivo directed evolution to fashion an AAV vector that delivers wild-type versions of defective genes throughout the retina after noninjurious injection into the eyes' vitreous humor.
- Dalkaradescribes a 7 mer peptide display library and an AAV library constructed by DNA shuffling of cap genes from AAV1, 2, 4, 5, 6, 8, and 9.
- the rcAAV libraries and rAAV vectors expressing GFP under a CAG or Rho promoterwere packaged and deoxyribonuclease-resistant genomic titers were obtained through quantitative PCR.
- the librarieswere pooled, and two rounds of evolution were performed, each consisting of initial library diversification followed by three in vivo selection steps.
- P30 rho-GFP micewere intravitreally injected with 2 ml of iodixanol-purified, phosphate- buffered saline (PBS) -dialyzed library with a genomic titer of about 1. times.10. sup.12 vg/ml.
- PBSphosphate- buffered saline
- the AAV vectors of Dalkara et al.may be applied to the nucleic acid-targeting system of the present disclosure, contemplating a dose of about 1 x 10 15 to about 1 x 10 16 vg/ml administered to a human.
- Lentiviral vectorsare retroviral vectors that are able to transduce or infect non-dividing cells and typically produce high viral titers. Selection of a retroviral gene transfer system would therefore depend on the target tissue. Retroviral vectors are comprised of cis-acting long terminal repeats with packaging capacity for up to 6-10 kb of foreign sequence. The minimum cis-acting LTRs are sufficient for replication and packaging of the vectors, which are then used to integrate the therapeutic gene into the target cell to provide permanent transgene expression.
- Widely used retroviral vectorsinclude those based upon murine leukemia virus (MuLV), gibbon ape leukemia virus (GaLV), Simian Immuno deficiency virus (SW), human immuno deficiency virus (HIV), and combinations thereof (see, e.g., Buchscher et al., J. Virol. 66:2731-2739 (1992); Johann et al., J. Virol. 66:1635-1640 (1992); Sommnerfelt et al., Virol. 176:58-59 (1990); Wilson et al., J. Virol. 63:2374-2378 (1989); Miller et al., J. Virol.
- MiLVmurine leukemia virus
- GaLVgibbon ape leukemia virus
- SWSimian Immuno deficiency virus
- HAVhuman immuno deficiency virus
- adenoviral based systemsmay be used.
- Adenoviral based vectorsare capable of very high transduction efficiency in many cell types and do not require cell division. With such vectors, high titer and levels of expression have been obtained. This vector can be produced in large quantities in a relatively simple system.
- Adeno-associated virus vectorsmay also be used to transduce cells with target nucleic acids, e.g., in the in vitro production of nucleic acids and peptides, and for in vivo and ex vivo gene therapy procedures (see, e.g., West et al., Virology 160:38-47 (1987); U.S. Pat. No. 4,797,368; WO 93/24641; Kotin, Human Gene Therapy 5:793-801 (1994); Muzyczka, J. Clin. Invest. 94:1351 (1994). Construction of recombinant AAV vectors are described in a number of publications, including U.S. Pat. No.
- Packaging cellsare typically used to form virus particles that are capable of infecting a host cell. Such cells include 293 cells, which package adenovirus, and yr2 cells or PA317 cells, which package retrovirus.
- Viral vectors used in gene therapyare usually generated by producing a cell line that packages a nucleic acid vector into a viral particle. The vectors typically contain the minimal viral sequences required for packaging and subsequent integration into a host, other viral sequences being replaced by an expression cassette for the polynucleotide(s) to be expressed. The missing viral functions are typically supplied in trans by the packaging cell line. For example, AAV vectors used in gene therapy typically only possess ITR sequences from the AAV genome which are required for packaging and integration into the host genome.
- Viral DNAis packaged in a cell line, which contains a helper plasmid encoding the other AAV genes, namely rep and cap, but lacking ITR sequences.
- the cell linemay also be infected with adenovirus as a helper.
- the helper viruspromotes replication of the AAV vector and expression of AAV genes from the helper plasmid.
- the helper plasmidis not packaged in significant amounts due to a lack of ITR sequences. Contamination with adenovirus can be reduced by, e.g., heat treatment to which adenovirus is more sensitive than AAV. Additional methods for the delivery of nucleic acids to cells are known to those skilled in the art. See, for example, US20030087817, incorporated herein by reference.
- a host cellis transiently or non-transiently transfected with one or more vectors described herein.
- a cellis transfected as it naturally occurs in a subject.
- a cell that is transfectedis taken from a subject.
- Cells taken from a subjectinclude, but are not limited to, hepatocytes or cells isolated from muscle, the CNS, eye or lung.
- Immunological cellsare also contemplated, such as but not limited to T cells, HSCs, B-cells and NK cells.
- mRNA delivery methods and compositionsthat may be utilized in the present disclosure including, for example, PCT/US2014/028330, US8822663B2, NZ700688A, ES2740248T3, EP2755693A4, EP2755986A4, WO2014152940A1, EP3450553B1, BR112016030852A2, and EP3362461A1.
- Expression of CRISPR systems in particularis described by W02020014577.
- Each of these publicationsare incorporated herein by reference in their entireties. Additional disclosure hereby incorporated by reference can be found in Kowalski et al., “Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery,” Mol Therap., 2019; 27(4): 710-728.
- the cellis derived from cells taken from a subject, such as a cell line.
- a cell lineA wide variety of cell lines for tissue culture are known in the art. Examples of cell lines include, but are not limited to, C8161, CCRF-CEM, MOLT, mIMCD-3, NHDF, HeLa-S3, Huhl, Huh4, Huh7, HUVEC, HASMC, HEKn, HEKa, MiaPaCell, Panel, PC-3, TF1, CTLL-2, CIR, Rat6, CV1, RPTE, A10, T24, J82, A375, ARH-77, Calul, SW480, SW620, SKOV3, SK-UT, CaCo2, P388D1, SEM-K2, WEHI-231, HB56, TIB55, Jurkat, J45.01, LRMB, Bcl-1, BC-3, IC21, DLD2, Raw264.7, NRK, NRK-52E, MRC5, MEF, He
- a cell transfected with one or more vectors described hereinis used to establish a new cell line comprising one or more vector-derived sequences.
- one or more vectors described hereinare used to produce a nonhuman transgenic animal or transgenic plant.
- the transgenic animalis a mammal, such as a mouse, rat, or rabbit.
- the organism or subjectis a plant.
- the organism or subject or plantis algae. Methods for producing transgenic plants and animals are known in the art, and generally begin with a method of cell transfection, such as described herein.
- the disclosureprovides for methods of modifying a target polynucleotide in a prokaryotic or eukaryotic cell, which may be in vivo, ex vivo or in vitro.
- the methodcomprises sampling a cell or population of cells from a human or non-human animal or plant (including micro-algae) and modifying the cell or cells. Culturing may occur at any stage ex vivo. The cell or cells may even be re-introduced into the non-human animal or plant (including micro-algae).
- pathogensare often host-specific.
- Fusariumn oxysporumf. sp. lycopersicicauses tomato wilt but attacks only tomato
- Plantshave existing and induced defenses to resist most pathogens. Mutations and recombination events across plant generations lead to genetic variability that gives rise to susceptibility, especially as pathogens reproduce with more frequency than plants.
- there can be non-host resistancee.g., the host and pathogen are incompatible.
- Horizontal Resistancee.g., partial resistance against all races of a pathogen, typically controlled by many genes
- Vertical Resistancee.g., complete resistance to some races of a pathogen but not to other races, typically controlled by a few genes.
- Plant and pathogensevolve together, and the genetic changes in one balance changes in other. Accordingly, using Natural Variability, breeders combine most useful genes for Yield. Quality, Uniformity, Hardiness, Resistance.
- the sources of resistance genesinclude native or foreign Varieties, Heirloom Varieties, Wild Plant Relatives, and Induced Mutations, e.g., treating plant material with mutagenic agents.
- plant breedersare provided with a new tool to induce mutations. Accordingly, one skilled in the art can analyze the genome of sources of resistance genes, and in Varieties having desired characteristics or traits employ the present disclosure to induce the rise of resistance genes, with more precision than previous mutagenic agents and hence accelerate and improve plant breeding programs.
- target polynucleotidesinclude a sequence associated with a signaling biochemical pathway, e.g., a signaling biochemical pathway-associated gene or polynucleotide.
- target polynucleotidesinclude a disease associated gene or polynucleotide.
- a “disease-associated” gene or polynucleotiderefers to any gene or polynucleotide which is yielding transcription or translation products at an abnormal level or in an abnormal form in cells derived from a disease-affected tissues compared with tissues or cells of a non disease control.
- a disease-associated genealso refers to a gene possessing mutation(s) or genetic variation that is directly responsible or is in linkage disequilibrium with a gene(s) that is responsible for the etiology of a disease.
- the transcribed or translated productsmay be known or unknown, and may be at a normal or abnormal level.
- compositionscomprising LNP formulations capable of delivering a cargo into a liver sinusoidal endothelial cell (LSEC).
- LSECliver sinusoidal endothelial cell
- a compositioncomprises: a lipid nanoparticle (LNP) formulation comprising a plurality of LNPs, wherein each LNP comprises: (i) an ionizable lipid; (ii) a cholesterol; (iii) a polyethyleneglycol (PEG)-modified lipid; (iv) optionally a neutral lipid, or (v) a combination of (i) - (iv); each LNP encapsulates a cargo; wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (LSEC) the encapsulated cargo.
- LNPlipid nanoparticle
- the cargois selected from: an mRNA, an siRNA, an antisense oligonucleotide, a DNA, a guide RNA (gRNA), a protein, a peptide, a chemotherapeutic agent, a small molecule drug, or a combination thereof.
- the cargois an mRNA and a gRNA.
- the mRNAis a gene editor polynucleotide encoding a gene editor polypeptide and the gRNA is an attachment site-containing guide RNA (atgRNA).
- the compositionincludes: a lipid nanoparticle (LNP) formulation comprising a plurality of LNPs, wherein each LNP comprises: (i) a polyethyleneglycol (PEG)- modified lipid; (ii) a neutral lipid, or (iii) both (i) and (ii); wherein each LNP encapsulates: (i) a gene editor polynucleotide; (ii) at least a first attachment-site containing guide RNA (atgRNA); (iii) or both (i) and (ii), wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (LSEC) the (i) a gene editor polynucleotide; (ii) at least a first attachment- site containing guide RNA (atgRNA); (iii) or both (i) and (ii).
- LNPlipid nanoparticle
- the lipid nanoparticle formulationincludes: (a) a gene editor polynucleotide; (b) at least a first attachment- site containing guide RNA (atgRNA); (c) an ionizable lipid; (d) a cholesterol; (e) a polyethyleneglycol (PEG) -modified lipid; and (f) optionally a neutral lipid, wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (a), (b), (c), (d), (e), (f), or a combination thereof.
- atgRNAguide RNA
- PEGpolyethyleneglycol
- the ionizable lipidis an ionizable cationic lipid.
- the ionizable cationic lipidis selected from N-(2,3dioleyloxy)propyl)- N,N,Ntrimethylammonium chloride (DOTMA); N-(2,3dioleoyloxy)propyl)-N,N,N- trimethylamntonium chloride (DODAP); N,N-dioleyl-N,N-dimethylammonium chloride (DODAC); 1,2 bis (oleoyloxy)-3-(trimethylammonio) propane (DOTAP); N,NdistearylN,N- dimethylammonium bromide (DDAB); 3-(N — (N,N-dimethylaminoethane)-carbam- oyl)cholesterol (DC-Choi); diheptadecylamidoglycylspermidine (DHGS)
- DOTMAN-(2,3d
- the anionic lipidis selected from: a phospholipid, phospholipid derivative, a sterol, a fatty acid, or a combination thereof.
- the PEG-modified lipidis select from 1,2-Dimyristoyl-sn-glycerol methoxypolyethylene glycol (PEG-DMG), PEG-disteryl glycerol (PEG-DSG), PEG- dipalmetoleyl, PEG-dioleyl, PEG-distearyl, PEG-diacylglycamide (PEG-DAG), PEG-dipalmitoyl phosphatidylethanolamine (PEG-DPPE), and PEG-l,2-dimyristyloxlpropyl-3-amine (PEG-c- DMA), or a combination thereof.
- PEG-DMG1,2-Dimyristoyl-sn-glycerol methoxypolyethylene glycol
- PEG-DSGPEG-disteryl glycerol
- PEG- dipalmetoleylPEG-dioleyl
- PEG-distearylPEG-diacylg
- the PEG-modified lipidcomprises a moiety capable binding to a receptor on a liver sinusoidal endothelial cell (LSEC).
- the moietyis capable of binding to a mannose receptor, a stabilin receptor, an Fc receptor, or a combination thereof.
- the moietyis capable of binding to a mannose receptor, a stabilin receptor, an Fc receptor, or a combination thereof.
- the moietyis a mannose.
- the PEG-modified lipidis a mannose-PEG lipid.
- the neutral lipidis a phospholipid.
- the phospholipidis a natural phospholipid selected from: phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylglycerol (PG), phosphatidylserine (PS), phosphatidylinositol (PI), Phosphatidic acid (phosphatidate) (PA), dipalmitoylphosphatidylcholine, monoacyl-phosphatidylcholine (lyso PC), l-palmitoyl-2-oleoyl- sn-glycero-3-phosphocholine (POPC), N-Acyl-PE, phosphoinositides, and phospho sphingolipids, or a combination thereof.
- PCphosphatidylcholine
- PEphosphatidylethanolamine
- PGphosphatidylglycerol
- PSphosphatidylserine
- PIphosphatidylinositol
- PAP
- the phospholipidis a phospholipid derivative selected from: phosphatidic acid (DMPA, DPPA, DSPA), phosphatidylcholine (DDPC, DLPC, DMPC, DPPC, DSPC, DOPC, POPC, DEPC), phosphatidylglycerol (DMPG, DPPG, DSPG, DOPG), phosphatidylethanolamine (DMPE, DPPE, DSPE DOPE), and phosphatidylserine (DOPS), or a combination thereof.
- DMPAphosphatidic acid
- DDPCphosphatidylcholine
- DDPCDLPC
- DMPCDPPC
- DSPCDOPC
- POPCPOPC
- DEPCphosphatidylglycerol
- DMPEDPPG, DSPG, DOPG
- DOPGphosphatidylethanolamine
- DOPSphosphatidylserine
- the phospholipidis selected from: l,2-dioleoyl-sn-glycero-3- phospho-rac-(l-glycerol) sodium salt (DOPG); l,2-dilinoleoyl-sn-glycero-3-phosphocholine (DLPC); 1,2-dimyristoyl-sn-glycero-phosphocholine (DMPC); 1 ,2-dioleoyl-sn-glycero-3- phosphocholine (DOPC); l,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC); 1,2-distearoyl- sn-glycero-3-phosphocholine (DSPC); 1,2-diundecanoyl-sn-glycero-phosphocholine (DUPC); 1- palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC); l,2-di-
- the phospholipidis DOPG.
- each LNPcomprises: (i) modified PEGylated lipids; and (ii) neutral lipids.
- each LNPcomprises (i) PEG-modified lipids comprising a moiety capable binding to a receptor on a liver sinusoidal endothelial cell (LSEC); and (ii) phospholipids.
- LSECliver sinusoidal endothelial cell
- each LNPcomprises mannose-PEG lipids and DOPGs.
- the LNPfurther comprises: cholesterol and an ionizable lipid.
- each LNP in the LNP formulationranges from about 100 nm to about 200 nm.
- each LNP in the LNP formulationranges from about 85 nm to about 120 nm.
- each LNP in the LNP formulationranges from about 45 nm to about 65 nm.
- each LNP in the LNP formulationranges from about 40 nm to about 50 nm.
- the encapsulation efficiencyis about 80% to about 100%. [0283] In some embodiments, the encapsulation efficiency is no less than 84%.
- the encapsulation efficiencyis no less than 90%.
- the polydispersity index of the compositionranges from about 0.005 to 0.5.
- the polydispersity index of the compositionranges from about 0.025 to 0.4.
- the LNP componentsare provided in the following ratios (e.g., molar ratios) 15-60% ionizable lipid, 4-40% phospholipid, 10-60% cholesterol, and 0-5% PEG- lipid.
- the LNP componentsare provided in the following ratios: 15-60% cKKE12, 4-40% DOPE or DOPG, 10-60% cholesterol, and 0-5% PEG2K-DMG. In some embodiments, the LNP components are provided in the following ratios: 40% cKKE12, 30% DOPE or DOPG, 28% Cholesterol, and 2% PEG2K-DMG.
- the LNP componentsare provided in the following ratios: 15-60% cKKE12, 4-40% DOPE or DOPG, 10-60% cholesterol, and 0-5% PEG2K-DMG/ Mannose- PEG2K-DSPE. In some embodiments, the LNP components are provided in the following ratios: 40% cKKE12, 30% DOPE or DOPG, 27% cholesterol, 0.5% PEG2K-DMG, and 2.5% Mannose- PEG2K-DSPE.
- the LNP componentsare provided in the following ratios: 15-60% ALC0315, 4-40% DSPC or DSPG, 10-60% Cholesterol, and 0-5% PEG2K-DMG. In some embodiments, the LNP components are provided in the following ratios: 46.3% ALC0315, 9.4% DSPC or DSPG, 42.7% Cholesterol, and 1.6% PEG2K-DMG.
- the LNP componentsare provided in the following ratios: 15-60% ALC0315, 4-40% DSPC or DSPG, 10-60% Cholesterol, and 0-5% PEG2K-DMG/Mannose- PEG2K-DSPE. In some embodiments, the LNP components are provided in the following ratios: 46.3% ALC0315, 9.4% DSPC or DSPG, 41.3% Cholesterol, 0.5% PEG2K-DMG, and 2.5% Mannose-PEG2K-DSPE.
- the LNP componentsare provided in the following ratios: 15- 60% 246C10, 4-40% DOPE or DOPG, 10-60% Cholesterol, and 0-5% C16-PEG2K-ceramide. In some embodiments, the LNP components are provided in the following ratios: 26.5% 246C10, 20% DOPE or DOPG, 50.5% Cholesterol, and 3% C16-PEG2K-ceramide.
- the LNP componentsare provided in the following ratios: 15- 60% 246C10, 4-40% DOPE or DOPG, 10-60% Cholesterol, and 0-5% C16-PEG2K- ceramide/Mannose-PEG2K-DSPE. In some embodiments, the LNP components are provided in the following ratios: 26.5% 246C10, 20% DOPE or DOPG, 50.5% Cholesterol, 0.5% C16- PEG2K-ceramide, and 2.5% Mannose-PEG2K-DSPE
- the gene editor polypeptide or polynucleotide encoding the gene editor polypeptideis packaged in a LNP and administered intravenously. In some embodiments, the gene editor polypeptide or polynucleotide encoding the gene editor polypeptide is packaged in a LNP and administered intrathecally. In some embodiments, the gene editor polypeptide or polynucleotide encoding the gene editor polypeptide is packaged in a LNP and administered by intracerebral ventricular injection. In some embodiments, the gene editor polypeptide or polynucleotide encoding the gene editor polypeptide is packaged in a LNP and administered by intracisternal magna administration. In some embodiments, the gene editor polypeptide or polynucleotide encoding the gene editor polypeptide is packaged in a LNP and administered by intravitreal injection.
- lipidmucleic acid complexesincluding targeted liposomes such as immunolipid complexes
- Boese et al.Cancer Gene Ther. 2:291-297 (1995); Behr et al., Bioconjugate Chem. 5:382-389 (1994); Remy et al., Bioconjugate Chem. 5:647-654 (1994); Gao et al., Gene Therapy 2:710-722 (1995); Ahmad et al., Cancer Res. 52:4817-4820 (1992); U.S. Pat. Nos. 4,186,183, 4,217,344, 4,235,871, 4,261,975, 4,485,054, 4,501,728, 4,774,085, 4,837,028, and 4,946,787).
- LNP dosesof about 0.01 to about 1 mg per kg of body weight administered intravenously are contemplated.
- Medications to reduce the risk of infusion-related reactionsare contemplated, such as dexamethasone, acetampinophen, diphenhydramine or cetirizine, and ranitidine are contemplated.
- Multiple doses of about 0.3 mg per kilogram every 4 weeks for five dosesare also contemplated.
- the charge of the LNPmust be taken into consideration. As cationic lipids combined with negatively charged lipids to induce nonbilayer structures that facilitate intracellular delivery. Because charged LNPs are rapidly cleared from circulation following intravenous injection, ionizable cationic lipids with pKa values below 7 were developed (see, e.g., Rosin et al, Molecular Therapy, vol. 19, no. 12, pages 1286-2200, December 2011). Negatively charged polymers such as RNA may be loaded into LNPs at low pH values (e.g., pH 4) where the ionizable lipids display a positive charge. However, at physiological pH values, the LNPs exhibit a low surface charge compatible with longer circulation times.
- pH 4e.g., pH 4
- ionizable cationic lipidsFour species of ionizable cationic lipids have been focused upon, namely l,2-dilineoyl-3-dimethylammonium-propane (DLinDAP), 1,2- dilinoleyloxy-3-N,N-dimethylaminopropane (DLinDMA), 1 ,2-dilinoleyloxy-keto-N,N-dimethyl- 3-aminopropane (DLinKDMA), and l,2-dilinoleyl-4-(2-dimethylaminoethyl)-[l,3]-dioxolane (DlinKC2-DMA).
- DLinDAP1,2- dilinoleyloxy-3-N,N-dimethylaminopropane
- DLinKDMA1,2-dilinoleyloxy-keto-N,N-dimethyl- 3-aminopropane
- DlinKC2-DMA
- LNP siRNA systems containing these lipidsexhibit remarkably different gene silencing properties in hepatocytes in vivo, with potencies varying according to the series DlinKC2-DMA>DLinKDMA>DLinDMA»DLinDAP employing a Factor VII gene silencing model (see, e.g., Rosin et al, Molecular Therapy, vol. 19, no. 12, pages 1286-2200, December 2011).
- a dosage of 1 pg/ml of LNP in or associated with the LNPmay be contemplated, especially for a formulation containing DlinKC2-DMA.
- the LNP compositioncomprises one or more one or more ionizable lipids.
- ionizable lipidhas its ordinary meaning in the art and may refer to a lipid comprising one or more charged moieties.
- an ionizable lipidmay be positively charged or negatively charged.
- the one or more ionizable lipidsare selected from the group consisting of 3-(didodecylamino)- Nl,Nl,4-tridodecyl-l-piperazineethanamine (KL10), Nl-[2-(didodecylamino)ethyl]-Nl,N4,N4- tridodecyl-l,4-piperazinediethanami- ne (KL22), 14,25-ditridecyl-15,18,21,24-tetraaza- octatriacontane (KL25), l,2-dilinoleyloxy-N,N-dimethylaminopropane (Dlin-DMA), 2,2- dilinoleyl-4-dimethylaminomethyl-[ 1 ,3]-dioxolane (Dlin-K-DMA), heptatriaconta-6,9,28,31 -
- KL10
- the ionizable lipidmay be selected from, but not limited to, an ionizable lipid described in International Publication Nos. WO2013086354 and WO2013116126.
- the lipid nanoparticlemay include one or more (e.g., 1, 2, 3, 4, 5, 6, 7, or 8) cationic and/or ionizable lipids.
- cationic and/or ionizable lipidsinclude, but are not limited to, 3-(didodecylamino)-Nl,Nl,4-tridodecyl-l-piperazineethanamine (KL10), Nl-[2- (didodecylamino)ethyl]-Nl,N4,N4-tridodecyl-l,4-piperazinediethanami- ne (KL22), 14,25- ditridecyl- 15, 18,21 ,24-tetraaza-octatriacontane (KL25), 1 ,2-dilinoleyloxy-N,N- dimethylaminopropane (Dlin-DMA) , 2,2-dilinoleyl-4-dimethyl
- lipidse.g., LIPOFECTIN.RTM. (including DOTMA and DOPE, available from GIBCO/BRL), and LIPOFECT AMINE. RTM. (including DOSPA and DOPE, available from GIBCO/BRL).
- LIPOFECTIN.RTMincluding DOTMA and DOPE, available from GIBCO/BRL
- LIPOFECT AMINE. RTM.including DOSPA and DOPE, available from GIBCO/BRL
- KL10, KL22, and KL25are described, for example, in U.S. Pat. No. 8,691,750.
- the LNP compositioncomprises one or more amino lipids.
- amino lipidand “cationic lipid” are used interchangeably herein to include those lipids and salts thereof having one, two, three, or more fatty acid or fatty alkyl chains and a pH-titratable amino head group (e.g., an alkylamino or dialkylamino head group).
- a pH-titratable amino head groupe.g., an alkylamino or dialkylamino head group.
- the cationic lipidis typically protonated (i.e., positively charged) at a pH below the pKa of the cationic lipid and is substantially neutral at a pH above the pKa.
- the cationic lipidscan also be termed titratable cationic lipids.
- the one or more cationic lipidsinclude: a protonatable tertiary amine (e.g., pH-titratable) head group; alkyl chains, wherein each alkyl chain independently has 0 to 3 (e.g., 0, 1, 2, or 3) double bonds; and ether, ester, or ketal linkages between the head group and alkyl chains.
- Such cationic lipidsinclude, but are not limited to, DSDMA, DODMA, DOTMA, DLinDMA, DLenDMA, .gamma.-DLenDMA, Dlin-K-DMA, Dlin-K-C2- DMA (also known as Dlin-C2K-DMA, XTC2, and C2K), Dlin-K-C3-DMA, Dlin-K-C4-DMA, Dlen-C2K-DMA, y-Dlen-C2-DMA, C12-200, cKK-E12, cKK-A12, cKK-012, Dlin-MC2-DMA (also known as MC2), and Dlin-MC3-DMA (also known as MC3).
- DSDMADSDMA
- DODMADODMA
- DOTMADLinDMA
- DLenDMADLenDMA
- .gamma.-DLenDMADlin-
- Anionic lipids suitable for use in lipid nanoparticlesinclude, but are not limited to, phosphatidylglycerol, cardiolipin, diacylphosphatidylserine, diacylphosphatidic acid, N- dodecanoyl phosphatidylethanoloamine, N-succinyl phosphatidylethanolamine, N-glutaryl phosphatidylethanolamine, lysylphosphatidylglycerol, and other anionic modifying groups joined to neutral lipids.
- Neutral lipidssuitable for use in lipid nanoparticles include, but are not limited to, diacylphosphatidylcholine, diacylphosphatidylethanolamine, ceramide, sphingomyelin, dihydrosphingomyelin, cephalin, sterols (e.g., cholesterol) and cerebrosides.
- the lipid nanoparticlecomprises cholesterol.
- Lipids having a variety of acyl chain groups of varying chain length and degree of saturationare available or may be isolated or synthesized by well-known techniques. Additionally, lipids having mixtures of saturated and unsaturated fatty acid chains and cyclic regions can be used.
- the neutral lipids used in the disclosureare DOPE, DSPC, DPPC, POPC, or any related phosphatidylcholine.
- the neutral lipidmay be composed of sphingomyelin, dihydrosphingomyeline, or phospholipids with other head groups, such as serine and inositol.
- amphipathic lipidsare included in nanoparticles.
- Exemplary amphipathic lipids suitable for use in nanoparticlesinclude, but are not limited to, sphingolipids, phospholipids, fatty acids, and amino lipids.
- the lipid composition of the pharmaceutical compositionmay comprise one or more phospholipids, for example, one or more saturated or (poly)unsaturated phospholipids or a combination thereof.
- phospholipidscomprise a phospholipid moiety and one or more fatty acid moieties.
- a phospholipid moietycan be selected, for example, from the non-limiting group consisting of phosphatidyl choline, phosphatidyl ethanolamine, phosphatidyl glycerol, phosphatidyl serine, phosphatidic acid, 2-lysophosphatidyl choline, and a sphingomyelin.
- a fatty acid moietycan be selected, for example, from the non-limiting group consisting of lauric acid, myristic acid, myristoleic acid, palmitic acid, palmitoleic acid, stearic acid, oleic acid, linoleic acid, alpha-linolenic acid, erucic acid, phytanoic acid, arachidic acid, arachidonic acid, eicosapentaenoic acid, behenic acid, docosapentaenoic acid, and docosahexaenoic acid.
- Particular amphipathic lipidscan facilitate fusion to a membrane.
- a cationic phospholipidcan interact with one or more negatively charged phospholipids of a membrane (e.g., a cellular or intracellular membrane). Fusion of a phospholipid to a membrane can allow one or more elements (e.g., a therapeutic agent) of a lipid-containing composition (e.g., LNPs) to pass through the membrane permitting, e.g., delivery of the one or more elements to a target tissue.
- a cationic phospholipidcan interact with one or more negatively charged phospholipids of a membrane (e.g., a cellular or intracellular membrane). Fusion of a phospholipid to a membrane can allow one or more elements (e.g., a therapeutic agent) of a lipid-containing composition (e.g., LNPs) to pass through the membrane permitting, e.g., delivery of the one or more elements to a target tissue.
- elementse.g., a
- Non-natural amphipathic lipid speciesincluding natural species with modifications and substitutions including branching, oxidation, cyclization, and alkynes are also contemplated.
- a phospholipidcan be functionalized with or cross-linked to one or more alkynes (e.g., an alkenyl group in which one or more double bonds is replaced with a triple bond).
- alkynese.g., an alkenyl group in which one or more double bonds is replaced with a triple bond.
- an alkyne groupcan undergo a copper-catalyzed cycloaddition upon exposure to an azide.
- Such reactionscan be useful in functionalizing a lipid bilayer of a nanoparticle composition to facilitate membrane permeation or cellular recognition or in conjugating a nanoparticle composition to a useful component such as a targeting or imaging moiety (e.g., a dye).
- a targeting or imaging moietye.g., a dye
- Phospholipidsinclude, but are not limited to, glycerophospholipids such as phosphatidylcholines, phosphatidylethanolamines, phosphatidylserines, phosphatidylinositols, phosphatidy glycerols, and phosphatidic acids. Phospholipids also include phospho sphingolipid, such as sphingomyelin.
- the LNP compositioncomprises one or more phospholipids.
- the phospholipidis selected from the group consisting of 1,2-dilinoleoyl-sn- glycero-3-phosphocholine (DLPC), 1,2-dimyristoyl-sn-glycero-phosphocholine (DMPC), 1,2- dioleoyl-sn-glycero-3-phosphocholine (DOPC), l,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), l,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), 1,2-diundecanoyl-sn-glycero- phosphocholine (DUPC), l-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1,2-di-O- octadecenyl-sn-glycero-3-phosphocholine (POPC), 1,2-d
- phosphorus-lacking compoundssuch as sphingolipids, glyco sphingolipid families, diacylglycerols, and .beta. -acyloxyacids, may also be used. Additionally, such amphipathic lipids can be readily mixed with other lipids, such as triglycerides and sterols.
- the LNP compositioncomprises one or more helper lipids.
- helper lipidrefers to lipids that enhance transfection (e.g., transfection of an LNP comprising an mRNA that encodes a site-directed endonuclease, such as a SpCas9 polypeptide).
- the mechanism by which the helper lipid enhances transfectionincludes enhancing particle stability.
- the helper lipidenhances membrane fusogenicity.
- helper lipid of the LNP compositions disclosure hereincan be any helper lipid known in the art.
- helper lipids suitable for the compositions and methodsinclude steroids, sterols, and alkyl resorcinols.
- helper lipids suitable for use in the present disclosureinclude, but are not limited to, saturated phosphatidylcholine (PC) such as distearoyl-PC (DSPC) and dipalymitoyl-PC (DPPC), dioleoylphosphatidylethanolamine (DOPE), 1,2-dilinoleoyl-sn- glycero-3-phosphocholine (DLPC), cholesterol, 5-heptadecylresorcinol, and cholesterol hemisuccinate.
- PCsaturated phosphatidylcholine
- DSPCdistearoyl-PC
- DPPCdipalymitoyl-PC
- DOPEdioleoylphosphatidylethanolamine
- DLPC1,2-dilinoleoyl-sn- glycero-3-phosphocholine
- cholesterol5-heptadecylresorcinol
- cholesterol hemisuccinatehemisuccinate.
- the helper lipid of the LNP composition
- the LNP compositioncomprises one or more structural lipids.
- structural lipidrefers to sterols and also to lipids containing sterol moieties. Without being bound to any particular theory, it is believed that the incorporation of structural lipids into the LNPs mitigates aggregation of other lipids in the particle.
- Structural lipidscan be selected from the group including but not limited to, cholesterol, fecosterol, sitosterol, ergosterol, campesterol, stigmasterol, brassicasterol, tomatidine, tomatine, ursolic acid, alpha-tocopherol, hopanoids, phytosterols, steroids, and mixtures thereof.
- the structural lipidis a sterol.
- sterolsare a subgroup of steroids consisting of steroid alcohols.
- the structural lipidis a steroid.
- the structural lipidis cholesterol.
- the structural lipidis an analog of cholesterol.
- the lipid component of a lipid nanoparticle compositionmay include one or more molecules comprising polyethylene glycol, such as PEG or PEG-modified lipids.
- the LNP composition disclosed hereincomprise one or more polyethylene glycol (PEG) lipid.
- PEG-lipidrefers to polyethylene glycol (PEG)-modified lipids. Such lipids are also referred to as PEGylated lipids.
- PEG-lipidsinclude PEG- modified phosphatidylethanolamine and phosphatidic acid, PEG-ceramide conjugates (e.g., PEG- CerC14 or PEG-CerC20), PEG-modified dialkylamines and PEG-modified 1,2-diacyloxypropan- 3-amines
- a PEG lipidcan be PEG-c-DOMG, PEG-DMG, PEG-DLPE, PEG-DMPE, PEG-DPPC, or a PEG-DSPE lipid.
- the PEG-lipidincludes, but not limited to 1,2-dimyristoyl-sn-glycerol methoxypolyethylene glycol (PEG-DMG), 1,2-distearoyl-sn- glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)] (PEG-DSPE), PEG-disteryl glycerol (PEG-DSG), PEG-dipalmetoleyl, PEG-dioleyl, PEG-distearyl, PEG-diacylglycamide (PEG-DAG), PEG-dipalmitoyl phosphatidylethanolamine (PEG-DPPE), or PEG- 1,2- dimyristyloxlpropyl-3-amine (PEG-c-DMA).
- PEG-DMG1,2-dimyristoyl-sn-glycerol methoxypolyethylene glycol
- PEG-DSPE1,2-distearoyl-sn-
- the PEG-lipidis selected from the group consisting of a PEG-modified phosphatidylethanolamine, a PEG-modified phosphatidic acid, a PEG-modified ceramide, a PEG-modified dialkylamine, a PEG-modified diacylglycerol, a PEG-modified dialkylglycerol, and mixtures thereof.
- the lipid moiety of the PEG-lipidsincludes those having lengths of from about C. sub.14 to about C.sub.22, preferably from about C. sub.14 to about C. sub.16.
- a PEG moietyfor example a mPEG-NH.sub.2, has a size of about 1000, 2000, 5000, 10,000, 15,000 or 20,000 daltons.
- the PEG-lipidis PEG2k-DMG.
- the one or more PEG lipids of the LNP compositioncomprises PEG-DMPE.
- the one or more PEG lipids of the LNP compositioncomprises PEG-DMG.
- the ratio between the lipid components and the nucleic acid molecules of the LNP compositionis sufficient for (i) formation of LNPs with desired characteristics, e.g., size, charge, and (ii) delivery of a sufficient dose of nucleic acid at a dose of the lipid component(s) that is tolerable for in vivo administration as readily ascertained by one of skill in the art.
- a nanoparticlee.g., a lipid nanoparticle
- a targeting moietythat is specific to a cell type and/or tissue type.
- a nanoparticlemay be targeted to a particular cell, tissue, and/or organ using a targeting moiety.
- a nanoparticlecomprises a targeting moiety.
- targeting moietiesinclude ligands, cell surface receptors, glycoproteins, vitamins (e.g., riboflavin) and antibodies (e.g., full-length antibodies, antibody fragments (e.g., Fv fragments, single chain Fv (scFv) fragments, Fab’ fragments, or F(ab’)2 fragments), single domain antibodies, camelid antibodies and fragments thereof, human antibodies and fragments thereof, monoclonal antibodies, and multispecific antibodies (e.g., bispecific antibodies)).
- the targeting moietymay be a polypeptide.
- the targeting moietymay include the entire polypeptide (e.g., peptide or protein) or fragments thereof.
- a targeting moietyis typically positioned on the outer surface of the nanoparticle in such a manner that the targeting moiety is available for interaction with the target, for example, a cell surface receptor.
- a variety of different targeting moieties and methodsare known and available in the art, including those described, e.g., in Sapra et al., Prog. Eipid Res. 42(5):439-62, 2003 and Abra et al., J. Eiposome Res. 12:1-3, 2002.
- a lipid nanoparticlemay include a surface coating of hydrophilic polymer chains, such as polyethylene glycol (PEG) chains (see, e.g., Allen et al., Biochimica et Biophysica Acta 1237: 99-108, 1995; DeFrees et al., Journal of the American Chemistry Society 118: 6101-6104, 1996; Blume et al., Biochimica et Biophysica Acta 1149: ISO- 184, 1993; Klibanov et al., Journal of Eiposome Research 2: 321-334, 1992; U.S. Pat. No.
- PEGpolyethylene glycol
- a targeting moiety for targeting the lipid nanoparticleis linked to the polar head group of lipids forming the nanoparticle.
- the targeting moietyis attached to the distal ends of the PEG chains forming the hydrophilic polymer coating (see, e.g., Klibanov et al., Journal of Eiposome Research 2: 321-334, 1992; Kirpotin et al., FEBS Letters 388: 115-118, 1996).
- Standard methods for coupling the targeting moiety or moietiesmay be used.
- phosphatidylethanolaminewhich can be activated for attachment of targeting moieties
- derivatized lipophilic compoundssuch as lipid-derivatized bleomycin
- Antibody- targeted liposomescan be constructed using, for instance, liposomes that incorporate protein A (see, e.g., Renneisen et al., J. Bio. Chem., 265:16337-16342, 1990 and Leonetti et al., Proc. Natl. Acad. Sci. (USA), 87:2448-2451, 1990).
- Other examples of antibody conjugationare disclosed in U.S. Pat. No.
- targeting moietiescan also include other polypeptides that are specific to cellular components, including antigens associated with neoplasms or tumors.
- Polypeptides used as targeting moietiescan be attached to the liposomes via covalent bonds (see, for example Heath, Covalent Attachment of Proteins to Liposomes, 149 Methods in Enzymology 111-119 (Academic Press, Inc. 1987)).
- Other targeting methodsinclude the biotin-avidin system.
- a lipid nanoparticleincludes a targeting moiety that targets the lipid nanoparticle to a cell including, but not limited to, hepatocytes, colon cells, epithelial cells, hematopoietic cells, epithelial cells, endothelial cells, lung cells, bone cells, stem cells, mesenchymal cells, neural cells, cardiac cells, adipocytes, vascular smooth muscle cells, cardiomyocytes, skeletal muscle cells, beta cells, pituitary cells, synovial lining cells, ovarian cells, testicular cells, fibroblasts, B cells, T cells, reticulocytes, leukocytes, granulocytes, and tumor cells (including primary tumor cells and metastatic tumor cells).
- the targeting moietytargets the lipid nanoparticle to a hepatocyte.
- the lipid nanoparticles described hereinmay be lipidoid-based.
- the synthesis of lipidoidshas been extensively described and formulations containing these compounds are particularly suited for delivery of polynucleotides (see Mahon et al., Bioconjug Chem. 2010 21:1448-1454; Schroeder et al., J Intern Med. 2010 267:9-21; Akinc et al., Nat. Biotechnol. 2008 26:561-569; Love et al., Proc Natl Acad Sci USA. 2010 107: 1864-1869; Siegwart et al., Proc Natl Acad Sci USA. 2011 108:12996-3001).
- lipidoid formulations for intramuscular or subcutaneous routesmay vary significantly depending on the target cell type and the ability of formulations to diffuse through the extracellular matrix into the blood stream. While a particle size of less than 150 nm may be desired for effective hepatocyte delivery due to the size of the endothelial fenestrae (see e.g., Akinc et al., Mol Ther. 2009 17:872-879), use of lipidoid oligonucleotides to deliver the formulation to other cells types including, but not limited to, endothelial cells, myeloid cells, and muscle cells may not be similarly size-limited.
- lipidoid formulationsmay have a similar component molar ratio.
- Different ratios of lipidoids and other componentsincluding, but not limited to, a neutral lipid (e.g., diacylphosphatidylcholine), cholesterol, a PEGylated lipid (e.g., PEG-DMPE), and a fatty acid (e.g., an omega-3 fatty acid) may be used to optimize the formulation of the mRNA or system for delivery to different cell types including, but not limited to, hepatocytes, myeloid cells, muscle cells, etc.
- a neutral lipide.g., diacylphosphatidylcholine
- cholesterole.g., a PEGylated lipid
- PEG-DMPEPEGylated lipid
- a fatty acide.g., an omega-3 fatty acid
- Exemplary lipidoidsinclude, but are not limited to, Dlin-DMA, Dlin-K-DMA, Dlin-KC2-DMA, 98N12-5, C 12-200 (including variants and derivatives), Dlin-MC3-DMA and analogs thereof.
- the use of lipidoid formulations for the localized delivery of nucleic acids to cells (such as, but not limited to, adipose cells and muscle cells) via either subcutaneous or intramuscular deliverymay also not require all of the formulation components which may be required for systemic delivery, and as such may comprise the lipidoid and the mRNA or system.
- a system described hereinmay be formulated by mixing the mRNA or system, or individual components of the system, with the lipidoid at a set ratio prior to addition to cells.
- In vivo formulationsmay require the addition of extra ingredients to facilitate circulation throughout the body.
- a system or individual components of a systemis added and allowed to integrate with the complex. The encapsulation efficiency is determined using a standard dye exclusion assays.
- In vivo delivery of systemsmay be affected by many parameters, including, but not limited to, the formulation composition, nature of particle PEGylation, degree of loading, oligonucleotide to lipid ratio, and biophysical parameters such as particle size (Akinc et al., Mol Ther. 2009 17:872-879; herein incorporated by reference in its entirety).
- particle sizeAkinc et al., Mol Ther. 2009 17:872-879; herein incorporated by reference in its entirety.
- small changes in the anchor chain length of poly(ethylene glycol) (PEG) lipidsmay result in significant effects on in vivo efficacy.
- Formulations with the different lipidoidsincluding, but not limited to penta[3-(l- laurylaminopropionyl)]-triethylenetetramine hydrochloride (TETA-5LAP; aka 98N12-5, see Murugaiah et al., Analytical Biochemistry, 401:61 (2010)), C12-200 (including derivatives and variants), MD1, Dlin-DMA, Dlin-K-DMA, Dlin-KC2-DMA and Dlin-MC3-DMA can be tested for in vivo activity.
- the lipidoid referred to herein as “98N12-5”is disclosed by Akinc et al., Mol Ther. 2009 17:872-879).
- the lipidoid referred to herein as “C12-200”is disclosed by Love et al., Proc Natl Acad Sci USA. 2010 107:1864-1869 and Liu and Huang, Molecular Therapy. 2010669- 670.
- LNPs in which a nucleic acid is entrapped within the lipid portion of the particle and is protected from degradationcan be formed by any method known in the art including, but not limited to, a continuous mixing method, a direct dilution process, and an in-line dilution process. Additional techniques and methods suitable for the preparation of the LNPs described herein include coacervation, microemulsions, supercritical fluid technologies, phase-inversion temperature (PIT) techniques.
- PITphase-inversion temperature
- the LNPs used hereinare produced via a continuous mixing method, e.g., a process that includes providing an aqueous solution a nucleic acid described herein in a first reservoir, providing an organic lipid solution in a second reservoir (wherein the lipids present in the organic lipid solution are solubilized in an organic solvent, e.g., a lower alkanol such as ethanol), and mixing the aqueous solution with the organic lipid solution such that the organic lipid solution mixes with the aqueous solution so as to substantially instantaneously produce a lipid vesicle (e.g., liposome) encapsulating the nucleic acid molecule within the lipid vesicle.
- a continuous mixing methode.g., a process that includes providing an aqueous solution a nucleic acid described herein in a first reservoir, providing an organic lipid solution in a second reservoir (wherein the lipids present in the organic lipid solution are solubilized in an organic solvent
- the LNPs used hereinare produced via a direct dilution process that includes forming a lipid vesicle (e.g., liposome) solution and immediately and directly introducing the lipid vesicle solution into a collection vessel containing a controlled amount of dilution buffer.
- the collection vesselincludes one or more elements configured to stir the contents of the collection vessel to facilitate dilution.
- the amount of dilution buffer present in the collection vesselis substantially equal to the volume of lipid vesicle solution introduced thereto.
- the LNPsare produced via an in-line dilution process in which a third reservoir containing dilution buffer is fluidly coupled to a second mixing region.
- the lipid vesicle (e.g., liposome) solution formed in a first mixing regionis immediately and directly mixed with dilution buffer in the second mixing region.
- This disclosureis not limited to systems and methods described herein. Any delivery method that is capable of delivering the systems described herein can be used as long as it is capable of site-specifically integrating a template polynucleotide into the genome of a cell.
- compositions, systems and methods for correcting or replacing genes or gene fragments (including introns or exons) or inserting genes in new locationscomprises recombination or integration into a safe harbor site (SHS).
- SHSsafe harbor site
- a frequently used human SHSis the AAVS1 site on chromosome 19q, initially identified as a site for recurrent adeno-associated virus insertion.
- Another locuscomprises the human homolog of the murine Rosa26 locus.
- Yet another SHScomprises the human Hl l locus on chromosome 22.
- a complete genemay be prohibitively large and replacement of an entire gene impractical.
- a method of the disclosurecomprises recombining corrective gene fragments into a defective locus.
- the methods and compositionscan be used to target, without limitation, stem cells for example induced pluripotent stem cells (iPSCs), HSCs, HSPCs, mesenchymal stem cells, or neuronal stem cells and cells at various stages of differentiation.
- stem cellsfor example induced pluripotent stem cells (iPSCs), HSCs, HSPCs, mesenchymal stem cells, or neuronal stem cells and cells at various stages of differentiation.
- methods and compositions of the disclosureare adapted to target organoids, including patient derived organoids.
- methods and compositions of the disclosureare adapted to treat muscle cells, not limited to cardiomyocytes for Duchene Muscular Dystrophy (DMD).
- the dystrophin geneis the largest gene in the human genome, spanning ⁇ 2.3 Mb of DNA. DMD is composed of 79 exons resulting in a 14-kb full-length mRNA. Common mutations include mutations that disrupt the reading frame of generate a premature stop codon.
- An aspect of DMD that lends it to gene editing as a therapeutic approachis the modular structure of the dystrophin protein. Redundancy in the central rod domain permits the deletion of internal segments of the gene that may harbor loss-of-function mutations, thereby restoring the open reading frame (ORFs).
- the methods and systems described hereinare used to treat DMD by site- specifically integrating in the genome a polynucleotide template that repairs or replaces all or a portion of the defective DMD gene.
- the followingare non-limiting diseases that may be treated utilizing the methods and compositions of the present disclosure:
- HCUHomocystinuria
- CFTRcystic fibrosis transmembrane conductance regulator
- the most common cystic fibrosis (CF) mutation F508delremoves a single amino acid.
- recombining human CFTR into an SHS of a cell that expresses CFTR F508delis a corrective treatment path.
- the methods and systems described hereinare used to CF by site-specifically integrating in the genome a polynucleotide template that corrects the mutation causing CF. Proposed validation is detection of persistent CFTR mRNA and protein expression in transduced cells.
- Sickle cell diseaseis caused by mutation of a specific amino acid — valine to glutamic acid at amino acid position 6.
- SCDis corrected by recombination of the HBB gene into a safe harbor site (SHS) and by demonstrating correction in a proportion of target cells that is high enough to produce a substantial benefit.
- the methods and systems described hereinare used to sickle cell disease by site-specifically integrating in the genome a polynucleotide template that corrects the mutation causing the disease.
- validationis detection of persistent HBB mRNA and protein expression in transduced cells.
- DMDDuchenne Muscular Dystrophy.
- the dystrophin geneis the largest gene in the human genome, spanning ⁇ 2.3 Mb of DNA. DMD is composed of 79 exons resulting in a 14- kb full-length mRNA. Common mutations include mutations that disrupt the reading frame of generate a premature stop codon.
- An aspect of DMD that lends it to gene editing as a therapeutic approachis the modular structure of the dystrophin protein. Redundancy in the central rod domain permits the deletion of internal segments of the gene that may harbor loss-of-function mutations, thereby restoring the open reading frame (ORFs).
- recombinationwill be into safe harbor sites (SHS).
- SHSsafe harbor sites
- a frequently used human SHSis the AAVS1 site on chromosome 19q, initially identified as a site for recurrent adeno-associated virus insertion.
- the siteis the human homolog of the e murine Rosa26 locus (pubmed.ncbi.nlm.nih.gov/18037879).
- the siteis the human Hl l locus on chromosome 22.
- Proposed target cells for recombinationinclude stem cells for example induced pluripotent stem cells (iPSCs) and cells at various stages of differentiation. In some cases, a complete gene may be prohibitively large and replacement of an entire gene impractical. In such instances, rescuing mutants by recombining in corrected gene fragments with the methods and systems described herein is a corrective option.
- iPSCsinduced pluripotent stem cells
- correcting mutations in exon 44 (or 51) by recombining in a corrective coding sequence downstream of exon 43 (or 50), using the methods and systems described hereinis a corrective option.
- Proposed validationis detection of persistent DMD mRNA and protein expression in transduced cells.
- F8Factor VIII
- F8Factor VIII
- F8A large proportion of severe hemophilia A patients harbor one of two types of chromosomal inversions in the FVIII gene.
- the recombinase technology and methods described hereinare well suited to correcting such inversions (and other mutations) by recombining of the FVIII gene into a SHS.
- correcting factor VIII deficiency by recombining the FVIII gene into an SHSis a corrective path.
- the methods and systems described hereinare used to correct factor VIII deficiency by site- specifically integrating in the genome a polynucleotide template that corrects the mutation causing the FIX deficiency. Proposed validation is detection of persistent FVIII mRNA and protein expression in transduced cells.
- Factor 9Hemophilia B, also called factor IX (FIX) deficiency is a genetic disorder caused by missing or defective factor IX, a clotting protein.
- the methods and systems described hereinare used to correct factor IX deficiency by site-specifically integrating in the genome a polynucleotide template that corrects the mutation causing the FIX deficiency.
- Proposed validationis detection of persistent FiX mRNA and protein expression in transduced cells.
- Ornithine transcarbamylase deficiencyis a rare genetic condition that causes ammonia to build up in the blood.
- the condition - more commonly called OTC deficiency -is more common in boys than girls and tends to be more severe when symptoms emerge shortly after birth.
- the methods and systems described hereinare used to correct OTC deficiency by site- specifically integrating in the genome a polynucleotide template that corrects the mutation causing the OTC deficiency or integrates a polynucleotide encoding a functional ornithine transcarbamylase enzyme.
- Proposed validationis detection of persistent OTC mRNA and protein expression in transduced cells.
- Phenylketonuriaalso called PKU, is a rare inherited disorder that causes an amino acid called phenylalanine to build up in the body. PKU is caused by a change in the phenylalanine hydroxylase (PAH) gene. This gene helps create the enzyme needed to break down phenylalanine.
- PKUphenylalanine hydroxylase
- the methods and systems described hereinare used to correct PKU by site-specifically integrating in the genome a polynucleotide template that corrects the mutation causing the PKU deficiency or integrates a polynucleotide encoding a functional phenylalanine hydroxylase (PAH) gene.
- Proposed validationis detection of persistent PAH mRNA and protein expression in transduced cells.
- Homocystinuriais elevation of the amino acid, homocysteine (protein building block coming from our diet) in the urine or blood.
- Common causes of HCUinclude: problems with the enzyme cystathionine beta synthase (CBS), which converts homocysteine to the amino acid cystathionine (which then becomes cysteine) and needs the vitamin B6 (pyridoxine); and roblems with converting homocysteine to the amino acid methionine.
- CBScystathionine beta synthase
- pyridoxinepyridoxine
- the methods and systems described hereinare used to correct HCU by site-specifically integrating in the genome a polynucleotide template that corrects the mutation causing the HCU or integrates a polynucleotide encoding a functional copy of a gene (e.g., CBS) able to reduce or prevent buildup of homocysteine in the urine.
- Proposed validationis detection of persistent CBS mRNA and protein expression in transduced cells.
- IgA Nephropathy(Berger’s disease). IgA nephropathy, also known as Berger's disease, is a kidney /autoimmune disease that occurs when an antibody called immunoglobulin A (IgA) builds up in the kidneys.
- IgAimmunoglobulin A
- the methods and systems described hereinare used to treat Berger’s disease by administering to a patient an iPSC-derived Natural Killer cell that includes a polynucleotide site-specifically integrated in the genome of the cell using the methods described herein.
- the iPSC-NK cellUpon administering the iPSC-NK cell to the patient, the iPSC-NK cell is capable of removing native cells (e.g., B cells) that are responsible, at least in part, for the symptoms of Berger’s disease.
- ANCA vasculitisis an autoimmune disease affecting small blood vessels in the body. It is caused by autoantibodies called ANCAs, or Anti-Neutrophilic Cytoplasmic Autoantibodies. ANCAs target and attack a certain kind of white blood cells called neutrophils.
- the methods and systems described hereinare used to treat ANCA vasculitis by administering to a patient an iPSC-derived Natural Killer cell that includes a polynucleotide site- specifically integrated in the genome of the cell using the methods described herein.
- the iPSC-NK cellUpon administering the iPSC-NK cell to the patient, the iPSC-NK cell is capable of removing native cells (e.g., B cells) that are responsible, at least in part, for the symptoms of ANCA vasculitis.
- Lupusis an autoimmune — a disorder in which the body’s immune system attacks the body’s own cells and organs.
- the methods and systems described hereinare used to treat SLE/LN by administering to a patient an iPSC-derived Natural Killer cell that includes a polynucleotide site- specifically integrated in the genome of the cell using the methods described herein.
- the iPSC-NK cellUpon administering the iPSC-NK cell to the patient, the iPSC-NK cell is capable of removing native cells (e.g., B cells) that are responsible, at least in part, for the symptoms of SLE/LN.
- MNMembranous Nephropathy
- the methods and systems described hereinare used to treat MN by administering to a patient an iPSC-derived Natural Killer cell that includes a polynucleotide site- specific ally integrated in the genome of the cell using the methods described herein.
- the iPSC-NK cellUpon administering the iPSC-NK cell to the patient, the iPSC-NK cell is capable of removing native cells (e.g., B cells) that are responsible, at least in part, for the symptoms of MN.
- native cellse.g., B cells
- C3 glomerulonephritisC3 glomerulopathy is a group of related conditions that cause the kidneys to malfunction.
- C3 glomerulopathyThe major features of C3 glomerulopathy include high levels of protein in the urine (proteinuria), blood in the urine (hematuria), reduced amounts of urine, low levels of protein in the blood, and swelling in many areas of the body. Affected individuals may have particularly low levels of a protein called complement component 3 (or C3) in the blood.
- complement component 3or C3
- the methods and systems described hereinare used to treat C3 glomerulopathy by administering to a patient an iPSC-derived Natural Killer cell that includes a polynucleotide site- specifically integrated in the genome of the cell using the methods described herein.
- the iPSC-NK cellUpon administering the iPSC-NK cell to the patient, the iPSC-NK cell is capable of removing native cells (e.g., B cells) that are responsible, at least in part, for the symptoms of C3 glomerulopathy.
- methods of treatmentcomprises administering an effective amount of the pharmaceutical composition comprising the nucleic acid construct or vectorized nucleic acid construct described above to a patient in need thereof.
- the systeme.g., any of the systems described herein
- the systemsare delivered to a patient, thereby delivering to a cell in vivo.
- DNA or RNA viral vectorscan be administered directly to patients (in vivo) or they can be used to treat cells in vitro, and the modified cells may optionally be administered to patients (ex vivo).
- Conventional viral based systems to be used hereincould include retroviral, lentivirus, adenoviral, adeno-associated and herpes simplex virus vectors for gene transfer. Integration in the host genome is possible with the retrovirus, lentivirus, and adeno-associated virus gene transfer methods, often resulting in long term expression of the inserted transgene. Additionally, high transduction efficiencies have been observed in many different cell types and target tissues.
- the co-delivery system described hereine.g., a gene editor construct packaged in a LNP and a donor template packaged in a vector
- the co-delivery system described hereinis administered intravenously.
- the co-delivery system described hereine.g., a gene editor construct packaged in a LNP and a donor template packaged in a vector
- the co-delivery system described hereine.g., a gene editor construct packaged in a LNP and a donor template packaged in a vector
- the co-delivery system described hereine.g., a gene editor construct packaged in a LNP and a donor template packaged in a vector
- the co-delivery system described hereinis administered by intracisternal magna administration.
- the co-delivery system described hereine.g., a gene editor construct packaged in a LNP and a donor template packaged in a vector
- Methods of non-viral delivery of the donor DNA template described hereininclude lipofection, nucleofection, microinjection, biolistics, virosomes, liposomes, immunoliposomes, polycation or lipidmucleic acid conjugates, naked DNA, artificial virions, and agent-enhanced uptake of DNA.
- Lipofectionis described in e.g., U.S. Pat. Nos. 5,049,386, 4,946,787; and 4,897,355) and lipofection reagents are sold commercially (e.g., TransfectamTM and LipofectinTM).
- Cationic and neutral lipids that are suitable for efficient receptor-recognition lipofection of polynucleotidesinclude those of Feigner, WO 91/17424; WO 91/16024. Delivery can be to cells (e.g. in vitro or ex vivo administration) or target tissues (e.g. in vivo administration).
- mRNA delivery methods and compositionsthat may be utilized in the present disclosure including, for example, PCT/US2014/028330, US8822663B2, NZ700688A, ES2740248T3, EP2755693A4, EP2755986A4, WO2014152940A1, EP3450553B1, BR112016030852A2, and EP3362461A1.
- Expression of CRISPR systems in particularis described by W02020014577.
- Each of these publicationsare incorporated herein by reference in their entireties. Additional disclosure hereby incorporated by reference can be found in Kowalski et al., “Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery,” Mol Therap., 2019; 27(4): 710-728.
- Example 1Delivery of gene editor polynucleotide sequence packaged in LNP and donor template packaged in AAV
- a gene editor polynucleotide constructis packaged into a LNP (FIG. 1), wherein the gene editor polynucleotide sequence comprises a polynucleotide sequence encoding a prime editor protein linked to an integrase via peptide linker a polynucleotide sequence encoding an attachment site-containing guide RNA (atgRNA); a polynucleotide sequence encoding a nickase guide RNA (ngRNA).
- atgRNAattachment site-containing guide RNA
- ngRNAnickase guide RNA
- a donor template polynucleotide constructis packaged in an AAV vector (FIG. 2).
- Co-administration of the gene editor construct packaged LNP and the donor template packaged AAVco-delivers the gene editor construct to a cell cytoplasm and the donor template to a cell nucleus.
- programmable genome editingto place integrase landing site at a desired location in the genome, the direct activity of the associated integrase to the specific genomic site is guided.
- Example 2Delivery of gene editor polynucleotide sequence packaged in LNP and donor template capable of self-circularization packaged in AAV
- a gene editor polynucleotide constructis packaged into a LNP (FIG. 1), wherein the gene editor polynucleotide sequence comprises a polynucleotide sequence encoding a prime editor protein linked to an integrase via peptide linker a polynucleotide sequence encoding an attachment site-containing guide RNA (atgRNA); a polynucleotide sequence encoding a nickase guide RNA (ngRNA).
- atgRNAattachment site-containing guide RNA
- ngRNAnickase guide RNA
- a donor template polynucleotide constructis packaged in an AAV vector (FIG. 2).
- Co-administration of the gene editor construct packaged LNP and the donor template packaged AAVco-delivers the gene editor construct to a cell cytoplasm and the donor template to a cell nucleus.
- Integrase-mediated self-circularization of donor templateoccurs at integration target recognition sites within the AAV genome (FIG. 3).
- an orthogonal integrase landing sitei.e., distinct att site from att sites used for selfcircularization
- Gene editor construct expression, with template co-delivery and integrase-mediated circularization of templateresults in integration of template “cargo” at a precisely defined target location.
- Example 3Delivery of gene editor polynucleotide sequence packaged in LNP and atgRNA, ngRNA, and donor template co-packaged in AAV
- a gene editor polynucleotide constructis packaged into a LNP (FIG. 4), wherein the gene editor polynucleotide sequence comprises a polynucleotide sequence encoding a prime editor protein linked to an integrase via peptide linker.
- a polynucleotide sequence encoding an attachment site-containing guide RNA (atgRNA), a polynucleotide sequence encoding a nicking guide RNA (ngRNA), and donor templateare packaged in an AAV vector (FIG. 4).
- atgRNAattachment site-containing guide RNA
- ngRNAnicking guide RNA
- donor template packaged AAVco-delivers the gene editor construct to a cell. Integrase- mediated self-circularization of donor template occurs at integration target recognition sites within the AAV genome (FIG. 3).
- Example 4Delivery of gene editor polynucleotide sequence and ngRNA packaged in LNP and atgRNA and donor template co-packaged in AAV
- a gene editor polynucleotide construct and a nicking guide RNA (ngRNA)are packaged into a LNP (FIG. 5), wherein the gene editor polynucleotide sequence comprises a polynucleotide sequence encoding a prime editor protein linked to an integrase via peptide linker.
- a polynucleotide sequence encoding an attachment site-containing guide RNA (atgRNA) and donor templateare packaged in an AAV vector (FIG. 5).
- Example 5Intramolecular circularization of plasmid and packaged AAV genomes
- scAAVself-complementary AAV genomes were designed and generated to verify recombinase/integrase-mediated intramolecular circularization of a DNA cargo from within a linear AAV genome (FIGs. 6A-6B). Circularization of a scAAV genome is mediated by one of Cre, FLPe (thermostable mutant), or Bxbl. Further, the scAAV genomes are comprised of a DNA cargo of interest (“payload”) and an attP site (GT central dinucleotide for circularization orthogonality) for gene insertion into a genome placed attB beacon site.
- payloada DNA cargo of interest
- attP siteGT central dinucleotide for circularization orthogonality
- FIG. 7A universal ddPCR probe capable of binding any linear or circularized AAV genome was designed, wherein the universal ddPCR probe is designed to only give signal upon cognate recombinase/integrase mediated circularization (FIGs. 8A-8B).
- Circularization productsare amplified by use of a circle junction PCR primer set that is designed to amplify only circular products due to primer direction constraints.
- an attR scar quencher-fluorophore probewas designed.
- a template reference primer setwas designed and generated to quantify total template DNA (linear or circular confirmation) (FIGs. 8A-8B).
- FIG. 10demonstrates circularization of AAV pDNA and packaged AAV genomic DNA for both IX Bxbl and 2X Bxbl conditions (confirmed by use of attR ddPCR primer set). Further, replicates that lacked either Bxbl or AAV pDNA substrate demonstrated insignificant circularization. All three of the Cre-, FEPe-, and Bxbl -targeted AAV pDNA substrates demonstrated circularization upon cognate recombinase/integrase introduction, as confirmed by using the universal ddPCR probe (FIG. 11). Moreover, Cre-, FEPe-, and Bxbl -mediated circularization of packaged AAV DJ genomes substrates were demonstrated and confirmed using the universal ddPCR probe (FIG. 12).
- the Bxbl-mediated attR scar probeprovided similar percent circularization quantification compared to the universal probe.
- Example 6In vitro beacon placement in primary mouse hepatocytes and primary human hepatocytes using mRNA and AAV for co-delivery
- This exampleassessed the efficiency of in vitro beacon placement in primary human hepatocytes using mRNA delivering of a polynucleotide encoding a gene editor polynucleotide construct and AAV to deliver the first and second atgRNA. See FIG. 15 for a non-limiting example of a dual atgRNA-mediated insertion of an integration recognition site.
- the mRNA and AAVwere delivered into the primary mouse hepatocytes (PMH) using (i) concurrent delivery (“co-dose”), (ii) AAV delivery followed by a “1- day delay” before delivery of the mRNA, or (iii) AAV delivery followed by a “2-day delay” before delivery of the mRNA.
- Co-doseconcurrent delivery
- AAV delivery followed by a “2-day delay” before delivery of the mRNABeacon placement was then assessed using next-generation sequencing of DNA isolated from cells subjected to the delivery conditions mentioned above.
- the mRNA encoding the gene editor polynucleotide constructwas delivered in various amounts per well: 2000 ng, 1000 ng, 500 ng, 250 ng, 125 ng, 62.5 ng, and 31.25 ng.
- AAV encoding the first and second atgRNAsee Table 12
- the primary mouse hepatocyte datais shown in FIG. 16 and the human primary hepatocyte data is shown in FIG. 17.
- PMHprimary mouse hepatocytes
- SEQ ID NO: 543the first atgRNAs
- SEQ ID NO: 544the second atgRNA
- a 2 day delayresulted in greater than 10% beacon placement for each amount of mRNA tested.
- a 2 day delayresulted in greater beacon placement than either no delay (“co-dose) or a 1 day delay.
- Example 7In vivo beacon placement with mRNA + AAV guide
- micewere administered AAV containing the first atgRNA (SEQ ID NO: 543; Table 12) and the second atgRNA (SEQ ID NO: 544) targeting the Nolcl locus at 3E11 to 1E12 vector genomes (vg) per animal two 2 weeks prior to administration of the mRNA containing the gene editing polynucleotide construct (see FIG. 18).
- mRNAwas delivered using various LNP formulations (e.g., LNP #F1 , LNP #F2, and LNP #F3) at concentrations ranging from 5 mg/kg to 0.5 mg/kg via intravenous injection (see FIG. 18).
- LNP formulationse.g., LNP #F1 , LNP #F2, and LNP #F3
- this dataprovided proof-of-concept for successful in vivo beacon placement using AAV to deliver the first and second atgRNA and LNPs to deliver the mRNA encoding the gene editor polynucleotide construct.
- Example 8In vivo integration in mice using AAV to deliver the template polynucleotide and Adenovirus to deliver BxBl
- adenovirusto deliver an integrase (e.g., Bxbl) and an AAV to deliver the template polynucleotide.
- the adenovirusi.e., adenovirus containing polynucleotide encoding the integrase
- the AAVi.e., AAV containing the template polynucleotide and an attB site
- micecontaining dual AttP sites integrated in to the Rosa26 locus (B6.RosaBxb-GT/GA; female, Strain# 036152).
- the Rosa26 locusincluded a first AttP site comprising a GT dinucleotide and a second AttP site comprising a GA dinucleotide.
- the AAVwas a scAAV8 containing a vector having a template polynucleotide and a 38 bp GT AttB site.
- the Adenoviruswas an adenovirus-type 5 (Ad5) containing a polynucleotide encoding Bxbl (“Bxbl AdV”) (SEQ ID NO: 563; Table 14). Mice were administered the adenovirus and AAV according to the experimental details in Table 13.
- this dataestablishes proof-of-concept for in vivo integration using an adenovirus to deliver and drive expression of Bxbl and an AAV to deliver the template polynucleotide to be integrated into a mammalian genome, in this case, the mouse genome.
- the first LNPcontained mRNA encoding a prime editing system and a first synthetic atgRNA (atgRNAl).
- the mRNA and atgRNAlwere included at 1:1 ratio in the first LNP.
- the second LNPcontained mRNA encoding a prime editing system and a second synthetic atgRNA (atgRNA2).
- the mRNA and atgRNA2were included at a 1:1 ratio in the second LNP.
- Each of the first and second atgRNAstargeted the mouse Nolcl locus and each encoded a portion of an integration recognition site (a “beacon”).
- AtgRNAl and atgRNA2together included a 6bp overlap.
- the first and second LNPswere combined 1:1 as mixture prior to administration.
- the first atgRNA and second atgRNAare provide in Table 15, where the atgRNA include one or more 2’O-methyl modifications and one or more phosphorothioate linkages.
- the LNP mixturewas administered to the neonatal mice (2-5 day old CD-I mice) according to the experimental details in Table 16.
- liver sampleseither whole liver for groups 1-3 or liver punches from each lobe for groups 4-6 (see Table 13) were collected and genomic DNA was isolated. Beacon placement was detected using ddPCR and NGS.
- Neonateswere also assessed at six weeks after administration of the LNP mixture. Beacon placement was detected using ddPCR and NGS. As shown in FIG. 21A., at six weeks post administration, ddPCR revealed about 4% beacon placement (in Nolcl alleles) for a 3mg/kg dose of the LNP mixture. Confirmation of beacon placement using NGS showed about 15% beacon placement (in Nolcl alleles) for a 3 mg/kg dose of the LNP mixture (see FIG. 21B). Assessment of the percent of integrated beacons that included the expected integration recognition site (“perfect beacon”) revealed that about 3.5% of beacons were comprised of perfect beacons (see FIG. 21C).
- this datademonstrated successful in vivo site-specific integration of an integration recognition site.
- this datashowed that a split LNP approach can be used for site- specifically integrating an integration recognition site in vivo in a mammalian genome, in this case neonatal mice.
- the first LNPcontained mRNA encoding a prime editing system and a first synthetic atgRNA (atgRNAl).
- the mRNA and atgRNAlwere included at different ratios (e.g., 1:0.5, 1:1, and 1 :2) ratio in the first LNP.
- the second LNPcontained mRNA encoding a prime editing system and a second synthetic atgRNA (atgRNA2).
- the mRNA and atgRNA2were included at different ratios (e.g., 1:0.5, 1:1, and 1:2) ratio in the second LNP.
- the first and second atgRNAstargeted mouse Factor IX (“mF9”) locus and each encoded a portion of an integration recognition site (“beacon”). Similar to Example 9, atgRNAl and atgRNA2 together included a 6bp overlap and were combined 1:1 as mixture prior to administration.
- the first atgRNA and second atgRNAare provide in Table 17, where the atgRNA include one or more 2’0-methyl modifications and one or more phosphorothioate linkages.
- liver samplesi.e., liver punches of each lobe (see Table 14)
- genomic DNAwas isolated. Beacon placement was detected using ddPCR and NGS.
- LNPlipoprotein
- Table 19show the LNP formulations and the modifications tested. Table 19 also shows the size, polydispersity (PDI), and encapsulation efficiency (EE% as determined by a Ribogreen assay) for the LNP formulations.
- LNPwere diluted 100-fold in lx PBS in cuvettes and analyzed using Dynamic Light Scattering (DLS) with a Malvern Zetasizer instrument and software to determine size and PDI.
- Encapsulation efficiencywas calculated by analyzing the free and encapsulated RNA using the Ribogreen dye (ThermoFisher) according to manufacturer’s protocol.
- Control LNPsinclude the unmodified four lipid components while the DOPG, Mannose, and DOPG/Mannose groups indicate a phospholipid, PEG-lipid, or both substitutions in the LNP.
- LNP compositions for these experimentsincluded components at the following ratios: included: “cKKE12”: 40% cKKE12, 30% DOPE or DOPG, 28% Cholesterol, 2% PEG2K-DMG; “cKKE12-Mannose”: 40% cKKE12, 30% DOPE or DOPG, 27% cholesterol, 0.5% PEG2K-DMG, 2.5% Mannose-PEG2K-DSPE; “ALC0315”: 46.3% ALC0315, 9.4% DSPC or DSPG, 42.7% Cholesterol, 1.6% PEG2K-DMG; “ALC-Mannose”: 46.3% ALC0315, 9.4% DSPC or DSPG, 41.3% Cholesterol, 0.5% PEG2K-DMG, 2.5% Mannose-PEG2K-DSPE; “246C10”: 26.5% 246C10, 20% DOPE or DOPG, 50.5% Cholesterol, 3% C16-PEG2K-ceramide; and “cKKE
- FIGs. 24A-24Bshow representative images of liver sinusoidal endothelial cells (LSECs) contacted with the indicated LNP formulations: cKK (500 ng GFP); cKK-DOPG (500 ng GFP).
- cKK500 ng GFP
- cKK-DOPG500 ng GFP
- FIG. 25shows representative images of LSECs contacted with the indicated LNP formulations: cKK-Mannose (500 ng GFP); or cKK-DOPG/Mannose (500 ng GFP).
- FIG. 26shows representative images of the controls (i.e., LSECs transfected using with GFP lipofectamine or untreated LSECs).
- a set of experimentswas performed in Hepa 1-6 cells to screen for guide RNA (gRNA) that target human factor VIII (“F8”).
- gRNAguide RNA
- F8human factor VIII
- This experimentserved, in part, as a proof of concept for using the indicated LNP formulations to deliver guide RNAs and polynucleotides encoding gene editor polynucleotides to LSECs.
- F8was targeted because it is expressed exclusively expressed in LSECs and when missing or defective, thereby causing Hemophilia A. Therefore, targeting F8 provided an opportunity to target a disease relevant gene in a disease relevant tissue using the optimized LNP formulations to do so.
- Hepa 1-6 cellswere plated at 15,000 cells per well and transfected one day post plating with five different F8 knockout (KO) guide RNAs (gRNAs) (see FIG. 27).
- F8 gRNAswere transfected at a concentration of 60 nM per guide.
- Cas9was transfected at a concentration of 200 ng.
- gRNAs usedare shown in Table 20.
- FIG. 28shows results of initial knockout guides screened in Hepa 1-6 cells. Successful F8 knockout was observed with all guides tested (see FIG. 28). Similar efficiencies were observed in guides 2, 3, and 4 (see FIG. 28). Guides 2, 3, and 4 were selected for further formulations studies (see Example 13).
- Example 13Assessment of LNP formulations to deliver gene editing reagents to LSECs in vitro
- a proof-of-concept studywas performed to assess use of various LNP formulations for delivering gRNA 2 (“G2”), gRNA 3 (“G3”) (data not shown) and gRNA 4 (“G4”) to LSECs.
- G2gRNA 2
- G3gRNA 3
- G4gRNA 4
- LNPs encapsulating G2, G3, or G4were added to the LSEC culture at four different concentrations (500 ng, 250 ng, 125 ng, and 62.5 ng). The cells were harvested 3 days after introduction of the LNPs and assessed for % indel formation.
- LNP formulations having a DOPG modificatione.g., a cKK LNP formulation comprising a DOPG modification or a ALC LNP formulation comprising a DOPG modification
- LNPs with a modificationare more efficient than unmodified LNPs at producing indels in LSEC.
- LNPs with a modificationare clearly more efficient than unmodified LNPs at targeting LSECs.
- FIG. 32shows that the LNP modification (mod 1), a complete substitution/replacement of a neutrally charged phospholipid (DSPC) with a negatively charged phospholipid (DOPG), resulted much higher LSEC targeting efficiency compared to the hepatocyte targeting with the modified LNP.
Landscapes
- Health & Medical Sciences (AREA)
- Life Sciences & Earth Sciences (AREA)
- Genetics & Genomics (AREA)
- Chemical & Material Sciences (AREA)
- Engineering & Computer Science (AREA)
- Organic Chemistry (AREA)
- Biotechnology (AREA)
- Molecular Biology (AREA)
- Zoology (AREA)
- General Health & Medical Sciences (AREA)
- Biomedical Technology (AREA)
- Wood Science & Technology (AREA)
- Bioinformatics & Cheminformatics (AREA)
- General Engineering & Computer Science (AREA)
- Biochemistry (AREA)
- Biophysics (AREA)
- Medicinal Chemistry (AREA)
- Plant Pathology (AREA)
- Physics & Mathematics (AREA)
- Microbiology (AREA)
- Hematology (AREA)
- Toxicology (AREA)
- Proteomics, Peptides & Aminoacids (AREA)
- Gastroenterology & Hepatology (AREA)
- Veterinary Medicine (AREA)
- Animal Behavior & Ethology (AREA)
- Public Health (AREA)
- Pharmacology & Pharmacy (AREA)
- Epidemiology (AREA)
- Virology (AREA)
- Cell Biology (AREA)
- Mycology (AREA)
- Medicines That Contain Protein Lipid Enzymes And Other Medicines (AREA)
Abstract
Description
SYSTEMS, COMPOSITIONS, AND METHODS FOR TARGETING LIVER SINUSODIAL ENDOTHELIAL CELLS (LSECS)
1. CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] This application claims the benefit of U.S. Provisional Patent Application No. 63/501,633, filed May 11, 2023, which is hereby incorporated in its entirety by reference.
2. BACKGROUND
[0002] Liver sinusoidal endothelial cells (LSECs) are of interest because Human factor VIII (FVIII), which deficiency leads to hemophilia A, is largely synthesized and secreted by these cells. Current gene therapies target hepatocytes in order to treat hemophilia A. However, the hepatocyte is not the natural site for FVIII synthesis and current gene therapy protocols are eliciting immune responses that require immune suppression (El-Maarri et al., Hamostaseologie Nov;40(S 01):S26- S31. doi: 10.1055/a- 1282-2286 (2020)). Therefore, there is a need in the art that addresses and overcome these shortcomings and enables the delivery of cargo into LSECs for therapeutic purposes.
3. SUMMARY
[0003] The present disclosure features compositions and LNP formulations capable of delivering a cargo into a liver sinusoidal endothelial cell (LSEC). In one embodiment, the LNP formulations provided herein are capable of delivering the components needed for genetic engineering (e.g., components needed for Programmable Addition via Site-Specific Targeting) into an LSEC. The liver is made up of parenchymal and nonparenchymal cells, like hepatocytes, LSECs, Kupffer, and stellate cells. To deliver RNA to specific liver cells, LNPs need to be precisely engineered due to cellular physiological differences. LSECs, the most permeable liver cells, facilitate macromolecule transport between the sinusoidal lumen and hepatocytes due to their high endocytic capacity and numerous fenestrae. Small nanoparticles can pass through these fenestrae, given their diameter (-100 nm in humans, -140 nm in mice). Therefore, LNP size and composition can influence endothelium- specific delivery. This disclosure provides compositions and formulations aimed at facilitating delivery to LSECs. In particular, this disclosure features a composition includes: a lipid nanoparticle (LNP) formulation comprising a plurality of LNPs, wherein each LNP comprises: (i) a PEG-modified lipid; (ii) a neutral lipid, or (iii) both (i) and (ii); wherein each LNP encapsulates: (i) the gene editor polynucleotide; (ii) the at least a first attachment- site containing guide RNA (atgRNA); or (iii) both (i) and (ii), wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (LSEC) the (i) a gene editor polynucleotide; (ii) at least a first attachment- site containing guide RNA (atgRNA); (iii) or both (i) and (ii). In another embodiment, the LNP composition includes: (a) a gene editor polynucleotide; (b) at least a first attachment- site containing guide RNA (atgRNA); (c) an ionizable lipid; (d) a cholesterol; (e) a PEG-modified lipid; and (f) a neutral lipid, wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (a), (b), (c), (d), (e), (f), or a combination thereof. In some embodiments, replacement of helper lipids with charged alternatives in lipid nanoparticles facilitates targeted mRNA delivery to the LSECs.
[0004] The present disclosure provides a compositions and formulations for site-specific genetic engineering of LSECs using Programmable Addition via Site-Specific Targeting Elements (PASTE) (see lonnidi et al. doi: 10.1101/2021.11.01.466786; the entirety of lonnidi et al. is incorporated by reference), transposon-mediated gene editing, or other suitable gene editing or gene incorporation technology.
[0005] The present disclosure also provides compositions and formulations for site-specific genetic engineering of LSECs using Programmable Addition via Site-Specific Targeting Elements (PASTE) as described in U.S. Patent No. 11,572,556 and PCT Publication No. WO 2022/087235A; each of which is hereby incorporated by reference in its entirety.
[0006] In one aspect, this disclosure features a composition comprising: a lipid nanoparticle (LNP) formulation comprising a plurality of LNPs, wherein each LNP comprises: (i) an ionizable lipid; (ii) a cholesterol; (iii) a polyethylene glycol (PEG) -modified lipid; (iv) optionally a neutral lipid, or (v) a combination of (i) - (iv); each LNP encapsulates a cargo; wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (LSEC) the encapsulated cargo.
[0007] In another aspect, the present disclosure provides a composition comprising: a lipid nanoparticle (LNP) formulation comprising a plurality of LNPs, wherein each LNP comprises: (i) an ionizable lipid; (ii) a cholesterol; and (iii) a polyethyleneglycol (PEG)-modified lipid; and (iv) optionally a neutral lipid, each LNP encapsulates a cargo; wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (LSEC) the encapsulated cargo.
[0008] In some embodiments, the cargo is selected from: an mRNA, an siRNA, an antisense oligonucleotide, a DNA, a guide RNA (gRNA), a protein, a peptide, a chemotherapeutic agent, a small molecule drug, or a combination thereof. In some embodiments, the cargo is an mRNA and a gRNA. In some embodiments, the mRNA is a gene editor polynucleotide encoding a gene editor polypeptide and the gRNA is an attachment site-containing guide RNA (atgRNA). [0009] In another aspect, this disclosure features a composition comprising: a lipid nanoparticle (LNP) formulation comprising a plurality of LNPs, wherein each LNP comprises: (i) a polyethylene glycol (PEG) -modified lipid; (ii) a neutral lipid, or (iii) both (i) and (ii); wherein each LNP encapsulates: (i) a gene editor polynucleotide; (ii) at least a first attachment-site containing guide RNA (atgRNA); (iii) or both (i) and (ii), wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (LSEC) (i) the gene editor polynucleotide; (ii) the at least a first attachment- site containing guide RNA (atgRNA); or (iii) both (i) and (ii).
[0010] In another aspect, this disclosure features a lipid nanoparticle (LNP) composition comprising: (a) a gene editor polynucleotide; (b) at least a first attachment- site containing guide RNA (atgRNA); (c) an ionizable lipid; (d) a cholesterol; (e) a polyethylene glycol (PEG)-modified lipid; and (f) optionally a neutral lipid, wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (a), (b), (c), (d), (e), (f), or a combination thereof.
[0011] In another aspect, the present disclosure provides a lipid nanoparticle (LNP) composition comprising: (a) a gene editor polynucleotide; (b) at least a first attachment- site containing guide RNA (atgRNA); (c) an ionizable lipid; (d) a cholesterol; and (e) a polyethylene glycol (PEG)- modified lipid; and (f) optionally a neutral lipid, wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (LSEC) the gene editor polynucleotide and the at least first attachment- site containing guide RNA (atgRNA).
[0012] In some embodiments, the ionizable lipid is an ionizable cationic lipid.
[0013] In some embodiments, the ionizable cationic lipid is selected from N- (2,3dioleyloxy)propyl)-N,N,Ntrimethylammonium chloride (DOTMA); N- (2,3dioleoyloxy)propyl)-N,N,N-trimethylamntonium chloride (DODAP); N,N-dioleyl-N,N- dimethylammonium chloride (DODAC); 1,2 bis (oleoyloxy)-3-(trimethylammonio) propane (DOTAP); N,NdistearylN,N-dimethylammonium bromide (DDAB); 3-(N — (N,N- dimethylaminoethane)-carbam-oyl)cholesterol (DC-Choi); diheptadecylamidoglycylspermidine (DHGS) and N-(l,2-dimyristyloxyprop-3-yl)-N,N-dimethyl-N-hydoxyethyl ammonium bromide (DMRIE), or a combination thereof
[0014] In some embodiments, the composition or LNP composition further comprising a anionic lipid.
[0015] In some embodiments, the ionizable lipid is replaced with an anionic lipid. [0016] In some embodiments, the anionic lipid is selected from: a phospholipid, phospholipid derivative, a sterol, a fatty acid, or a combination thereof.
[0017] In some embodiments, the PEG-modified lipid is select from 1,2-Dimyristoyl-sn-glycerol methoxypolyethylene glycol (PEG-DMG), PEG-disteryl glycerol (PEG-DSG), PEG- dipalmetoleyl, PEG-dioleyl, PEG-distearyl, PEG-diacylglycamide (PEG-DAG), PEG- dipalmitoyl phosphatidylethanolamine (PEG-DPPE), and PEG-l,2-dimyristyloxlpropyl-3-amine (PEG-c-DMA), or a combination thereof.
[0018] In some embodiments, the PEG-modified lipid comprises a moiety capable binding to a receptor on a liver sinusoidal endothelial cell.
[0019] In some embodiments, the moiety is capable of binding to a mannose receptor, a stabilin receptor, an Fc receptor, or a combination thereof.
[0020] In some embodiments, the moiety is a mannose.
[0021] In some embodiments, the PEG-modified lipid is a mannose-PEG lipid.
[0022] In some embodiments, the neutral lipid is a phospholipid.
[0023] In some embodiments, the phospholipid is a natural phospholipid selected from: phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylglycerol (PG), phosphatidylserine (PS), phosphatidylinositol (PI), Phosphatidic acid (phosphatidate) (PA), dipalmitoylphosphatidylcholine, monoacyl-phosphatidylcholine (lyso PC), l-palmitoyl-2-oleoyl- sn-glycero-3-phosphocholine (POPC), N-Acyl-PE, phosphoinositides, and phospho sphingolipids, or a combination thereof.
[0024] In some embodiments, the phospholipid is a phospholipid derivative selected from: phosphatidic acid (DMPA, DPPA, DSPA), phosphatidylcholine (DDPC, DLPC, DMPC, DPPC, DSPC, DOPC, POPC, DEPC), phosphatidylglycerol (DMPG, DPPG, DSPG, DOPG), phosphatidylethanolamine (DMPE, DPPE, DSPE DOPE), and phosphatidylserine (DOPS), or a combination thereof.
[0025] In some embodiments, the phospholipid is selected from: l,2-dioleoyl-sn-glycero-3- phospho-rac-(l-glycerol) sodium salt (DOPG); l,2-dilinoleoyl-sn-glycero-3-phosphocholine (DLPC); 1,2-dimyristoyl-sn-glycero-phosphocholine (DMPC); 1 ,2-dioleoyl-sn-glycero-3- phosphocholine (DOPC); l,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC); 1,2-distearoyl- sn-glycero-3-phosphocholine (DSPC); 1,2-diundecanoyl-sn-glycero-phosphocholine (DUPC); 1- palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC); l,2-di-0-octadecenyl-£-glycero-3- phosphocholine (18:0 Diether PC); l-oleoyl-2-cholesterylhemisuccinoyl-glycero-3- phosphocholine (OChemsPC); l-hexadecyl-sn-glycero-3-phosphocholine (C16 Lyso PC); 1,2- dilinolenoyl-sn-glycero-3-phosphocholine; l,2-diarachidonoyl-sn-glycero-3-phosphocholine; 1,2- didocosahexaenoyl-sn-glycero-3-phosphocholine; l,2-dioleoyl-sn-glycero-3- phosphoethanolamine (DOPE); l,2-diphytanoyl-sn-glycero-3-phosphoethanolamine (ME 16.0 PE); l,2-distearoyl-sn-glycero-3-phosphoethanolamine; l,2-dilinoleoyl-sn-glycero-3- phosphoethanolamine; 1 ,2-dilinolenoyl-sn-glycero-3-phosphoethanolamine; 1 ,2-diarachidonoyl- sn-glycero-3-phosphoethanolamine; and l,2-didocosahexaenoyl-sn-glycero-3- phosphoethanolamine; and mixtures thereof.
[0026] In some embodiments, the phospholipid is DOPG.
[0027] In some embodiments, each LNP in the LNP formulation comprises: (i) modified PEGylated lipids; and (ii) neutral lipids.
[0028] In some embodiments, each LNP in the LNP formulation comprises (i) PEG-modified lipids comprising a moiety capable binding to a receptor on a liver sinusoidal endothelial cell (LSEC); and (ii) phospholipids.
[0029] In some embodiments, each LNP in the LNP formulation comprises mannose-PEG lipids and DOPGs.
[0030] In some embodiments, the LNP further comprises: cholesterol and an ionizable lipid.
[0031] In some embodiments, each LNP in the LNP formulation ranges from about 100 nm to about 200 nm. In some embodiments, each LNP in the LNP formulation ranges from about 85 nm to about 120 nm. In some embodiments, each LNP in the LNP formulation ranges from about 45 nm to about 65 nm. In some embodiments, each LNP in the LNP formulation ranges from about 40 nm to about 50 nm.
[0032] In some embodiments, the encapsulation efficiency is about 80% to about 100%. In some embodiments, the encapsulation efficiency is no less than 84%. In some embodiments, the encapsulation efficiency is no less than 90%. [0033] In some embodiments, the polydispersity index of the composition ranges from about 0.005 to 0.5. In some embodiments, the polydispersity index of the composition ranges from about 0.025 to 0.4.
[0034] In some embodiments, the gene editor polynucleotide comprises a nucleotide sequence encoding a nickase and a nucleotide sequence encoding a reverse transcriptase.
[0035] In some embodiments, the nickase is linked to the reverse transcriptase by a linker.
[0036] In some embodiments, the first atgRNA comprises: (i) a domain that is capable of guiding the nickase to a target sequence; and (ii) a reverse transcriptase (RT) template that comprises at least a portion of an at least first integration recognition site, whereby the at least portion of the at least first integration recognition site is integrated into the genome of the cell at the target sequence.
[0037] In some embodiments, the LNPs further comprises a nicking gRNA.
[0038] In some embodiments, the LNPs further comprise a second atgRNA.
[0039] In some embodiments, the first atgRNA and the second atgRNA are an at least first pair of atgRNAs, wherein the at least first pair of atgRNAs have domains that are capable of guiding the prime editor system to a target sequence; the first atgRNA further includes a first RT template that comprises at least a portion of the first integration recognition site; the second atgRNA further includes a second RT template that comprises at least a portion of the first integration recognition site, and the first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site, whereby the first integration recognition site is integrated into the genome of the cell at the target sequence.
[0040] In some embodiments, the LNP or LNP composition further comprising an integration enzyme or a polynucleotide encoding an integration enzyme.
[0041] In some embodiments, the integration enzyme or the polynucleotide encoding the integration enzyme is encapsulated in an LNP along with (i) a gene editor polynucleotide; (ii) at least a first attachment- site containing guide RNA (atgRNA); or (iii) both (i) and (ii),
[0042] In some embodiments, the LNP or LNP composition further comprising a donor polynucleotide template. [0043] In another aspect, this disclosure features a liver sinusoidal endothelial cell (LSEC) comprising any of the compositions or LNP compositions described herein.
[0044] In another aspect, this disclosure features a genetically engineered liver sinusoidal endothelia cell (LSEC) made with any of the compositions or LNP compositions described herein.
[0045] In another aspect, this disclosure features a method for site-specifically incorporating an integration recognition site into a liver sinusoidal endothelial cell (LSEC) genome at a target DNA sequence, comprising: incorporating an integration target recognition site into the genome by delivering into the cell: any of the compositions or LNP compositions described herein.
[0046] In another aspect, this disclosure features a method for site-specifically integrating a donor polynucleotide template into a liver sinusoidal endothelial cell (LSEC) genome at a target DNA sequence, comprising: incorporating an integration recognition site into the genome by delivering into the cell: any of the compositions or LNP compositions described herein; and integrating the donor polynucleotide template into the genome by delivering into the cell: an integration enzyme; and a donor polynucleotide template, wherein the donor polynucleotide template is linked to a sequence that is an integration cognate of the integration recognition site present in the atgRNA, and wherein the donor polynucleotide template is integrated into the genome at the incorporated genomic integration recognition site by the integration enzyme.
[0047] In another aspect, this disclosure features a genetically engineered liver sinusoidal endothelia cell (LSEC) made by using any of the composition or LNP compositions described herein.
4. BRIEF DESCRIPTION OF THE DRAWINGS
[0048] These and other features, aspects, and advantages of the present disclosure will become better understood with regard to the following description, and accompanying drawings, where:
[0049] FIG. l shows a non-limiting illustration of a gene editor construct packaged within a lipid nanoparticle (LNP).
[0050] FIG. 2 illustrates the donor template (i.e., “cargo” or “payload” or “template polynucleotide”)) packaged within a vector. [0051] FIG. 3 illustrates integrase-mediated self-circularization of the donor template (template polynucleotide) within viral genome. The circularized donor template is capable of being genomically incorporated into an orthogonal integrase target recognition site (i.e., “beacon”).
[0052] FIG. 4 shows non-limiting illustrations of a gene editor construct packaged within a lipid nanoparticle and an atgRNA, ngRNA, and donor template (i.e., template polynucleotide encoding a gene of interest) packaged within a vector. GOI = gene of interest. PGI = programmable gene insertion. U6 = U6 promoter. atgRNA = attachment site-containing guide RNA (atgRNA).
[0053] FIG. 5 shows non-limiting illustrations of a gene editor construct (e.g., mRNA encoding PE2-BxBl) and a nicking guide RNA (ngRNA) packaged within a lipid nanoparticle (LNP) and an atgRNA and donor template (i.e., template polynucleotide encoding a gene of interest) packaged within a vector.
[0054] FIGs. 6A-6B show non-limiting illustrations of three self-complementary AAV (scAAV) genomes capable of recombinase/integrase-mediated self-circularization. FIG. 6A shows the structure of the three self-complementary AAV (scAAV) genomes capable of recombinase/integrase-mediated self-circularization. FIG. 6B shows non-limiting examples of sequences that enable self-circularization (e.g., LoxP AttP GT (SEQ ID NO: 568 and SEQ ID NO: 569); FRT AttP GT (SEQ ID NO: 570 and SEQ ID NO: 571); and AttB CC AttP GT (SEQ ID NO: 572 and SEQ ID NO: 573)). GT indicates an AttP site with a GT dinucleotide. AttB CC indicates an AttB site with a CC dinucleotide. LoxP = a LoxP recombinase recognition site. FRT = a FRT recombinase recognition site.
[0055] FIG. 7 shows a non-limiting illustration of recombinase/integrase-mediated intramolecular circularization products.
[0056] FIGs. 8A-8B show non-limiting illustrations of a ddPCR assay and intramolecular circularization ddPCR detection probes. FIG. 8A shows a non-limiting illustration of the ddPCR strategy. FIG. 8B shows non-limiting examples of the universal probe (SEQ ID NO: 574 and SEQ ID NO: 575) and an AttR probe (SEQ ID NO: 576 and SEQ ID NO: 577) that can be used in the assay shown in FIG. 8A.
[0057] FIG. 9 shows a non-limiting illustration of a pDNA genome and AAV transfection and screening protocol. [0058] FIG. 10 shows data for circularization of AAV pDNA and packaged AAV genomic DNA with Bxbl.
[0059] FIG. 11 shows data for Cre-, FLPe-, and Bxbl-mediated circularization of AAV pDNA confirmed by ddPCR.
[0060] FIG. 12 shows Cre-, FLPe-, and Bxbl-mediated circularization of packaged AAV confirmed by ddPCR
[0061] FIG. 13 shows percent circularization between the Bxbl-mediated attR scar ddPCR probe (“attR probe” described in FIG. 8B) and the universal ddPCR probe (“universal probe” described in FIG. 8B).
[0062] FIGs. 14A-14E shows analysis of AttP variants. FIG. 14A shows a non-limiting schematic of AttP mutations tested for improving integration efficiency (SEQ ID NOS: 394 and 540-542, respectively, in order of appearance). FIG. 14B shows integration efficiencies of wildtype and mutant AttP sites across a panel of AttB lengths. FIG. 14C shows a non-limiting schematic of multiplexed integration of different cargo sets at specific genomic loci. Three fluorescent cargos (GFP, mCherry, and YFP) are inserted orthogonally at three different loci (ACTB, LMNB1, NOLC1) for in-frame gene tagging. FIG. 14D shows orthogonality of top 4 AttB/AttP dinucleotide pairs evaluated for GFP integration with PASTE at the ACTB locus. FIG. 14E shows efficiency of multiplexed PASTE insertion of combinations of fluorophores at ACTB, LMNB1, and NOLC1 loci. Data are mean (n= 3) ± s.e.m.
[0063] FIG. 15 illustrates a schematic of single atgRNA and dual atgRNA approaches for beacon placement (“integration recognition site”).
[0064] FIG. 16 shows percent beacon placement in primary mouse hepatocytes (PMH) following delivery of mRNA to deliver a polynucleotide encoding a gene editor polynucleotide construct and an AAV to deliver the first and second atgRNA according to the following conditions: (i) concurrent delivery (“co-dose”), (ii) AAV delivery followed by a “1-day delay” before delivery of the mRNA, or (iii) AAV delivery followed by a “2-day delay” before delivery of the mRNA.
[0065] FIG. 17 shows percent beacon placement in primary human hepatocytes (PHH) following delivering of mRNA to deliver a polynucleotide encoding a gene editor polynucleotide construct and an AAV to deliver the first and second atgRNA. The mRNA and AAV were delivered concurrently. [0066] FIG. 18 shows percent in vivo beacon placement in the Nolcl locus of mice following delivery of a polynucleotide encoding a gene editor polynucleotide construct using a lipid nanoparticle (LNP) and a first atgRNA and second atgRNA using an AAV. %BP = % beacon placement. LNP were administered at doses of 0.5 mg/kg, 1.5 mg/kg, 3 mg/kg, and 5 mg/kg. AAV was administered at 1E11, 3E11, or 1E12 viral genomes (vg) per animal. LNP #F1 = LNP formulation #1. LNP #F2 = LNP formulation #F2. LNP #F3 = LNP formulation #F3.
[0067] FIG. 19 show percent in vivo integration of a template polynucleotide in AttP mice following delivering of the Bxbl using adenovirus (AdV) and the template polynucleotide using an AAV (“AAV Cargo”). Bxbl Adv was administered to the mice at a dose of either 3E10 or 1E11 vector genomes (vg) per animal. AAV Cargo was administered to the mice at a dose of 1E12.
[0068] FIG. 20A shows ddPCR data for percent in vivo beacon placement in the Nolcl locus of neonatal mice at eight days post-delivery of a single dose of a mixture of two LNPs. First LNP contained mRNA encoding a prime editing system and a first synthetic atgRNA (atgRNA 1) at a 1:1 ratio. Second LNP contained mRNA encoding a prime editing system and a second synthetic atgRNA (atgRNA2) at a 1:1 ratio. Each of the first and second atgRNAs targeted the mouse Nolcl locus, encoded a portion of an integration recognition site (“beacon”), and together included a 6bp overlap. The first and second LNPs were combined 1:1 as mixture and administered at either 1 mg/kg or 3 mg/kg. LNP #F2 = LNP formulation #F2.
[0069] FIG. 20B show NGS data for percent in vivo beacon placement in the Nolcl locus of the same neonatal mice and treatment conditions as described in FIG. 20A. NGS data shows beacon placement eight days after administration of the LNP mixture. LNP #F2 = LNP formulation #F2.
[0070] FIG. 20C shows NGS data for percentage of in vivo beacons placed in the Nolcl NGS data is for the same mice with the same treatment conditions as described in FIG. 20A. NGS data shows data for eight days after administration of the LNP mixture. LNP #F2 = LNP formulation #F2.
[0071] FIG. 21A shows ddPCR data for percent in vivo beacon placement in the Nolcl locus of neonatal mice at 6 weeks post-delivery of a single dose of a mixture of two LNPs. First LNP contained mRNA encoding a prime editing system and a first synthetic atgRNA (atgRNA 1) at a 1:1 ratio. Second LNP contained mRNA encoding a prime editing system and a second synthetic atgRNA (atgRNA2) at a 1:1 ratio. Each of the first and second atgRNAs targeted the mouse Nolcl locus, encoded a portion of an integration recognition site (“beacon”), and together included a 6bp overlap. The first and second LNPs were combined 1:1 as mixture and administered at either 1 mg/kg or 3 mg/kg. LNP #F2 = LNP formulation #F2.
[0072] FIG. 21B shows NGS data for percent in vivo beacon placement in the Nolcl locus of the same neonatal mice and treatment conditions as described in FIG. 21A. NGS data shows beacon placement 6 weeks after administration of the LNP mixture. LNP #F2 = LNP formulation #F2.
[0073] FIG. 21C shows NGS data for percentage of in vivo beacons placed in the Nolcl locus that included the expected integration recognition site. Data is from the same mice with the same treatment conditions as described in FIG. 22A. NGS data shows data at 6 weeks after administration of the LNP mixture. LNP #F2 = LNP formulation #F2.
[0074] FIG. 22A shows ddPCR data for percent in vivo beacon placement in the Factor IX (“mF9”) locus of 6-8 week old mice at day 8 post-delivery of a single dose of a mixture of two LNPs. First LNP contained mRNA encoding a prime editing system and a first synthetic atgRNA (atgRNAl) at a ratio of 1:0.5, 1:1, or 1:2. Second LNP contained mRNA encoding a prime editing system and a second synthetic atgRNA (atgRNA2) at a ratio of 1:1, 1:0.5, or 1:2. Each of the first and second atgRNAs targeted the mouse Factor IX locus, encoded a portion of an integration recognition site (“beacon”), and together included a 6bp overlap. The first and second LNPs were combined 1:1 as mixture with the final ratio of mRNA:atgRNAl:atgRNA2 at 1:0.25:0.25; 1:0.5:0.5, or 1:1:1. LNP #F2 = LNP formulation #F2.
[0075] FIG. 22B shows NGS data for percent in vivo beacon placement in the mF9 locus of the same neonatal mice and treatment conditions as described in FIG. 22A. NGS data shows beacon placement 8 days after administration of the LNP mixture. LNP #F2 = LNP formulation #F2.
[0076] FIG. 22C shows NGS data for percent of in vivo beacons placed in the mF9 locus that included the expected integration recognition site. Data is from the same mice with the same treatment conditions as described in FIG. 22A. NGS data shows data at 8 days after administration of the LNP mixture. LNP #F2 = LNP formulation #F2.
[0077] FIGs. 23A-23B show the experimental conditions tested in Example 11. FIG. 23A shows LNP formulations tested in Example 11. FIG. 23B shows control conditions for Example 11.
[0078] FIGs. 24A-24B show representative images of liver sinusoidal endothelial cells (LSECs) contacted with the indicated LNP formulations: cKK (500 ng GFP) or cKK-DOPG (500 ng GFP). Top panels are GFP images. Bottom panels are brightfield. FIG. 24A shows 10X magnification of LSECs contacted with cKK (500 ng GFP) or cKK-DOPG (500 ng GFP). FIG. 24B shows 20X magnification of LSEC contacted with cKK (500 ng GFP) or cKK-DOPG (500 ng GFP).
[0079] FIG. 25 shows representative images of LSECs contacted with the indicated LNP formulations: cKK-Mannose (500 ng GFP) or cKK-DOPG/Mannose (500 ng GFP).
[0080] FIG. 26 shows representative images of control LSECs: transfected with GFP mRNA at 1000 ng per well or untreated LSECs.
[0081] FIG. 27 shows the experimental conditions tested in Example 12.
[0082] FIG. 28 shows a plot of percent (%) Indels in genomic DNA harvested from LSECs following introduction of the indicated guide RNAs (gRNAs). F8 refers to Factor VIII.
[0083] FIG. 29 shows a plot of percent (%) Indels in genomic DNA harvested from LSECs following introduction of guide RNA 2 (G2) using the indicated LNP formulations at the indicated amounts.
[0084] FIG. 30 shows a plot of percent (%) Indels in genomic DNA harvested from LSECs following introduction of guide RNA 4 (G4) using the indicated LNP formulations at the indicated amounts.
[0085] FIG. 31 shows a plot of percent (%) Indels in genomic DNA harvested from LSEC and hepatocytes. LSEC are treated with LNPs with and without modifications e.g., a PEG-modified lipid or a phospholipid (e.g., one of the indicated phospholipids)). Hepatocytes are treated with LNPs with and without modifications.
[0086] FIG. 32 shows a plot of ratio of LSEC targeting to hepatocyte targeting (“LSEC:Hepatocyte targeting”) comparing LNPs with and without modifications e.g., a PEG- modified lipid or a phospholipid (e.g., one of the indicated phospholipids)).
5. DETAILED DESCRIPTION
5.1. Terminology
[0087] Unless defined otherwise, all technical and scientific terms used herein have the meaning commonly understood by a person skilled in the art to which this disclosure belongs. As used herein, the following terms have the meanings ascribed to them below. [0088] “Gene editor’’ as used herein, is a protein that that can be used to perform gene editing, gene modification, gene insertion, gene deletion, or gene inversion. As used herein, the terms “gene editor polynucleotide” refers to polynucleotide sequence encoding the gene editor protein. Such an enzyme or enzyme fusion may contain DNA or RNA targetable nuclease protein (i.e., Cas protein, ADAR, or AD AT), wherein target specificity is mediated by a complexed nucleic acid (i.e., guide RNA). Such an enzyme or enzyme fusion may be a DNA/RNA targetable protein, wherein target specificity is mediated by internal, conjugated, fused, or linked amino acids, such as within TALENs, ZFNs, or meganucleases. The skilled person in the art would appreciate that the gene editor can demonstrate targeted nuclease activity, targeted binding with no nuclease activity, or targeted nickase activity (or cleavase activity). A gene editor comprising a targetable protein may be fused, linked, complexed, operate in cis or trans to one or more proteins or protein fragment motifs. Gene editors may be fused or linked to one or more integrase, recombinase, polymerase, telomerase, reverse transcriptase, or invertase. A gene editor can be a prime editor fusion protein or a gene writer fusion protein.
[0089] “Prime editor fusion protein” as used herein, describes a protein that is used in prime editing. “Prime editor system” as used herein describes the components used in prime editing. Prime editing uses CRISPR enzyme that nicks or cuts only single strand of double stranded DNA, i.e., a nickase; the nickase can occur either naturally or by mutation or modification of a nuclease that makes double stranded cuts. The nickase is programmed (directed) with a prime-editing guide RNA (pegRNA). The skilled person in the art would appreciate that the pegRNA both specifies the target site and encodes the desired edit. Described herein are attachment site containing guide RNA (atgRNA) that both specifies the target and encodes for the desired integrase target recognition site. The nickase may be programmed (directed) with an atgRNA. Advantageously the nickase is a catalytically impaired Cas9 endonuclease, a Cas9 nickase, that is fused to the reverse transcriptase. During genetic editing, the Cas9 nickase part of the protein is guided to the DNA target site by the atgRNA (or pegRNA), whereby a nick or single stranded cut occurs. The reverse transcriptase domain then uses the atgRNA (or pegRNA) to template reverse transcription of the desired edit, directly polymerizing DNA onto the nicked target DNA strand. The edited DNA strand replaces the original DNA strand, creating a heteroduplex containing one edited strand and one unedited strand. Afterward, optionally, the prime editor (PE) guides resolution of the heteroduplex to favor copying the edit onto the unedited strand, completing the process (typically achieved with a nickase gRNA). Other enzymes that can be used to nick or cut only a single strand of double stranded DNA includes a cleavase (e.g., cleavase I enzyme). [0090] In some embodiments, an additional agent or agents may be added that improve the efficiency and outcome purity of the prime edit. In some embodiments, the agent may be chemical or biological and disrupt DNA mismatch repair (MMR) processes at or near the edit site (i.e., PE4 and PE5 and PEmax architecture by Chen et al. Cell, 184, 1-18, October 28, 2021; Chen et al. is incorporated herein by reference). In typical embodiments, the agent is a MMR-inhibiting protein. In certain embodiments, the MMR-inhibiting protein is dominant negative MMR protein. In certain embodiments, the dominant negative MMR protein is MLHldn. In particular embodiments, the MMR-inhibiting agent is incorporated into the co-delivery method described herein. In some embodiments, the MMR-inhibiting agent is linked or fused to the prime editor protein fusion, which may or may not have a linked or fused integrase. In some embodiments, the MMR-inhibiting agent is linked or fused to the Gene Writer™ protein, which may or may not have a linked or fused integrase.
[0091] The prime editor or gene editor system can be used to achieve DNA deletion and replacement. In some embodiments, the DNA deletion replacement is induced using a pair of atgRNAs or pegRNA that target opposite DNA strands, programming not only the sites that are nicked but also the outcome of the repair (i.e., PrimeDel by Choi et al. Nat. Biotechnology, October 14, 2021; Choi et al. is incorporated herein by reference and TwinPE by Anzalone et al. BioRxiv, November 2, 2021; Anzalone et al. is incorporated herein by reference). In some embodiments described herein, the DNA deletion is induced using a single atgRNA. In some embodiments, the DNA deletion and replacement is induced using a wild type Cas9 prime editor (PE-Cas9) system (i.e., PED AR by Jiang et al. Nat. Biotechnology, October 14, 2021; Jiang et al. is incorporated herein by reference in its entirety). In some embodiments, the DNA replacement is an integrase target recognition site or recombinase target recognition site. In certain embodiments, the constructs and methods described herein may be utilized to incorporate the pair of pegRNAs (or atgRNAs) used in PrimeDel, TwinPE (WO2021226558 incorporated by reference herein in its entirety), or PED AR, the prime editor fusion protein or Gene Writer protein, optionally a nickase guide RNA (ngRNA), an integrase, a nucleic acid cargo, and optionally a recombinase into a LNP delivery system or vector delivery system (e.g., AAV or Adenovirus). The integrase may be directly linked, for example by a peptide linker, to the prime editor fusion or gene writer protein.
[0092] In some embodiments, the prime editors can refer to a retrovirus or lentivirus reverse transcriptase such as a Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase (RT) fused to a CRISPR enzyme nickase such as a Cas9 H840A nickase, a Cas9nickase. In some embodiments, the prime editors can refer to a retrovirus or lentivirus reverse transcriptase such as a Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase (RT) fused to a cleavase. In some embodiments the RT can be fused at, near or to the C-terminus of a Cas9nickase, e.g., Cas9 H840A. Fusing the RT to the C-terminus region, e.g., to the C-terminus, of the Cas9 nickase may result in higher editing efficiency. Such a complex is called PEI. In some embodiments, the CRISPR enzyme nickase, e.g., Cas9(H840A), i.e., a Cas9nickase, can be linked to a non-M-MLV reverse transcriptase such as an AMV-RT or XRT (Cas9(H840A)-AMV-RT or XRT). In some embodiments, instead of the CRISPR enzyme nickase being a Cas9 (H840A), i.e., instead of being a Cas9 nickase, the CRISPR enzyme nickase instead can be a CRISPR enzyme that naturally is a nickase or cuts a single strand of double stranded DNA; for instance, the CRISPR enzyme nickase can be Casl2a/b. Alternatively, the CRISPR enzyme nickase can be another mutation of Cas9, such as Cas9(D10A). A CRISPR enzyme, such as a CRISPR enzyme nickase, such as Cas9 (wild type), Cas9(H840A), Cas9(D10A) or Cas 12a/b nickase can be fused in some embodiments to a pentamutant of M-MLV RT (D200N/ L603W/ T330P/ T306K/ W313F), whereby there can be up to about 45-fold higher efficiency, and this is called PE2. In some embodiments, the M-MLV RT comprise one or more of the mutations Y8H, P51L, S56A, S67R, E69K, V129P, L139P, T197A, H204R, V223H, T246E, N249D, E286R, Q2911, E302K, E302R, F309N, M320L, P330E, L435G, L435R, N454K, D524A, D524G, D524N, E562Q, D583N, H594Q, E607K, D653N, and L671P. Specific M-MLV RT mutations are shown in Table 1.
[0093] In some embodiments, the reverse transcriptase can also be a wild-type or modified transcription xenopolymerase (RTX), avian myeloblastosis virus reverse transcriptase (AMV RT), Feline Immunodeficiency Virus reverse transcriptase (FIV-RT), FeLV-RT (Feline leukemia virus reverse transcriptase), HIV-RT (Human Immunodeficiency Virus reverse transcriptase). In some embodiments, the reverse transcriptase can be a fusion of MMuLV to the Sto7d DNA binding domain (see lonnidi et al.; https://doi.org/10.1101/2021.ll.01.466786). The fusion of MMuLV to the Sto7d DNA binding domain sequence is given in Table 2. Table 2
[0094] PE3, PE3b, PE4, PE5, and/or PEmax, which a skilled person can incorporate into the co-delivery system described herein, involves nicking the non-edited strand, potentially causing the cell to remake that strand using the edited strand as the template to induce HR. The nicking of the non-edited strand can involve the use of a nicking guide RNA (ngRNA).
[0095] The skilled person can readily incorporate into the co-delivery system described herein described herein a prime editing or CRISPR system. Examples of prime editors can be found in the following: W02020/191153, W02020/191171, WO2020/191233,
WO2020/191234, WO2020/191239, W02020/191241, WO2020/191242, WO2020/191243, WO2020/191245, WO2020/191246, WO2020/191248, WO2020/191249, each of which is incorporated by reference herein in its entirety. In addition, mention is made, and can be used herein, of CRISPR Patent Applications and Patents of the Zhang laboratory and/or Broad Institute, Inc. and Massachusetts Institute of Technology and/or Broad Institute, Inc.,
Massachusetts Institute of Technology and President and Fellows of Harvard College and/or
Editas Medicine, Inc. Broad Institute, Inc., The University of Iowa Research Foundation and
Massachusetts Institute of Technology, including those claiming priority to US Application 61/736,527, filed December 12, 2012, including US Patents 11,104,937, 11,091,798,
11,060,115, 11,041,173, 11,021,740, 11,008,588, 11,001,829, 10,968,257, 10,954,514,
10,946,108, 10,930,367, 10,876,100, 10,851,357, 10,781,444, 10,711,285, 10,689,691,
10,648,020, 10,640,788, 10,577,630, 10,550,372, 10,494,621, 10,377,998, 10,266,887,
10,266,886, 10,190,137, 9,840,713, 9,822,372, 9,790,490, 8,999,641, 8,993,233, 8,945,839,
8,932,814, 8,906,616, 8,895,308, 8,889,418, 8,889,356, 8,871,445, 8,865,406, 8,795,965, 8,771,945, and 8,697,359; CRISPR Patent Applications and Patents of the Doudna laboratory and/or of Regents of the University of California, the University of Vienna and Emmanuelle Charpentier, including those claiming priority to US application 61/652,086, filed May 25, 2012, and/or 61/716,256, filed October 19, 2012, and/or 61/757,640, filed January 28, 2013, and/or 61/765,576, filed February 15, 2013 and/or 13/842,859, including US Patents
11,028,412, 11,008,590, 11,008,589, 11,001,863, 10,988,782, 10,988,780, 10,982,231,
10,982,230, 10,900,054, 10,793,878, 10,774,344, 10,752,920, 10,676,759, 10,669,560,
10,640,791, 10,626,419, 10,612,045, 10,597,680, 10,577,631, 10,570,419, 10,563,227,
10,550,407, 10,533,190, 10,526,619, 10,519,467, 10,513,712, 10,487,341, 10,443,076,
10,428,352, 10,421,980, 10,415,061, 10,407,697, 10,400,253, 10,385,360, 10,358,659,
10,358,658, 10,351,878, 10,337,029, 10,308,961, 10,301,651, 10,266,850, 10,227,611,
10,113,167, and 10,000,772; CRISPR Patent Applications and Patents of Vilnius University and/or the Siksnys laboratory, including those claiming priority to US application 62/046384 and/or 61/625,420 and/or 61/613,373 and/or PCT/IB2015/056756, including US Patent 10,385,336; CRISPR Patent Applications and Patents of the President and Fellows of Harvard
College, including those of George Church’s laboratory and/or claiming priority to US application 61/738,355, filed December 17, 2012, including 11,111,521, 11,085,072,
11,064,684, 10,959,413, 10,925,263, 10,851,369, 10,787,684, 10,767,194, 10,717,990,
10,683,490, 10,640,789, 10,563,225, 10,435,708, 10,435,679, 10,375,938, 10,329,587,
10,273,501, 10,100,291, 9,970,024, 9,914,939, 9,777,262, 9,587,252, 9,267,135, 9,260,723,
9,074,199, 9,023,649; CRISPR Patent Applications and Patents of the President and Fellows of Harvard College, including those of David Liu’s laboratory, including 11,111,472,
11,104,967, 11,078,469, 11,071,790, 11,053,481, 11,046,948, 10,954,548, 10,947,530,
10,912,833, 10,858,639, 10,745,677, 10,704,062, 10,682,410, 10,612,011, 10,597,679,
10,508,298, 10,465,176, 10,323,236, 10,227,581, 10,167,457, 10,113,163, 10,077,453,
9,999,671, 9,840,699, 9,737,604, 9,526,784, 9,388,430, 9,359,599, 9,340,800, 9,340,799, 9,322,037, 9,322,006, 9,228,207, 9,163,284, and 9,068,179; and CRISPR Patent Applications and Patents of Toolgen Incorporated and/or the Kim laboratory and/or claiming priority to US application 61/717,324, filed October 23, 2012 and/or 61/803,599, filed March 20, 2013 and/or 61/837,481, filed June 20, 2013 and/or 62/033,852, filed August 6, 2014 and/or PCT/KR2013/009488 and/or PCT/KR2015/008269, including US Patent 10,851,380, and 10,519,454; and CRISPR Patent Applications and Patents of Sigma and/or Millipore and/or the Chen laboratory and/or claiming priority to US application 61/734,256, filed December 6, 2012 and/or 61/758,624, filed January 30, 2013 and/or 61/761,046, filed February 5, 2013 and/or 61/794,422, filed March 15, 2013, including US Patent 10,731,181, each of which is hereby incorporated herein by reference, and from the disclosures of the foregoing, the skilled person can readily make and use a prime editing or CRISPR system, and can especially appreciate impaired endonucleases, such as a mutated Cas9 that only nicks a single strand of DNA and is hence a nickase, or a CRISPR enzyme that only makes a single-stranded cut that can be employed in a PASTE system of the disclosure. Further, from the disclosures of the foregoing, the skilled person can incorporate the selected CRISPR enzyme, as part of the prime editor fusion or gene editor fusion, into the co-delivery method described herein.
[0096] Prior to RT-mediated edit incorporation, the prime editor protein (or system) (1) site-specifically targets a genomic locus and (2) performs a catalytic cut or nick. These steps are typically performed by a CRISPR-Cas. However, in some embodiments the Cas protein may be substituted by other nucleic acid programmable DNA binding proteins (napDNAbp) such as zinc finger nucleases (ZFNs), transcription activator- like effector nucleases (TAEENs), or meganucleases. In addition, to the extent the “targeting rules” of other napDNAbp are known or are newly determined, it becomes possible to use new napDNAbp, beyond Cas9, to site specifically target and modify genomic sites of interest.
[0097] Similar to a prime editor protein, a Gene Writer can introduce novel DNA elements, such as an integration target site, into a DNA locus. A Gene Writer protein comprises: (A) a polypeptide or a nucleic acid encoding a polypeptide, wherein the polypeptide comprises (i) a reverse transcriptase domain, and either (x) an endonuclease domain that contains DNA binding functionality or (y) an endonuclease domain and separate DNA binding domain; and (B) a template RNA comprising (i) a sequence that binds the polypeptide and (ii) a heterologous insert sequence. Examples of such Gene Writer™ proteins and related systems can be found in US20200109398, which is incorporated by reference herein in its entirety.
[0098] In some embodiments, the prime editor or Gene Writer protein fusion or prime editor protein linked or fused to an integrase is expressed as a split construct. In typical embodiments, the split construct in reconstituted in a cell. In some embodiments, the split construct can be fused or ligated via intein protein splicing. In some embodiments, the split construct can be reconstituted via protein-protein inter- molecular bonding and/or interactions. In some embodiments, the split construct can be reconstituted via chemical, biological, or environmental induced oligomerization. In certain embodiments, the split construct can be adapted into one or more delivery vectors described herein.
[0099] In some embodiments, an integrase or recombinase is directly linked or fused, for example by a peptide linker, which may be cleavable or non-cleavable, to the prime editor fusion protein (i.e., fused Cas9 nickase-reverse transcriptase) or Gene Writer protein. Suitable linkers, for example between the Cas9, RT, and integrase, may be selected from Table 3:
[0100] In some embodiments, the prime editor or Gene Writer protein fusion or prime editor protein linked or fused to an integrase is expressed as a split construct. In typical embodiments, the split construct in reconstituted in a cell. In some embodiments, the split construct can be fused or ligated via intein protein splicing. In some embodiments, the split construct can be reconstituted via protein-protein inter- molecular bonding and/or interactions. In some embodiments, the split construct can be reconstituted via chemical, biological, or environmental induced oligomerization. In certain embodiments, the split construct can be adapted into one or more nucleic acid constructs described herein.
5.2. Type II CRISPR proteins
[0101] The skilled person can incorporate a selected CRISPR enzyme, described below, as part of the prime editor fusion, into the co-delivery method described herein. Streptococcus pyogenes Cas9 (SpCas9), the most common enzyme used in genome-editing applications, is a large nuclease of 1368 amino acid residues. Advantages of SpCas9 include its short, 5'-NGG-3' PAM and very high average editing efficiency. SpCas9 consists of two lobes: a recognition (REC) lobe and a nuclease (NUC) lobe. The REC lobe can be divided into three regions, a long a helix referred to as the bridge helix (residues 60-93), the RECI (residues 94-179 and 308-713) domain, and the REC2 (residues 180-307) domain. The NUC lobe consists of the RuvC (residues 1-59, 718-769, and 909-1098), HNH (residues 775-908), and PAM-interacting (PI) (residues 1099- 1368) domains. The negatively charged sgRNA:target DNA heteroduplex is accommodated in a positively charged groove at the interface between the REC and NUC lobes. In the NUC lobe, the RuvC domain is assembled from the three split RuvC motifs (RuvC I— III) and interfaces with the PI domain to form a positively charged surface that interacts with the 30 tail of the sgRNA. The HNH domain lies between the RuvC II— III motifs and forms only a few contacts with the rest of the protein. Structural aspects of SpCas9 are described by Nishimasu et al., Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA, Cell 156, 935-949, February 27, 2014.
[0102] REC lobe'. The REC lobe includes the RECI and REC2 domains. The REC2 domain does not contact the bound guide:target heteroduplex, indicating that truncation of REC lobe may be tolerated by SpCas9. Further, SpCas9 mutant lacking the REC2 domain (D175-307) retained -50% of the wild-type Cas9 activity, indicating that the REC2 domain is not critical for DNA cleavage. In striking contrast, the deletion of either the repeat-interacting region (D97-150) or the anti-repeat-interacting region (D312-409) of the RECI domain abolished the DNA cleavage activity, indicating that the recognition of the repeat:anti-repeat duplex by the RECI domain is critical for the Cas9 function.
[0103] PAM-Interacting domain'. The NUC lobe contains the PAM-interacting (PI) domain that is positioned to recognize the PAM sequence on the noncomplementary DNA strand. The PI domain of SpCas9 is required for the recognition of 5’-NGG-3’ PAM, and deletion of the PI domain (A1099-1368) abolished the cleavage activity, indicating that the PI domain is critical for SpCas9 function and a major determinant for the PAM specificity.
[0104] RuvC domain'. The RuvC nucleases of SpCas9 have an RNase H fold and four catalytic residues, AsplO (Ala), Glu762, His983, and Asp986, that are critical for the two-metal cleavage of the noncomplementary strand of the target DNA. In addition to the conserved RNase H fold, the Cas9 RuvC domain has other structural elements involved in interactions with the guide:target heteroduplex (an end-capping loop between a42 and a43) and the PI domain/stem loop 3 (P hairpin formed by P3 and [34).
[0105] HNH domain'. SpCas9 HNH nucleases have three catalytic residues, Asp839, His840, and Asn863 and cleave the complementary strand of the target DNA through a single-metal mechanism.
[0106] sgRNA. DNA recognition'. The sgRNA guide region is primarily recognized by the REC lobe. The backbone phosphate groups of the guide region (nucleotides 2, 4-6, and 13-20) interact with the RECI domain (Argl65, Glyl66, Arg403, Asn407, Lys510, Tyr515, and Arg661) and the bridge helix (Arg63, Arg66, Arg70, Arg71, Arg74, and Arg78). The 20-hydroxyl groups of Gl, C15, U16, and G19 hydrogen bond with Vall009, Tyr450, Arg447/Ile448, and Thr404, respectively. [0107] A mutational analysis demonstrated that the R66A, R70A, and R74A mutations on the bridge helix markedly reduced the DNA cleavage activities, highlighting the functional significance of the recognition of the sgRNA “seed” region by the bridge helix. Although Arg78 and Argl65 also interact with the “seed” region, the R78A and R165A mutants showed only moderately decreased activities. These results are consistent with the fact that Arg66, Arg70, and Arg74 form multiple salt bridges with the sgRNA backbone, whereas Arg78 and Argl65 form a single salt bridge with the sgRNA backbone. Moreover, the alanine mutations of the repeat:anti- repeat duplex-interacting residues (Arg75 and Lysl63) and the stemloop- 1 -interacting residue (Arg69) resulted in decreased DNA cleavage activity, confirming the functional importance of the recognition of the repeat: anti-repeat duplex and stem loop 1 by Cas9.
[0108] RNA-guided DNA targeting'. SpCas9 recognizes the guide:target heteroduplex in a sequence-independent manner. The backbone phosphate groups of the target DNA (nucleotides 1, 9-11, 13, and 20) interact with the RECI (Asn497, Trp659, Arg661, and Gln695), RuvC (Gln926), and PI (Glul 108) domains. The C2’ atoms of the target DNA (nucleotides 5, 7, 8, 11, 19, and 20) form van der Waals interactions with the RECI domain (Leul69, Tyr450, Met495, Met694, and His698) and the RuvC domain (Ala728). The terminal base pair of the guide:target heteroduplex (GEC20’) is recognized by the RuvC domain via end-capping interactions; the sgRNA G1 and target DNA C20’ nucleobases interact with the Tyrl013 and Vall015 side chains, respectively, whereas the 20-hydroxyl and phosphate groups of sgRNA G1 interact with Vall009 and Gln926, respectively.
[0109] Repeat:Anti-Repeat duplex recognition'. The nucleobases of U23/A49 and A42/G43 hydrogen bond with the side chain of Argl l22 and the main-chain carbonyl group of Phe351, respectively. The nucleobase of the flipped U44 is sandwiched between Tyr325 and His328, with its N3 atom hydrogen bonded with Tyr325, whereas the nucleobase of the unpaired G43 stacks with Tyr359 and hydrogen bonds with Asp364.
[0110] The nucleobases of G21 and U50 in the G2EU50 wobble pair stack with the terminal C20:G10 pair in the guide:target heteroduplex and Tyr72 on the bridge helix, respectively, with the U5004 atom hydrogen bonded with Arg75. Notably, A51 adopts the syn conformation and is oriented in the direction opposite to U50. The nucleobase of A51 is sandwiched between Phel 105 and U63, with its Nl, N6, and N7 atoms hydrogen bonded with G62, Glyl l03, and Phel 105, respectively. [0111] Stem-loop recognition'. Stem loop 1 is primarily recognized by the REC lobe, together with the PI domain. The backbone phosphate groups of stem loop 1 (nucleotides 52, 53, and 59- 61) interact with the RECI domain (Leu455, Ser460, Arg467, Thr472, and Ile473), the PI domain (Lysl l23 and Lysl l24), and the bridge helix (Arg70 and Arg74), with the 20-hydroxyl group of G58 hydrogen bonded with Leu455. A52 interacts with Phel l05 through a face-to-edge p-p stacking interaction, and the flipped U59 nucleobase hydrogen bonds with Asn77.
[0112] The single-stranded linker and stem loops 2 and 3 are primarily recognized by the NUC lobe. The backbone phosphate groups of the linker (nucleotides 63-65 and 67) interact with the RuvC domain (Glu57, Lys742, and Lysl097), the PI domain (Thrl l02), and the bridge helix (Arg69), with the 20-hydroxyl groups of U64 and A65 hydrogen bonded with Glu57 and His721, respectively. The C67 nucleobase forms two hydrogen bonds with Vail 100.
[0113] Stem loop 2 is recognized by Cas9 via the interactions between the NUC lobe and the non-Watson-Crick A68:G81 pair, which is formed by direct (between the A68 N6 and G81 06 atoms) and water-mediated (between the A68 N1 and G81 N1 atoms) hydrogen-bonding interactions. The A68 and G81 nucleobases contact Serl351 and Tyrl356, respectively, whereas the A68:G81 pair interacts with Thrl358 via a water-mediated hydrogen bond. The 20-hydroxyl group of A68 hydrogen bonds with His 1349, whereas the G81 nucleobase hydrogen bonds with Lys33.
[0114] Stem loop 3 interacts with the NUC lobe more extensively, as compared to stem loop 2. The backbone phosphate group of G92 interacts with the RuvC domain (Arg40 and Lys44), whereas the G89 and U90 nucleobases hydrogen bond with Glnl272 and Glul225/Alal227, respectively. The A88 and C91 nucleobases are recognized by Asn46 via multiple hydrogenbonding interactions.
[0115] Cas9 proteins smaller than SpCas9 allow more efficient packaging of nucleic acids encoding CRISPR systems, e.g., Cas9 and sgRNA into one rAAV (“all-in-one-AAV”) particle. In addition, efficient packaging of CRISPR systems can be achieved in other viral vector systems (i.e., lentiviral, integration deficient lentiviral, hd-AAV, etc.) and non-viral vector systems (i.e., lipid nanoparticle). Small Cas9 proteins can be advantageous for multidomain-Cas-nuclease-based systems for prime editing. Well characterized smaller Cas9 proteins include Staphylococcus aureus (SauCas9, 1053 amino acid residues) and Campylobacter jejuni (CjCas9, 984 amino residues). However, both recognize longer PAMs, 5'-NNGRRT-3' for SauCas9 (R = A or G) and 5'-NNNNRYAC-3' for CjCas9 (Y = C or T), which reduces the number of uniquely addressable target sites in the genome, in comparison to the NGG SpCas9 PAM. Among smaller Cas9s, Schmidt et al. identified Staphylococcus lugdunensis (Siu) Cas9 as having genome-editing activity and provided homology mapping to SpCas9 and SauCas9 to facilitate generation of nickases and inactive (“dead”) enzymes (Schmidt et al., 2021, Improved CRISPR genome editing using small highly active and specific engineered RNA-guided nucleases. Nat Commun 12, 4219. doi.org/10.1038/s41467-021-24454-5) and engineered nucleases with higher cleavage activity by fragmenting and shuffling Cas9 DNAs. The small Cas9s and nickases are useful in the instant disclosure.
[0116] Besides dead Cas9 and Cas9 nickase variants, the Cas9 proteins used herein may also include other “Cas9 variants” having at least about 70% identical, at least about 80% identical, at least about 90% identical, at least about 95% identical, at least about 96% identical, at least about 97% identical, at least about 98% identical, at least about 99% identical, at least about 99.5% identical, or at least about 99.9% identical to any reference Cas9 protein, including any wild type Cas9, or mutant Cas9 (e.g., a dead Cas9 or Cas9 nickase), or fragment Cas9, or circular permutant Cas9, or other variant of Cas9 disclosed herein or known in the art. In some embodiments, a Cas9 variant may have 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 21, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50 or more amino acid changes compared to a reference Cas9. In some embodiments, the Cas9 variant comprises a fragment of a reference Cas9 (e.g., a gRNA binding domain or a DNA-cleavage domain), such that the fragment is at least about 70% identical, at least about 80% identical, at least about 90% identical, at least about 95% identical, at least about 96% identical, at least about 97% identical, at least about 98% identical, at least about 99% identical, at least about 99.5% identical, or at least about 99.9% identical to the corresponding fragment of wild type Cas9. In some embodiments, the fragment is at least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least 95% identical, at least 96%, at least 97%, at least 98%, at least 99%, or at least 99.5% of the amino acid length of a corresponding wild type Cas9 (e.g., SEQ ID NO: 18).
[0117] In some embodiments, the disclosure also may utilize Cas9 fragments that retain their functionality and that are fragments of any herein disclosed Cas9 protein. In some embodiments, the Cas9 fragment is at least 100 amino acids in length. In some embodiments, the fragment is at least 100, 150, 200, 250, 300, 350, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, 1100, 1150, 1200, 1250, or at least 1300 amino acids in length. [0118] In various embodiments, the prime editors disclosed herein may comprise one of the Cas9 variants described as follows, or a Cas9 variant thereof having at least about 70% identical, at least about 80% identical, at least about 90% identical, at least about 95% identical, at least about 96% identical, at least about 97% identical, at least about 98% identical, at least about 99% identical, at least about 99.5% identical, or at least about 99.9% identical to any reference Cas9 variants.
[0119] In some embodiments, prime editors utilized herein comprise CRISPR-Cas system enzymes other than type II enzymes. In certain embodiments, prime editors comprise type V or type VI CRISPR-Cas system enzymes. It will be appreciated that certain CRISPR enzymes exhibit promiscuous ssDNA cleavage activity and appropriate precautions should be considered. In certain embodiments, prime editors comprise a nickase or a dead CRISPR with nuclease function comprised in a different component. [0120] In various embodiments, the nucleic acid programmable DNA binding proteins utilized herein include, without limitation, Cas9 (e.g., dCas9 and nCas9), Casl2a (Cpfl), Casl2bl (C2cl), Casl2b2, Casl2c (C2c3), Casl2d (CasY), Casl2e (CasX), C2c4, C2c5, C2c8, C2c9, C2cl0, Casl3a (C2c2), Casl3b (C2c6), Casl3c (C2c7), Casl3d, and Argonaute. Cas-equivalents further include those described in Makarova et al., “C2c2 is a single-component programmable RNA- guided RNA-targeting CRISPR effector,” Science 2016; 353(6299) and Makarova et al., “Classification and Nomenclature of CRISPR-Cas Systems: Where from Here?,” The CRISPR Journal, Vol.l. No.5, 2018, the contents of which are incorporated herein by reference. One example of a nucleic acid programmable DNA-binding protein that has different PAM specificity than Cas9 is Clustered Regularly Interspaced Short Palindromic Repeats from Prevotella and Francisella 1 (i.e, Casl2a (Cpfl)). Similar to Cas9, Casl2a (Cpfl) is also a Class 2 CRISPR effector, but it is a member of type V subgroup of enzymes, rather than the type II subgroup. It has been shown that Casl2a (Cpfl) mediates robust DNA interference with features distinct from Cas9. Casl2a (Cpfl) is a single RNA-guided endonuclease lacking tracrRNA, and it utilizes a T- rich protospacer-adjacent motif (TTN, TTTN, or YTN). Moreover, Cpfl cleaves DNA via a staggered DNA double- stranded break. Out of 16 Cpfl -family proteins, two enzymes from Acidaminococcus and Lachnospiraceae are shown to have efficient genome-editing activity in human cells. Cpfl proteins are known in the art and have been described previously, for example Yamano et al., “Crystal structure of Cpfl in complex with guide RNA and target DNA.” Cell (165) 2016, p.949-962; the entire contents of which is hereby incorporated by reference.
5.3. Type V CRISPR proteins
[0121] In some embodiments, prime editors used herein comprise the type V CRISPR family includes Francisella novicida U112 Cpfl (FnCpfl) also known as FnCasl2a. FnCpfl adopts a bilobed architecture with the two lobes connected by the wedge (WED) domain. The N-terminal REC lobe consists of two a-helical domains (RECI and REC2) that have been shown to coordinate the crRNA-target DNA heteroduplex. The C-terminal NUC lobe consists of the C-terminal RuvC and Nuc domains involved in target cleavage, the arginine-rich bridge helix (BH), and the PAM- interacting (PI) domain. The repeat-derived segment of the crRNA forms a pseudoknot stabilized by intra-molecular base-pairing and hydrogen-bonding interactions. The pseudoknot is coordinated by residues from the WED, RuvC, and REC2 domains, as well as by two hydrated magnesium cations. Notably, nucleotides 1-5 of the crRNA are ordered in the central cavity of FnCasl2a and adopt an A-form-like helical conformation. Conformational ordering of the seed sequence is facilitated by multiple interactions between the ribose and phosphate moieties of the crRNA backbone and FnCpfl residues in the WED and RECI domains. These include residues Thrl6, Lys595, His804, and His881 from the WED domain and residues Tyr47, Lys51, Phel82, and Argl86 from the RECI domain. The structure of the FnCasl2a-crRNA complex further reveals that the bases of the seed sequence are solvent exposed and poised for hybridization with target DNA. Structural aspects of FnCpfl are described by Swarts et al., Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRTS PR-Cas 12a, Molecular Cell 66, 221-233, April 20, 2017.
[0122] Pre-crRNA processing'. Essential residues for crRNA processing include His843, Lys852, and Lys869. Structural observations are consistent with an acid-base catalytic mechanism in which Lys869 acts as the general base catalyst to deprotonate the attacking 2’ -hydroxyl group of U(-19), while His843 acts as a general acid to protonate the 5’-oxygen leaving group of A(-18). In turn, the side chain of Lys852 is involved in charge stabilization of the transition state. Collectively, these interactions facilitate the intra-molecular attack of the 20-hydroxyl group of U(-19) on the scissile phosphate and promote the formation of the 2’,3’-cyclic phosphate product.
[0123] R-loop formation'. The crRNA-target DNA strand heteroduplex is enclosed in the central cavity formed by the REC and NUC lobes and interacts extensively with the RECI and REC2 domains. The PAM-containing DNA duplex comprises target strand nucleotides dT0-dT8 and non-target strand nucleotides dA(8)*-dA0* and is contacted by the PI, WED, and RECI domains. The 5’-TTN-3’ PAM is recognized in FnCasl2a by a mechanism combining the shapespecific recognition of a narrowed minor groove, with base-specific recognition of the PAM bases by two invariant residues, Lys671 and Lys613. Directly downstream of the PAM, the duplex of the target DNA is disrupted by the side chain of residue Lys667, which is inserted between the DNA strands and forms a cation-7t stacking interaction with the dAO-dTO* base pair. The phosphate group linking target strand residues dT(- 1) and dTO is coordinated by hydrogen-bonding interactions with the side chain of Lys823 and the backbone amide of Gly826. Target strand residue dT(-l) bends away from residue TO, allowing the target strand to interact with the seed sequence of the crRNA. The non-target strand nucleotides dTl*-dT5* interact with the Arg692- Ser702 loop in FnCasl2a through hydrogen-bonding and ionic interactions between backbone phosphate groups and side chains of Arg692, Asn700, Ser702, and Gln704, as well as main-chain amide groups of Lys699, Asn700, and Ser702. Alanine substitution of Q704 or replacement of residues Thr698-Ser702 in FnCasl2a with the sequence Ala-Gly3 (SEQ ID NO: 115) substantially reduced DNA cleavage activity, suggesting that these residues contribute to R-loop formation by stabilizing the displaced conformation of the nontarget DNA strand. [0124] In the FnCasl2a R-loop complex, the crRNA-target strand heteroduplex is terminated by a stacking interaction with a conserved aromatic residue (Tyr410). This prevents base pairing between the crRNA and the target strand beyond nucleotides U20 and dA(-20), respectively. Beyond this point, the target DNA strand nucleotides re-engage the non-target DNA strand, forming a PAM-distal DNA duplex comprising nucleotides dC(-21)-dA(-27) and dG21*-dT27*, respectively. The duplex is confined between the REC2 and Nuc domains at the end of the central channel formed by the REC and NUC lobes.
[0125] Target DNA cleavage: FnCpfl can independently accommodate both the target and non-target DNA strands in the catalytic pocket of the RuvC domain. The RuvC active site contains three catalytic residues (D917, E1006, and D1255). Structural observations suggest that both the target and non-target DNA strands are cleaved by the same catalytic mechanism in a single active site in Cpfl/Casl2a enzymes.
[0126] Another type V CRISPR is AsCpfl from Acidaminococcus sp BV3L6 (Yamano et al., Crystal structure of Cpfl in complex with guide RNA and target DNA, Cell 165, 949-962, May 5, 2016)
[0127] In certain embodiments, the nuclease comprises a Casl2f effector. Small CRISPR- associated effector proteins belonging to the type V-F subtype have been identified through the mining of sequence databases and members classified into Casl2fl (Casl4a and type V-U3), Casl2f2 (Casl4b) and Casl2f3 (Casl4c, type V-U2 and U4). (See, e.g., Karvelis et al., PAM recognition by miniature CRISPR-Cas 12f nucleases triggers programmable double- stranded DNA target cleavage. Nucleic Acids Research, 21 May 2020, 48(9), 5016-23 doi.org/10.1093/nar/gkaa208). Xu et al. described development of a 529 amino acid Casl2f-based system for mammalian genome engineering through multiple rounds of iterative protein engineering and screening. (Xu, X. et al., Engineered Miniature CRISPR-Cas System for Mammalian Genome Regulation and Editing. Molecular Cell, October 21, 2021, 81(20): 4333-45, doi.org/10.1016/j.molcel.2021.08.008).
[0128] Exemplary CRISPR-Cas proteins and enzymes used in the prime editors herein include the following without limitation. Table 6 - Casl2b (C2cl) orthologs
5.4. Protospacer Adjacent Motif
[0129] As used herein, the term “protospacer adjacent sequence” or “protospacer adjacent motif’ or “PAM” refers to an approximately 2-6 base pair DNA sequence (or a 2-, 3-, 4-, 5-, 6-, 7-, 8-, 9-, 10-, 11-, 12-long nucleotide sequence) that is an important targeting component of a Cas9 nuclease. Typically, the PAM sequence is on either strand, and is downstream in the 5' to 3' direction of Cas9 cut site. The canonical PAM sequence (i.e., the PAM sequence that is associated with the Cas9 nuclease of Streptococcus pyogenes or SpCas9) is 5'-NGG-3' wherein “N” is any nucleobase followed by two guanine (“G”) nucleobases. Different PAM sequences can be associated with different Cas9 nucleases or equivalent proteins from different organisms. In addition, any given Cas9 nuclease may be modified to alter the PAM specificity of the nuclease such that the nuclease recognizes alternative PAM sequence.
[0130] For example, with reference to the canonical SpCas9 amino acid sequence, the PAM specificity can be modified by introducing one or more mutations, including (a) DI 135V, R1335Q, and T1337R “the VQR variant”, which alters the PAM specificity to NGAN or NGNG, (b) D1135E, R1335Q, and T1337R “the EQR variant”, which alters the PAM specificity to NGAG, and (c) DI 135V, G1218R, R1335E, and T1337R “the VRER variant”, which alters the PAM specificity to NGCG. In addition, the D1135E variant of canonical SpCas9 still recognizes NGG, but it is more selective compared to the wild type SpCas9 protein.
[0131] It will also be appreciated that Cas9 enzymes from different bacterial species (i.e., Cas9 orthologs) can have varying PAM specificities and some embodiments are therefore chosen based on the desired PAM recognition. For example, Cas9 from Staphylococcus aureus (SaCas9) recognizes NGRRT or NGRRN. In addition, Cas9 from Neisseria meningitis (NmCas) recognizes NNNNGATT. In another example, Cas9 from Streptococcus thermophilis (StCas9) recognizes NNAGAAW. In still another example, Cas9 from Treponema denticola (TdCas) recognizes NAAAAC. These examples are not meant to be limiting. It will be further appreciated that non- SpCas9s bind a variety of PAM sequences, which makes them useful to expand the range of sequences that can be targeted according to the disclosure. Furthermore, non-SpCas9s may have other characteristics that make them more useful than SpCas9. For example, Cas9 from Staphylococcus aureus (SaCas9) is about 1 kilobase smaller than SpCas9, so it can be packaged into adeno-associated virus (AAV). Further reference may be made to Shah et al., “Protospacer recognition motifs: mixed identities and functional diversity,” RNA Biology, 10(5): 891-899 (which is incorporated herein by reference). Gasiunas used cell-free biochemical screens to identify protospacer adjacent motif (PAM) and guide RNA requirements of 79 Cas9 proteins. (Gasiunas et al., A catalogue of biochemically diverse CRISPR-Cas9 orthologs, Nature Communications 11:5512 doi.org/10.1038/s41467-020-19344-l) The authors described 7 classes of gRNA and 50 different PAM requirement.
[0132] Oh, Y. et al. describe linking reverse transcriptase to a Francisella novicida Cas9 [FnCas9(H969A)] nickase module. (Oh, Y. et al., Expansion of the prime editing modality with Cas9 from Francisella novicida, bioRxiv 2021.05.25.445577; doi.org/10.1101/2021.05.25.445577). By increasing the distance to the PAM, the FnCas9(H969A) nickase module expands the region of a reverse transcription template (RTT) following the primer binding site.
5.5. Prime Editors
[0133] “Prime editor fusion protein” describes a protein that is used in prime editing. Prime editing uses CRISPR enzyme that nicks or cuts only single strand of double stranded DNA, i.e., a
I l l nickase; and a nickase can occur either naturally or by mutation or modification of a nuclease that makes double stranded cuts. Such an enzyme can be a catalytically-impaired Cas9 endonuclease (a nickase). Such an enzyme can be a Casl2a/b, MAD7, or variant thereof. The nickase is fused to an engineered reverse transcriptase (RT). The nickase is programmed (directed) with a primeediting guide RNA (pegRNA). The skilled person in the art would appreciate that the pegRNA both specifies the target site and encodes the desired edit. Advantageously the nickase is a catalytically-impaired Cas9 endonuclease, a Cas9 nickase, that is fused to the reverse transcriptase. During genetic editing, the Cas9 nickase part of the protein is guided to the DNA target site by the pegRNA, whereby a nick or single stranded cut occurs. The reverse transcriptase domain then uses the pegRNA to template reverse transcription of the desired edit, directly polymerizing DNA onto the nicked target DNA strand. The edited DNA strand replaces the original DNA strand, creating a heteroduplex containing one edited strand and one unedited strand. Afterward, optionally, the prime editor (PE) guides resolution of the heteroduplex to favor copying the edit onto the unedited strand, completing the process (typically achieved with a nickase gRNA).
[0134] As used herein, “PEI” refers to a PE complex comprising a fusion protein comprising Cas9(H840A) and a wild type MMLV RT having the following N-terminus to C-terminus structure: [NLS]-[Cas9(H840A)]- [linker] -[MMLV_RT(wt)] + a desired atgRNA (or PEgRNA). In various embodiments, the prime editors disclosed herein is comprised of PEI.
[0135] As used herein, “PE2” refers to a PE complex comprising a fusion protein comprising Cas9(H840A) and a variant MMLV RT having the following N-terminus to C-terminus structure:
[NLS]-[Cas9(H840A)]- [linker]-[MMLV_RT(D200N)(T330P)(L603W)(T306K)(W313F)] + a desired atgRNA (or PEgRNA). In various embodiments, the prime editors disclosed herein are comprised of PE2. In various embodiments, the prime editors disclosed herein is comprised of PE2 and co-expression of MMR protein MLHldn, that is PE4.
[0136] As used herein, “PE3” refers to PE2 plus a second-strand nicking guide RNA that complexes with the PE2 and introduces a nick in the non-edited DNA strand. The induction of the second nick increases the chances of the unedited strand, rather than the edited strand, to be repaired. In various embodiments, the prime editors disclosed herein are comprised of PE3. In various embodiments, the prime editors disclosed herein are comprised of PE3 and co-expression of MMR protein MLHldn, that is PE5. [0137] As used herein, “PE3b” refers to PE3 but wherein the second-strand nicking guide RNA is designed for temporal control such that the second strand nick is not introduced until after the installation of the desired edit. This is achieved by designing a gRNA with a spacer sequence with mismatches to the unedited original allele that matches only the edited strand. Using this strategy, mismatches between the protospacer and the unedited allele should disfavor nicking by the sgRNA until after the editing event on the PAM strand takes place.
5.6. Guides for Prime Editing
[0138] Anzalone et al., 2019 (Nature 576:149) describes prime editing and a prime editing complex using a type II CRISPR and can be used herein. A prime editing complex consists of a type II CRISPR PE protein containing an RNA-guided DNA-nicking domain fused to a reverse transcriptase (RT) domain and complexed with a pegRNA. The pegRNA comprises (5’ to 3’) a spacer that is complementary to the target sequence of a genomic DNA, a nickase (e.g. Cas9) binding site, a reverse transcriptase template including editing positions, and primer binding site (PBS). The PE-pegRNA complex binds the target DNA and the CRISPR protein nicks the PAM- containing strand. The resulting 3' end of the nicked target hybridizes to the primer-binding site (PBS) of the pegRNA, then primes reverse transcription of new DNA containing the desired edit using the RT template of the pegRNA. The overall structure of the pegRNA is like that of a typical type II sgRNA with a reverse transcriptase template/primer binding site appended to the 3’ end. The structure leaves the PBS at the 3’ end of the pegRNA free to bind to the nicked strand complementary to the target which forms the primer for reverse transcription.
[0139] Guide RNAs of CRISPRs differ in overall structure. For example, while the spacer of a type II gRNA is located at the 5’ end, the spacer of a type V gRNA is located towards the 3’ end, with the CRISPR protein (e.g. Casl2a) binding region located toward the 5’ end. Accordingly, the regions of a type V pegRNA are rearranged compared to a type II pegRNA. The overall structure of the pegRNA is like that of a typical type II sgRNA with a reverse transcriptase template/primer binding site appended to the 3’ end. The pegRNA comprises (5’ to 3’) a CRISPR protein-binding region, a spacer which is complementary to the target sequence of a genomic DNA, a reverse transcriptase template including editing positions, and primer binding site (PBS).
[0140] In typical embodiments, an atgRNA comprises a reverse transcriptase template that encodes, partially or in its entirety, an integration recognition site (also referred to as an integration target recognition site) or a recombinase recognition site (also referred to as a recombinase target recognition site). The integration target recognition site, which is to be placed at a desired location in the genome or intracellular nucleic acid, is referred to as a “beacon” site or an “attachment site” or a “landing pad” or “landing site.” An integration target recognition site or recombinase target recognition site incorporated into the pegRNA is referred to as an attachment site containing guide RNA (atgRNA).
5.7. Attachment Site-Containing Guide RNA (atgRNA)
[0141] As used herein, the term “attachment site-containing guide RNA” (atgRNA) and the like refer to an extended single guide RNA (sgRNA) comprising a primer binding site (PBS), a reverse transcriptase (RT) template sequence, and wherein the RT template encodes for an integration recognition site or a recombinase recognition site that can be recognized by a recombinase, integrase, or transposase. In some embodiments, the RT template comprises a clamp sequence and an integration recognition site. As referred to herein an atgRNA may be referred to as a guide RNA. An integration recognition site or recombinase target recognition site incorporated into the pegRNA is referred to as an attachment site containing guide RNA (atgRNA).
[0142] As used herein, the term “cognate integration recognition site” or “integration cognate” or “cognate pair” refers to a first integration recognition site (e.g., any of the integration recognition sites described herein) and a second integration recognition site (e.g., any of the integration recognition sites described herein) that can be recombined. Recombination between a first integration recognition site (e.g., any of the integration recognition sites described herein) and a second recognition site (e.g., any of the integration recognition sites described herein) is mediated by functional symmetry between the two integration recognition sites and the central dinucleotide of each of the two integration recognition sites. In some cases, a first integration recognition site (e.g., any of the integration recognition sites described herein) that can be recombined with a second integration recognition site (e.g., any of the integration recognition sites described herein) are referred to as a “cognate pair.” A non-limiting example of a cognate pair include an attB site and an attP site, whereby a serine integrase mediates recombination between the attB site and the attP site.
[0143] In typical embodiments, an atgRNA comprises a reverse transcriptase template that encodes, partially or in its entirety, an integration recognition site (also referred to as an integration target recognition site) or a recombinase recognition site (also referred to as a recombinase target recognition site). The integration target recognition site, which is to be placed at a desired location in the genome or intracellular nucleic acid, is referred to as a “beacon,” a “beacon” site or an “attachment site” or a “landing pad” or “landing site.” An integration target recognition site or recombinase target recognition site incorporated into the pegRNA is referred to as an attachment site containing guide RNA (atgRNA).
[0144] During genome editing, the primer binding site allows the 3’ end of the nicked DNA strand to hybridize to the atgRNA, while the RT template serves as a template for the synthesis of edited genetic information. The atgRNA is capable for instance, without limitation, of (i) identifying the target nucleotide sequence to be edited and (ii) encoding new genetic information that replaces (or in some cases adds) the targeted sequence. In some embodiments, the atgRNA is capable of (i) identifying the target nucleotide sequence to be edited and (ii) encoding an integration site that replaces (or inserts/deletes within) the targeted sequences.
[0145] In some embodiments, the co-delivery system described herein includes a polynucleotide sequence encoding an attachment site-containing guide RNA (atgRNA) packaged in an LNP. In some embodiments, the co-delivery system described herein includes a vector comprising a polynucleotide sequence encoding an atgRNA. In some embodiments, the atgRNA comprises a domain that is capable of guiding the prime editor fusion protein to a target sequence, thereby identifying the target nucleotide sequence to be edited; and a reverse transcriptase (RT) template that comprises a first integration recognition site. In some embodiments, the atgRNA comprises a domain that is capable of guiding the prime editor fusion protein (or prime editor system) to a target sequence, thereby identifying the target nucleotide sequence to be edited; and a reverse transcriptase (RT) template that comprises at least a portion first integration recognition site.
[0146] In some embodiments, the co-delivery system described herein includes a polynucleotide sequence encoding a first attachment site-containing guide RNA (atgRNA) and a polynucleotide nucleotide sequence encoding a second attachment site-containing guide RNA (atgRNA) packaged into the same LNP. In some embodiments, the co-delivery system described herein includes a polynucleotide sequence encoding a first attachment site-containing guide RNA (atgRNA) packaged into a first LNP and a polynucleotide nucleotide sequence encoding a second attachment site-containing guide RNA (atgRNA) packaged into a second LNP.
[0147] In some embodiments, the co-delivery system described herein includes a vector comprising a polynucleotide sequence encoding a first attachment site-containing guide RNA (atgRNA), a polynucleotide sequence encoding a second atgRNA, or both. [0148] In some embodiments, the co-delivery system described herein includes a polynucleotide sequence encoding a first attachment site-containing guide RNA (atgRNA) packaged into a first LNP and a vector comprising a polynucleotide sequence encoding a second atgRNA.
[0149] In some embodiments, where the co-delivery system contains a first atgRNA and a second atgRNA, the first atgRNA and the second atgRNA are an at least first pair of atgRNAs, where the at least first pair of atgRNAs have domains that are capable of guiding the gene editor protein or prime editor fusion protein to a target sequence, the first atgRNA further includes a first RT template that comprises at least a portion of the first integration recognition site; and the second atgRNA further includes a second RT template that comprises at least a portion of the first integration recognition site, and the first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site.
[0150] In some embodiments, the first atgRNA’ s reverse transcriptase template encodes for a first single-stranded DNA sequence (i.e., a first DNA flap) that contains a complementary region to a second single-stranded DNA sequence (i.e., a second DNA flap) encoded by a second atgRNA comprising a second reverse transcriptase template. In certain embodiments, the complementary region between the first and second single- stranded DNA sequences is comprised of more than 5 consecutive bases of an integrase target recognition site. In certain embodiments, the complementary region between the first and second single- stranded DNA sequences is comprised of more than 10 consecutive bases of an integrase target recognition site. In certain embodiments, the complementary region between the first and second single-stranded DNA sequences is comprised of more than 20 consecutive bases of an integrase target recognition site. In certain embodiments, the complementary region between the first and second single-stranded DNA sequences is comprised of more than 30 consecutive bases of an integrase target recognition site. Use of two guide RNAs that are (or encode DNA that is) partially complementarity to each other and comprised of consecutive bases of an integrase target recognition site are referred to as dual, paired, annealing, complementary, or twin attachment site-containing guide RNAs (atgRNAs). In certain embodiments, use of two guide RNAs that are (or encode DNA that is) full complementarity to each other and comprised of consecutive bases of an integrase target recognition site are referred to as dual, paired, annealing, complementary, or twin attachment sitecontaining guide RNAs (atgRNAs). [0151] In some embodiments, upon introducing the nucleic acid construct into a cell, the first atgRNA incorporates the first integration recognition site into the cell’s genome at the target sequence.
[0152] Table 9 includes atgRNAs, sgRNAs and nicking guides that can be used herein. Spacers are labeled in capital font (SPACER), RT regions in bold capital (RT REGION), AttB sites in bold lower case (attB site), and PBS in capital italics (PBS). Unless otherwise denoted, the AttB is for Bxbl.
5.8. Integrases/Recombinases and Integration/Recombination Sites
[0153] In typical embodiments, the co-delivery system described herein contains an integrase and/or a recombinase. In some embodiments, the co-delivery system includes an integrase and/or a recombinase packaged in a LNP. In one embodiment, the co-delivery system includes a polynucleotide encoding an integrase and/or a recombinase. In some embodiments, the co-delivery system includes an integrase or a recombinase packaged in a vector (e.g., a viral vector). In some embodiments, the co-delivery system includes at least a first integrase (e.g., a first integrase and a second integrase) and/or at least a first recombinase (e.g., a first recombinase and a second recombinase).
[0154] In some embodiments, the integration enzyme (e.g., the integrase or recombinase) is selected from the group consisting of Dre, Vika, Bxbl, cpC31, RDF, cpBTl, Rl, R2, R3, R4, R5, TP901-1, Al 18, (pFCl, (pCl, MR11, TGI, cp370.1, W , BL3, SPBc, K38, Peaches, Veracruz, Rebeuca, Theia, Benedict, KSSJEB, PattyP, Doom, Scowl, Lockley, Switzer, Bob3, Troube, Abrogate, Anglerfish, Sarfire, SkiPole, Concept!!, Museum, Severus, Airmid, Benedict, Hinder, ICleared, Sheen, Mundrea, BxZ2, cpRV, retrotransposases encoded by a Tcl/mariner family member including but not limited to retrotransposases encoded by LI, Tol2, Tel, Tc3, Himar 1 (isolated from the horn fly, Haematobia irritans), Mosl (Mosaic element of Drosophila mauritiana), and Minos, and any mutants thereof. As can be used herein, Xu et al describes methods for evaluating integrase activity in E. coli and mammalian cells and confirmed at least R4, (pC31, (pBTl, Bxbl, SPBc, TP901-1 and WP integrases to be active on substrates integrated into the genome of HT1080 cells (Xu et al., 2013, Accuracy and efficiency define Bxbl integrase as the best of fifteen candidate serine recombinases for the integration of DNA into the human genome. BMC Biotechnol. 2013 Oct 20;13:87. doi: 10.1186/1472-6750-13-87). Durrant describes new large serine recombinases (LSRs) divided into three classes distinguished from one another by efficiency and specificity, including landing pad LSRs which outperform wild-type Bxbl in episomal and chromosomal integration efficiency, LSRs that achieve both efficient and sitespecific integration without a landing pad, and multi-targeting LSRs with minimal site- specificity. Additionally, embodiments can include any serine recombinase such as BceINT, SSCINT, SACINT, and INT10 (see lonnidi et al., 2021; Drag-and-drop genome insertion without DNA cleavage with CRISPR directed integrases. bioRxiv 2021. ! 1.0 ! .466786, doi.org/10.1101/2021.11.01.466786). In some embodiments, the integration site can be selected from an attB site, an attP site, an attL site, an attR site, a lox71 site a Vox site, or a ERT site.
[0155] It will be appreciated that desired activity of integrases, transposases and the like can depend on nuclear localization. In certain embodiments, prokaryotic enzymes are adapted to modulate nuclear localization. In certain embodiments, eukaryotic or vertebrate enzymes are adapted to modulate nuclear localization. In certain embodiments, the disclosure provides fusion or hybrid proteins. Such modulation can comprise addition or removal of one or more nuclear localization signal (NLS) and/or addition or removal of one or more nuclear export signal (NES). Xu et al compared derivatives of fourteen serine integrases that either possess or lack a nuclear localization signal (NLS) to conclude that certain integrases benefit from addition of an NLS whereas others are transported efficiently without addition, and a major determinant of activity in yeast and vertebrate cells is avoidance of toxicity. (Xu et al., 2016, Comparison and optimization of ten phage encoded serine integrases for genome engineering in Saccharomyces cerevisiae. BMC Biotechnol. 2016 Eeb 9; 16:13. doi: 10.1186/s 12896-016-0241-5). Ramakrishnan et al. systematically studied the effect of different NES mutants developed from mariner-like elements (MLEs) on transposase localization and activity and concluded that nuclear export provides a means of controlling transposition activity and maintaining genome integrity. (Ramakrishnan et al. Nuclear export signal (NES) of transposases affects the transposition activity of mariner-like elements Ppmarl and. Ppmar2 of moso bamboo. Mob DNA. 2019 Aug 19; 10:35. doi:10.1186/sl3100-019-0179-y). The methods and constructs are used to modulate nuclear localization of system components of the disclosure.
[0157] Sequences of insertion sites (i.e., recognition target sites) suitable for use in embodiments of the disclosure are presented below (Table 11). FIGs. 14A-14E shows analysis of effect of variant AttP sites on integration efficiency.
5.9. Co-delivery of gene editor and donor DNA template
[0158] In some embodiments, provided herein are methods of delivering (e.g., co-delivery or dual delivery) a system capable of site- specifically integrating a template polynucleotide into the genome of a cell, where the methods includes delivering to a (i) gene editor construct and a (ii) template polynucleotide, and (iii) at least a first attachment site-containing guide (atgRNA).
[0159] In some embodiments, provided herein are methods for delivering a system capable of site- specifically integrating a template polynucleotide into the genome of a cell, where the method includes: delivering a lipid nanoparticle (LNP) comprising a gene editor polynucleotide (e.g., a gene editor polynucleotide construct) and a vector comprising a template polynucleotide and at least a first attachment site-containing guide RNA (atgRNA). In some embodiments, the first atgRNA comprises (i) a domain that is capable of guiding the prime editor system to a target sequence; and (ii) a reverse transcriptase (RT) template that comprises at least a portion of a first integration recognition site. In some embodiments, where the vector comprises a polynucleotide encoding a first atgRNA, the RT template comprises the entirety of the first integration recognition site. In some embodiments, where the vector comprises a polynucleotide encoding a first atgRNA, the vector also includes a sequence encoding a nicking guide RNA (ngRNA). [0160] In some embodiments, provided herein are methods for delivering a system capable of site- specifically integrating a template polynucleotide into the genome of a cell, where the method includes: delivering a lipid nanoparticle (LNP) comprising a gene editor polynucleotide (e.g., a gene editor polynucleotide construct) and a vector comprising a template polynucleotide and a first attachment site-containing guide RNA (atgRNA) and a second attachment site-containing guide RNA (atgRNA). In some embodiments, the first atgRNA and the second atgRNA are an at least first pair of atgRNAs, wherein the at least first pair of atgRNAs have domains that are capable of guiding the prime editor system to a target sequence, the first atgRNA further includes a first RT template that comprises at least a portion of the at least first integration recognition site; and the second atgRNA further includes a second RT template that comprises at least a portion of the first integration recognition site, and the first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site. In such embodiments, the first atgRNA and second atgRNA include at least a 6bp overlap (e.g., 6bp of complementarity).
[0161] In some embodiments, provided herein are methods for delivering a system capable of site- specifically integrating a template polynucleotide into the genome of a cell, where the method includes: delivering into a cell a lipid nanoparticle (LNP) comprising: (i) a gene editor polynucleotide (e.g., a gene editor polynucleotide construct), and (ii) a first attachment sitecontaining guide RNA (atgRNA); and a vector comprising: (i) a template polynucleotide, and (ii) a second atgRNA. In some embodiments, the first atgRNA and the second atgRNA are an at least first pair of atgRNAs, wherein the at least first pair of atgRNAs have domains that are capable of guiding the prime editor system to a target sequence, the first atgRNA further includes a first RT template that comprises at least a portion of the a first integration recognition site; and the second atgRNA further includes a second RT template that comprises at least a portion of the first integration recognition site, and the first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site. In such embodiments, the first atgRNA and second atgRNA include at least a 6bp overlap (e.g., 6bp of complementarity).
[0162] In some embodiments, provided herein are methods for delivering a system capable of site- specifically integrating a template polynucleotide into the genome of a cell, where the method includes delivering: a lipid nanoparticle (LNP) comprising: (i) a gene editor polynucleotide (e.g., a gene editor polynucleotide construct), (ii) a first attachment site-containing guide RNA (atgRNA), and (iii) a second atgRNA; and a vector comprising (i) a template polynucleotide. In some embodiments, the first atgRNA and the second atgRNA are an at least first pair of atgRNAs, wherein the at least first pair of atgRNAs have domains that are capable of guiding the prime editor system to a target sequence, the first atgRNA further includes a first RT template that comprises at least a portion of the at least first integration recognition site; and the second atgRNA further includes a second RT template that comprises at least a portion of the at least first integration recognition site, and the first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site. In such embodiments, the first atgRNA and second atgRNA include at least a 6bp overlap (e.g., 6bp of complementarity).
[0163] In some embodiments, provided herein are methods for delivering a system capable of site- specifically integrating a template polynucleotide into the genome of a cell, where the method includes delivering: a lipid nanoparticle (LNP) comprising: (i) a gene editor polynucleotide (e.g., a gene editor polynucleotide construct) and (ii) a first attachment site-containing guide RNA (atgRNA); and a vector comprising: (i) a template polynucleotide, and (ii) a nicking atgRNA. In some embodiments, the first atgRNA comprises (i) a domain that is capable of guiding the prime editor system to a target sequence; and (ii) a reverse transcriptase (RT) template that comprises at least a portion of a first integration recognition site. In some embodiments, where the vector comprises a polynucleotide encoding a first atgRNA, the RT template comprises the entirety of the first integration recognition site.
[0164] In some embodiments, where the method includes delivering an LNP and a first vector, the LNP and the first vector are delivered at least 1 day, 2 days, 3 days, 4 days, 5 days, 6 days, 7 days, 2 weeks, 3 weeks, 4 weeks, 5 weeks, 6 weeks, 7 weeks, or 8 weeks apart. In some embodiments, where the method includes delivering an LNP and a second vector, the LNP and the second vector are delivered at least 1 day, 2 days, 3 days, 4 days, 5 days, 6 days, 7 days, 2 weeks, 3 weeks, 4 weeks, 5 weeks, 6 weeks, 7 weeks, or 8 weeks apart.
[0165] In some embodiments, provided herein are systems capable of site-specifically integrating a template polynucleotide into the genome of a cell, the system comprising: a lipid nanoparticle (LNP) comprising a gene editor polynucleotide (e.g., a gene editor polynucleotide construct) and a vector comprising a template polynucleotide and at least a first attachment site-containing guide RNA (atgRNA). In some embodiments, the first atgRNA comprises (i) a domain that is capable of guiding the prime editor system to a target sequence; and (ii) a reverse transcriptase (RT) template that comprises at least a portion of a first integration recognition site. In some embodiments, where the vector comprises a polynucleotide encoding a first atgRNA, the RT template comprises the entirety of the first integration recognition site. In some embodiments, where the vector comprises a polynucleotide encoding a first atgRNA, the vector also includes a sequence encoding a nicking guide RNA (ngRNA).
[0166] In some embodiments, provided herein are systems capable of site-specifically integrating a template polynucleotide into the genome of a cell, the system comprising: a lipid nanoparticle (LNP) comprising a gene editor polynucleotide (e.g., a gene editor polynucleotide construct) and a vector comprising a template polynucleotide and a first attachment site-containing guide RNA (atgRNA) and a second attachment site-containing guide RNA (atgRNA). In some embodiments, the first atgRNA and the second atgRNA are an at least first pair of atgRNAs, wherein the at least first pair of atgRNAs have domains that are capable of guiding the prime editor system to a target sequence, the first atgRNA further includes a first RT template that comprises at least a portion of the a first integration recognition site; and the second atgRNA further includes a second RT template that comprises at least a portion of the first integration recognition site, and the first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site. In such embodiments, the first atgRNA and second atgRNA include at least a 6bp overlap.
[0167] In some embodiments, provided herein are systems capable of site-specifically integrating a template polynucleotide into the genome of a cell, the system comprising: a lipid nanoparticle (LNP) comprising: (i) a gene editor polynucleotide (e.g., a gene editor polynucleotide construct), and (ii) a first attachment site-containing guide RNA (atgRNA); and a vector comprising: (i) a template polynucleotide, and (ii) a second atgRNA. In some embodiments, the first atgRNA and the second atgRNA are an at least first pair of atgRNAs, wherein the at least first pair of atgRNAs have domains that are capable of guiding the prime editor system to a target sequence, the first atgRNA further includes a first RT template that comprises at least a portion of the a first integration recognition site; and the second atgRNA further includes a second RT template that comprises at least a portion of the first integration recognition site, and the first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site. In such embodiments, the first atgRNA and second atgRNA include at least a 6bp overlap.
[0168] In some embodiments, provided herein are systems capable of site-specifically integrating a template polynucleotide into the genome of a cell, the system comprising: co-delivering: a lipid nanoparticle (LNP) comprising: (i) a gene editor polynucleotide (e.g., a gene editor polynucleotide construct), (ii) a first attachment site-containing guide RNA (atgRNA), and (iii) a second atgRNA; and a vector comprising (i) a template polynucleotide. In some embodiments, the first atgRNA and the second atgRNA are an at least first pair of atgRNAs, wherein the at least first pair of atgRNAs have domains that are capable of guiding the prime editor system to a target sequence, the first atgRNA further includes a first RT template that comprises at least a portion of the a first integration recognition site; and the second atgRNA further includes a second RT template that comprises at least a portion of the first integration recognition site, and the first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site. In such embodiments, the first atgRNA and second atgRNA include at least a 6bp overlap.
[0169] In some embodiments, provided herein are systems capable of site-specifically integrating a template polynucleotide into the genome of a cell, the system comprising: a lipid nanoparticle (LNP) comprising: (i) a gene editor polynucleotide (e.g., a gene editor polynucleotide construct) and (ii) a first attachment site-containing guide RNA (atgRNA); and a vector comprising: (i) a template polynucleotide, and (ii) a nicking atgRNA. In some embodiments, the first atgRNA comprises (i) a domain that is capable of guiding the prime editor system to a target sequence; and (ii) a reverse transcriptase (RT) template that comprises at least a portion of a first integration recognition site. In some embodiments, where the vector comprises a polynucleotide encoding a first atgRNA, the RT template comprises the entirety of the first integration recognition site.
[0170] In typical embodiments, the LNP comprising a gene editor polynucleotide construct is capable delivering to a cell cytoplasm the gene editor polynucleotide construct. In some embodiments, the LNP comprising a gene editor polynucleotide construct is capable delivering to a cell nucleus the gene editor polynucleotide construct. In some embodiments, the LNP comprises a gene editor protein and associated guide nucleic acids. In some embodiments, the LNP comprises a gene editor protein and associated guide nucleic acids that are capable of localizing to cell nucleus.
[0171] In some embodiments, a gene editor polynucleotide construct is delivered to a cell by a fusosome. In some embodiments, a gene editor polynucleotide construct is delivered to a cell cytoplasm by a fusosome. In some embodiments, the fusosome comprises a gene editor protein and associated guide nucleic acids.
[0172] In some embodiments, a gene editor polynucleotide construct is delivered to a cell by an exosome. In some embodiments, a gene editor polynucleotide construct is delivered to a cell cytoplasm by an exosome. In some embodiments, the exosome comprises a gene editor protein and associated guide nucleic acids. [0173] In some embodiments, the prime editor or Gene Writer protein fusion, either of which may have a fused/linked integrase, is incorporated (i.e., packaged) into LNP as protein. Further, associated atgRNA and optional ngRNAs may be co-packaged with gene editor proteins in LNP.
[0174] In some embodiments, the gene editor polynucleotide construct comprises (a) a polynucleotide sequence encoding a prime editor fusion protein or a Gene Writer™ protein, (b) a polynucleotide sequence encoding an attachment site-containing guide RNA (atgRNA), (c) optionally, a polynucleotide sequence encoding a nickase guide RNA (ngRNA), (d) a polynucleotide sequence encoding an integrase, (e) and optionally, a polynucleotide sequence encoding a recombinase.
[0175] In some embodiments, the prime editor or Gene Writer protein fusion, either of which may have a fused/linked integrase, is expressed as a split construct. In typical embodiments, the split construct in reconstituted in a cell. In some embodiments, the split construct can be fused or ligated via intein protein splicing. In some embodiments, the split construct can be reconstituted via protein-protein inter-molecular bonding and/or interactions. In some embodiments, the split construct can be reconstituted via chemical, biological, or environmental induced oligomerization. In certain embodiments, the split construct can be adapted into one or more nucleic acid constructs described herein.
5.9.1. Gene Editor Polynucleotide
[0176] In some embodiments, the systems described include a gene editor polynucleotide that is delivered to a cell using the methods described herein. In some embodiments, the gene editor polynucleotide is delivered as a polynucleotide (e.g., an mRNA). In some embodiments, the gene editor polynucleotide is delivered as a protein. In some embodiments, the gene editor polynucleotide or protein is packaged, and thereby vectorized, within a lipid nanoparticle (LNP). In some embodiments, the gene editor polynucleotide or protein is packaged in a LNP and is codelivered with a template polynucleotide (i.e., nucleic acid “cargo” or nucleic acid “payload”) packaged into a separate vector (e.g., a viral vector (e.g., an AAV or adenovirus)) or a second lipid nanoparticle (LNP).
[0177] In some embodiments, the gene editor polynucleotide is delivered to the cells as a polynucleotide. For example, the gene editor polynucleotide is delivered to the cells as an mRNA encoding the gene editor polynucleotide (e.g., the gene editor protein or the prime editor system). In some embodiments, the mRNA comprises one or more modified uridines. In some embodiments, the mRNA comprises a sequence where each of the uridines is a modified uridine. In some embodiments, the mRNA is uridine depleted. In some embodiments, the mRNA encoding the nickase comprises one or more modified uridines. In some embodiments, the mRNA encoding the reverse transcriptase comprises one or more modified uridines. In some embodiments, the mRNA encoding the nickase comprises one or more modified uridines, and the mRNA encoding the reverse transcriptase comprises one or more modified uridines. In some embodiments, where the integrase is encoded in an mRNA, the mRNA comprises modified uridines. In some embodiments, a modified uridine is a Nl-Methylpseudouridine-5’ -Triphosphate. In some embodiments, a modified uridine is a pseudouridine. In some embodiments, the mRNA comprises a 5’ cap. In some embodiments, the 5’ cap comprises a molecular formula of C32H43N15O24P4 (free acid).
[0178] In some embodiments, the gene editor polynucleotide (e.g., a gene editor polynucleotide construct) comprises a polynucleotide sequence encoding a primer editor system (e.g., any of the prime editor systems described herein). In some embodiments, the prime editor system comprises a nucleotide sequence encoding a nickase (e.g., any of the Cas proteins or variants thereof (e.g., nickases) and nickases described herein, see Tables 4-8) and a nucleotide sequence encoding a reverse transcriptase (e.g., any of the reverse transcriptases described herein). In some embodiments, the nucleotide sequence encoding the nickase and the nucleotide sequence encoding the reverse transcriptase are positioned in the construct such that when expressed the nickase is linked to the reverse transcriptase. In some embodiments, the nickase is linked to the reverse transcriptase by in-frame fusion. In some embodiments, the nickase is linked to the reverse transcriptase by a linker. In some embodiments, the linker is a peptide fused in-frame between the nickase and reverse transcriptase.
[0179] In some embodiments, the gene editor polynucleotide (e.g., a gene editor polynucleotide construct) further comprises a polynucleotide sequence encoding at least a first integrase (e.g., any of the integrases described herein, e.g., as described in Table 10 and also in Yamall et al., Nat. Biotechnol., 2022, doi.org/10.1038/s41587-022-01527-4 and Durrant et al., Nat. Biotechnol., 2022, doi.org/10.1038/s41587-022-01494-w, each of which are herein incorporated by reference in their entireties). In some embodiments, the linked nickase-reverse transcriptase are further linked to the first integrase.
[0180] In some embodiments, the gene editor polynucleotide construct further comprises a polynucleotide sequence encoding at least a first recombinase (e.g., any of the recombinases described herein). 5.9.2. Split Nickase and Reverse Transcriptase
[0181] Also provided herein are prime editor systems where the nucleotide sequence encoding the nickase and the nucleotide sequence encoding the reverse transcriptase are positioned in a construct such that when expressed the nickase is not linked to the reverse transcriptase. In a nonlimiting example, a sequence encoding a self-cleaving peptide (e.g., a P2A) is positioned between the nucleotide sequence encoding the nickase and the nucleotide sequence encoding the reverse transcriptase such that when expressed the nickase is not linked to the reverse transcriptase.
[0182] Also provided herein are prime editor systems where the nucleotide sequence encoding the nickase and the nucleotide sequence encoding the reverse transcriptase are on separate polynucleotides. In some embodiments where the prime editor system includes separate polynucleotides encoding the nickase and the reverse transcriptase, the nickase and/or the reverse transcriptase can be engineered such that when expressed they are linked to a binding domain, where the binding domain is part of a binding pair that enables recruitment of the reverse transcriptase to the genomic location of the nickase (e.g., nCas9). A binding pair can be any pair of molecules wherein upon forming the binding pair the binding domains enable recruitment of the reverse transcriptase to the genomic location of the nickase. Once in sufficient proximity with each other the binding domains can dimerize. This dimerization (or other type of interaction) brings the nickase and RT into proximity with each other, thereby enhancing the reverse transcription of the RT template in the atgRNA.
[0183] Non-limiting examples of binding pairs that can aid recruitment of the RT to the genomic location of the nickase include without limitation: dimerizing leucine zippers, an antigen and a corresponding antigen binding domains (e.g., antibody), RNA aptamers and corresponding RNA aptamer binding proteins, affinity tags and corresponding affinity tag binding proteins.
[0184] In some embodiments, dimerizing leucine zippers that can serve as a binding pair includes two binding domains: a Leucine Zip LZ1 domain having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 568 and a Leucine Zip LZ2 domain having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 569. In one embodiment, the Leucine Zip LZ1 domain is positioned either N- terminally or C-terminally to the nickase and the Leucine Zip LZ2 is positioned either N-terminally or C-terminally to the RT. In one embodiment, the Leucine Zip LZ1 domain is positioned either N-terminally or C-terminally to the RT and the Leucine Zip LZ2 domain is positioned either N- terminally or C-terminally to the Cas9.
[0185] In one embodiment, the system and methods described herein include a construct that encodes a polypeptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 576 (a Leucine Zip LZ1 domain positioned C-terminally to the nickase), and a construct that encodes a polypeptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 577 (a Leucine Zip LZ2 domain positioned N-terminally to the RT).
[0186] In one embodiment, the system and methods described herein include a construct that encodes a polypeptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 578 (a Leucine Zip LZ1 domain positioned N-terminally to the nickase), and a construct that encodes a polypeptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 577 (a Leucine Zip LZ2 domain positioned N-terminally to the RT).
[0187] In some embodiments, dimerizing leucine zippers that can serve as a binding pair includes two binding domains: a EE1234L peptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 570 and a RR1234L peptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 571. In one embodiment, the EE1234L peptide is positioned either N-terminally or C-terminally to the nickase and the RR1234L peptide is positioned either N-terminally or C-terminally to the RT. In one embodiment, the EE1234L peptide is positioned either N-terminally or C-terminally to the RT and the RR1234L peptide is positioned either N-terminally or C-terminally to the Cas9. [0188] In one embodiment, the system and methods described herein include a construct that encodes a polypeptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 579 (a EE1234L peptide positioned N-terminally to the nickase), and a construct that encodes a polypeptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 580 (a RR1234L peptide positioned N-terminally to the RT).
[0189] In some embodiments, an affinity tag is a SpyTag and the corresponding affinity tag binding protein is SpyCatcher, where upon binding of the SpyTag with the SpyCatcher the fusion protein is a covalently stabilized multi-protein complex. In one embodiment, the affinity tag (e.g., SpyTag) is positioned either N-terminally or C-terminally to the nickase and the affinity tag binding protein (e.g,. SpyCatcher) is positioned either N-terminally or C-terminally to the RT. In one embodiment, the affinity tag (e.g., SpyTag) is positioned either N-terminally or C-terminally to the RT and the affinity tag binding protein (e.g,. SpyCatcher) is positioned either N-terminally or C-terminally to the nickase.
[0190] In some embodiments, an antigen and corresponding antigen binding domain includes an antigen such as a peptide present within a tag or a tag itself and an antigen binding domain that binds specifically to the antigen. In one embodiments, an antigen is a GCN4 peptide within a SunTag and the antigen binding domain is an anti-GCN4 scFv. In one embodiment, the SunTag (GCN4 peptide) is positioned either N-terminally or C-terminally to the nickase and the anti-GCN4 antigen binding domain is positioned either N-terminally or C-terminally to the RT. In one embodiment, the SunTag (GCN4 peptide) is positioned either N-terminally or C-terminally to the RT and the anti-GCN4 antigen binding domain is positioned either N-terminally or C-terminally to the nickase.
[0191] In one embodiment, the system and methods described herein include a construct that encodes a polypeptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 574 (SunTag (GCN4 peptide) positioned C-terminally to the nickase), and a construct that encodes a polypeptide having a sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 575 (anti-GCN4 antigen binding domain positioned N-terminally to the RT).
[0192] In some embodiments, provided herein are prime editor systems where a construct includes a nucleotide sequence encoding a self-cleaving peptide (e.g., a P2A) positioned between the nucleotide sequence encoding the nickase and the nucleotide sequence encoding the RT. In such cases, the position of the nucleotide sequence encoding the nickase, the nucleotide sequence encoding the self-cleaving peptide (e.g., a P2A), and the nucleotide sequence encoding the RT is such that when expressed the nickase is not linked to the reverse transcriptase. In one embodiment, the construct includes the nucleotide sequences positioned 5 ’-3’: nucleotide sequence encoding the nickase, nucleotide sequence encoding the self-cleaving peptide (e.g., a P2A), and nucleotide sequence encoding the RT. In one embodiment, the construct includes the nucleotide sequences positioned 5 ’-3’: nucleotide sequence encoding the RT, nucleotide sequence encoding the selfcleaving peptide (e.g., a P2A), and nucleotide sequence encoding the nickase.
[0193] In one embodiment, the construct that includes a nucleotide sequence encoding a selfcleaving peptide (e.g., a P2A) positioned between the nucleotide sequence encoding the nickase and the nucleotide sequence encoding the RT has an amino acid sequence with at least 80%, 85%, 90%, 95%, or 99% sequence identity to a sequence of SEQ ID NO: 581 or 582.
5.9.3. Vector
[0194] In some embodiments, the systems and methods described herein include a vector that is capable of co-delivering a template polynucleotide, one or more attachment site-containing gRNA, one or more integrases, one or more recombinases, a gene editor polynucleotide, one or more integration recognition sites, one or more recombinase recognition sites, or a combination thereof.
[0195] Non-limiting examples of vectors that can be used in the methods or systems described herein include the vectors described in FIGs. 3-6.
5.9.3.1 AtgRNA and/or ngRNA
[0196] In some embodiments, the vector includes a polynucleotide sequence encoding an attachment site-containing guide RNA (atgRNA). In such embodiments, the polynucleotide sequence encoding the attachment site-containing guide RNA (atgRNA) is operably linked to a regulatory element (e.g., a U6 promoter) that is capable of driving expression of the atgRNA. In such embodiments, the atgRNA comprises (i) a domain that is capable of guiding the prime editor system to a target sequence; and (ii) a reverse transcriptase (RT) template that comprises at least a portion of a first integration recognition site. In some embodiments, where the system, and thereby the vector, include a polynucleotide encoding only a first atgRNA, the RT template comprises the entirety of the first integration recognition site. In such embodiments, the vector or the LNP includes a polynucleotide sequence encoding a nicking gRNA.
[0197] In some embodiments, the vector includes a polynucleotide sequence encoding a first attachment site-containing guide RNA (atgRNA) and a polynucleotide sequence encoding a second attachment site-containing guide RNA (atgRNA). In such embodiments, the first atgRNA and the second atgRNA are an at least first pair of atgRNAs, wherein the at least first pair of atgRNAs have domains that are capable of guiding the prime editor system to a target sequence, the first atgRNA further includes a first RT template that comprises at least a portion of the a first integration recognition site; the second atgRNA further includes a second RT template that comprises at least a portion of the first integration recognition site, and the first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site. In such embodiments, the first atgRNA and second atgRNA include at least a 6bp overlap.
5.9.3.2 Template Polynucleotide
[0198] In typical embodiments, the vector includes a template polynucleotide and a sequence that is an integration cognate of an integration recognition site site-specifically incorporated into the genome of a cell. For example, the vector includes a template polynucleotide and a second integration recognition site that is a cognate pair with the first integration recognition site site- specifically incorporated into the genome of the cell. In such embodiments, the sequence that is an integration cognate (e.g., a second integration recognition site) enables integration of the template polynucleotide or portion thereof when contacted with an integrase and the site-specifically incorporated first integration recognition site.
[0199] In typical embodiments, the vector comprising a template polynucleotide is a recombinant adenovirus, a helper dependent adenovirus, an AAV, a lentivirus, an HSV, an annelovirus, a retrovirus, a Doggybone™ DNA (dbDNA), a minicircle, a plasmid, a miniDNA, an exosome, a fusosome, or an nanoplasmid. In preferred embodiments, the vector is capable of localizing to the nucleus.
[0200] In certain embodiments, the template polynucleotide is delivered to the cytoplasm and localizes to the nucleus. In certain embodiments, the template polynucleotide is delivered to the cytoplasm by LNP. In certain embodiments, the donor template polynucleotide construct comprises a recognition sequence that is recognized by a DNA binding protein (DNA binding domain) or a transcription factor binding domain. In certain embodiments, the donor template polynucleotide construct is delivered to the nucleus by an integrase or recombinase.
[0201] In certain embodiments, the template polynucleotide is delivered to the mitochondria. In certain embodiments, the donor template polynucleotide construct comprises a mitochondria targeting sequence.
[0202] In certain embodiments, the vector comprising a template polynucleotide is AAV. In some embodiments, the AAV contains a 5’ inverted terminal repeat (ITR). In some embodiments, the AAV contains a 3’ inverted terminal repeat (ITR). In some embodiments, the AAV contains a 5’ and a 3’ ITR. In some embodiments, the 5’ and 3’ ITR are not derived from the same serotype of virus. In some embodiments, the ITRs are derived from adenovirus, AAV2, and/or AAV5.
[0203] In certain embodiments, the vector comprising a template polynucleotide is single stranded AAV (ssAAV). In certain embodiments, the vector comprising a donor template polynucleotide construct is self-complementary AAV (sc AAV).
[0204] In some embodiments, a vector comprises an attachment site-containing guideRNA (atgRNA), a nicking-guideRNA (ngRNA), and template polynucleotide. In typical embodiments, the vector comprising an attachment site-containing guideRNA (atgRNA), a nicking-guideRNA (ngRNA), and template polynucleotide is recombinant adenovirus, helper dependent adenovirus, AAV, lentivirus, HSV, annelovirus, retrovirus, Doggybone™ DNA (dbDNA), minicircle, plasmid, miniDNA, exosome, fusosome, or nanoplasmid. In preferred embodiments, the vector is capable of localizing to the nucleus. In typical embodiments, the attachment site-containing guideRNA (atgRNA) sequence and the nicking-guideRNA (ngRNA) sequence contain a terminal poly dT.
[0205] In some embodiments, a vector comprises an attachment site-containing guideRNA (atgRNA), and donor template. In typical embodiments, the vector comprising an attachment sitecontaining guideRNA (atgRNA) and donor template is recombinant adenovirus, helper dependent adenovirus, AAV, lentivirus, HSV, annelovirus, retrovirus, Doggybone™ DNA (dbDNA), minicircle, plasmid, miniDNA, exosome, fusosome, or nanoplasmid. In preferred embodiments, the vector is capable of localizing to the nucleus. In typical embodiments, the attachment sitecontaining guideRNA (atgRNA) sequence contain a terminal poly dT. [0206] In typical embodiments, the template polynucleotide is capable of being integrated into a genomic locus that contains an integrase target recognition site or a recombinase target recognition site.
[0207] In certain embodiments, the template polynucleotide comprises at least one of the following: a gene, a gene fragment, an expression cassette, a logic gate system, or any combination thereof. In some embodiments, the template polynucleotide comprises at least one intron or exon.
[0208] In typical embodiments, the template polynucleotide further comprises at least one integrase target recognition site or a recombinase target integrase site. In certain embodiments, at least one integrase target recognition site or a recombinase target integrase site is placed within the donor template vector inverted terminal repeat.
5.9.3.3 Integrase- or recombinase-mediated self-circularization of a subsequence of a vector delivered as part of the co-delivery system
[0209] In some embodiments, the delivery system (e.g., co-delivery system) includes a vector having a sub-sequence that is capable of self-circularizing to form a self-circular nucleic acid. In some embodiments, the vector comprises a physical portion or region of the vector that is capable of self-circularizing to form a circular construct. As used herein, the term “sub-sequence” refers to a portion of the vector that is capable of self-circularizing, where the sub-sequence is flanked by integration recognition sites or recombinase recognition sites positioned to enable selfcircularization. As used herein, the term “self-circular nucleic acid” refers to a double- stranded, circular nucleic acid construct produced as a result of recombination of a cognate pair of integrase or recombinase recognition sites present on the vector. Recombination occurs when the vector is contacted with an integrase or a recombinase under conditions that allow for recombination of the cognate pair of integrase or recombinase recognition sites.
[0210] In some embodiments, the sub-sequence of the vector includes a first recombinase recognition site and a second recombinase recognition site, wherein the first and second recombinase recognition sites are capable of being recombined by a recombinase. In some embodiments, the sub-sequence of the vector includes a first recombinase recognition site, a second recombinase recognition site, and a second integration recognition site (e.g., the second integration recognition site is a cognate pair of the first integration recognition site), where the first and second recombinase recognition sites flank the integration recognition site. In such cases, the first recombinase recognition site, the second recombinase recognition, and a recombinase enable the self-circularizing and formation of the circular construct.
[0211] In some embodiments, the sub-sequence of the vector includes a third integration recognition site and a fourth integration recognition site, wherein the third and fourth integration recognition sites are a cognate pair. In some embodiments, the subsequence of the vector includes the second integration recognition site, the third integration recognition site, the fourth integration recognition site, where the third and fourth integration recognition sites flank the second integration recognition site (where the second integration recognition site is a cognate pair of the first integration recognition site). In such cases, the third integration recognition site, the fourth integration recognition site, and an integrase enable self -circularization and formation of the circular construct. In such cases, the third integration recognition site and/or the fourth integration recognition sites cannot recombine with the first integration recognition site and/or the second integration recognition site due, in part, to having different central dinucleotides than the first and second integration recognition sites.
[0212] In some embodiments where the subsequence includes three or more integration recognition sites, each integration recognition site or each pair of integration recognition is capable of being recognized by a different integrase. In some embodiments where the subsequence includes three or more integration recognition sites, each integration recognition site or each pair of integration recognition comprises a different central dinucleotide.
[0213] In some embodiments, self-circularizing is mediated at the integration recognition sites or recombinase recognition sites. In some embodiments, the self-circularizing is mediated by an integrase or a recombinase.
[0214] In some embodiments, upon introducing the vector into a cell and after selfcircularizing to form the self-circular nucleic acid, the self-circular nucleic acid comprising the second integration recognition site is capable of being integrated into the cell’s genome at the target sequence that contains the first integration recognition site.
[0215] In some embodiments, following self-circularization, the self-circular nucleic acid comprises one or more additional integration recognition sites that enable integration of an additional nucleic acid cargo. In such cases, the additional nucleic acid cargo includes a sequence that is a cognate pair with one or more of the additional integration recognition sites in the selfcircular nucleic acid. For example, integration of the self-circular nucleic acid into the genome of a cell results in integration of the one or more additional integration recognition sites into the genome along with the nucleic acid cargo. The integrated one or more additional integration recognition sites serve as an integration recognition site (beacon) for placing the additional nucleic acid cargo. Upon contacting the cell harboring the integrated nucleic acid cargo and the one or more additional integration recognition sites with an integrase and the second additional nucleic acid cargo that includes a sequence that is an integration cognate to the one or more additional integration recognition sites the additional nucleic acid cargo is integrated into the cell’s genome.
[0216] In typical embodiments, the self-circularized nucleic acid comprises a DNA cargo, embodiments, the DNA cargo is a gene or gene fragment. In some embodiments the DNA cargo is an expression cassette. In some embodiments, the DNA cargo is a logic gate or logic gate system. The logic gate or logic gate system may be DNA based, RNA based, protein based, or a mix of DNA, RNA, and protein. In some embodiments, the nucleic acid cargo is a genetic, protein, or peptide tag and/or barcode.
5.9.3.4 A second vector
[0217] In some embodiments, the system or methods described herein include a second vector. In some embodiments, where the gene editor polynucleotide encodes a prime editor system comprising a nickase (e.g., any of the Cas proteins or variants thereof (e.g., nickases) and nickases described herein, see Tables 4-8) and a reverse transcriptase (e.g., any of the reverse transcriptase described herein), the second vector comprises a polynucleotide sequence encoding an integrase (e.g., any of the integrases described herein, e.g., as described in Table 10 and also in Yamall et al., Nat. Biotechnol., 2022, doi.org/10.1038/s41587-022-01527-4 and Durrant et al., Nat. Biotechnol., 2022, doi.org/10.1038/s41587-022-01494-w, each of which are herein incorporated by reference in their entireties).
[0218] In some embodiments, where the gene editor polynucleotide encodes a prime editor system comprising a nickase and a reverse transcriptase, the second vector comprises a polynucleotide sequence encoding at least a first recombinase. In some embodiments, where the gene editor polynucleotide encodes a prime editor system comprising a nickase, a reverse transcriptase, and an integrase, the second vector comprises a polynucleotide sequence encoding at least a first recombinase. In some embodiments, where the gene editor polynucleotide encodes a prime editor system comprising a nickase, a reverse transcriptase, and an integrase, the second vector comprises a polynucleotide sequence encoding at least a second integrase. [0219] In some embodiments, the second vector includes a template polynucleotide and a sequence that is an integration cognate of an integration recognition site site-specifically incorporated into the genome of a cell. For example, the second vector includes a template polynucleotide and a second integration recognition site that is a cognate pair with the first integration recognition site site-specifically incorporated into the genome of the cell. In such embodiments, the sequence that is an integration cognate (e.g., a second integration recognition site) enables integration of the template polynucleotide or portion thereof when contacted with an integrase and the site-specifically incorporated first integration recognition site.
[0220] In some embodiments, the second vector is a vector selected from: adenovirus, AAV, lentivirus, HSV, annelovirus, retrovirus, Doggybone™ DNA (dbDNA), minicircle, plasmid, miniDNA, exosome, fusosome, or nanoplasmid.
[0221] In some embodiments, the polynucleotide sequence encoding the prime editor system is encoded on at least two different vectors. In one embodiment, a first vector comprises a polynucleotide sequence encoding a nickase and a second vector comprises a polynucleotide sequence encoding a reverse transcriptase. In such cases, the first vector and second are delivered concurrently.
[0222] In some embodiments, the polynucleotide sequence(s) encoding the prime editor system is encoded on at least two (non-contiguous) polynucleotide sequences. In one embodiment, a first polynucleotide sequence encodes a nickase and a second polynucleotide sequence encodes a reverse transcriptase. In such cases, the first vector and second are delivered concurrently (e.g., in a first LNP).
5.9.4. Split Lipid Nanoparticles (LNPs)
[0223] Also provided herein are methods of co-delivering a system capable of site-specifically integrating at least a first integration recognition site into the genome of a cell, where the method includes delivering to a cell a mixture of a first LNP and a second LNP (“split LNPs”). In one embodiment, the method includes co-delivering to a cell a first gene editor polynucleotide construct and a first attachment site-containing guide RNA (atgRNA) are packaged, and thereby vectorized, within the first LNP, and a second gene editor polynucleotide construct and a second attachment site containing guide RNR (atgRNA) are packaged, and thereby vectorized, within the second LNP, where the first atgRNA and the second atgRNA are an at least first pair of atgRNA. The at least first pair of atgRNAs comprise domains that are capable of guiding the prime editor system to a target sequence. The first atgRNA further includes a first RT template that comprises at least a portion of a first integration recognition site. The second atgRNA further includes a second RT template that comprises at least a portion of the first integration recognition site. The first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site. In such embodiments, the first atgRNA and second atgRNA include at least a 6bp overlap.
[0224] In some embodiments, where the method includes delivering a first LNP (e.g., a first LNP comprising a first gene editor polynucleotide construct and a first atgRNA) and a second LNP (e.g., a second LNP comprising a second gene editor polynucleotide construct and a second atgRNA), the first LNP and the second LNP are mixed prior to delivering to a cell. In some embodiments, the first LNP and the second LNP are mixed at a ratio of first LNP to second LNP of 1:10, 1:9, 1:8, 1:7, 1:6, 1:5, 1:4, 1:3, 1:2, 1:1, 1:0.75, 0.75:1, 2:1, 3:1, 4:1, 5:1, 6:1, 7:1, 8:1, 9:1, or 10:1. In some embodiments, the first LNP and the second LNP are mixed at a ratio of 1:1.
[0225] In some embodiments, a first LNP comprising a first gene editor polynucleotide construct and a first attachment site-containing guide RNA (atgRNA 1) comprises a ratio of ratio of gene editor polynucleotide construct (e.g., mRNA) to atgRNAl of 1:10, 1:9, 1:8, 1:7, 1:6, 1:5, 1:4, 1:3, 1:2, 1:1, 1:0.75, 0.75:1, 2:1, 3:1, 4:1, 5:1, 6:1, 7:1, 8:1, 9:1, or 10:1. In some embodiments, the first LNP comprises a ratio of mRNA to atgRNAl of 2:1.
[0226] In some embodiments, a second LNP comprising a second gene editor polynucleotide construct and a second attachment site-containing guide RNA (atgRNA2) comprises a ratio of gene editor polynucleotide construct (e.g., mRNA) to atgRNA2 of 1:10, 1:9, 1:8, 1:7, 1:6, 1:5, 1:4, 1:3, 1:2, 1:1, 1:0.75, 0.75:1, 2:1, 3:1, 4:1, 5:1, 6:1, 7:1, 8:1, 9:1, or 10:1. In some embodiments, the second LNP comprises a ratio of mRNA to atgRNA2 of 2:1.
[0227] In some embodiments, where the method includes delivering a first LNP (e.g., a first LNP comprising a first gene editor polynucleotide construct and a first atgRNA) and a second LNP (e.g., a second LNP comprising a second gene editor polynucleotide construct and a second atgRNA), the first LNP and the second LNP are mixed such that the ratio of gene editor polynucleotide construct (e.g., mRNA) to first atgRNA (atgRNAl) to second atgRNA (atgRNA2) is 1:0.25:0.25, l:0.5:0.5, 1:0.75:0.75, or 1:1:1.
[0228] In some embodiments, the method of co-delivering to a cell a mixture of LNPs includes co-delivering three or more LNPs, four or more LNPs, five or more LNPs, six or more LNPs, seven or more LNPs, eight or more LNPs, nine or more LNPs, or ten or more LNPs. [0229] Also provided herein is a system capable of site-specifically integrating at least a first integration recognition site into the genome of a cell, the system comprising: a first gene editor polynucleotide construct and a first attachment site-containing guide RNA (atgRNA) are packaged, and thereby vectorized, within the first LNP, and a second gene editor polynucleotide construct and a second attachment site containing guide RNR (atgRNA) are packaged, and thereby vectorized, within the second LNP, where the first atgRNA and the second atgRNA are an at least first pair of atgRNA. The at least first pair of atgRNAs comprise domains that are capable of guiding the prime editor system to a target sequence. The first atgRNA further includes a first RT template that comprises at least a portion of a first integration recognition site. The second atgRNA further includes a second RT template that comprises at least a portion of the first integration recognition site. The first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site. In such embodiments, the first atgRNA and second atgRNA include at least a 6bp overlap.
[0230] In some embodiments, the system comprises a first LNP (e.g., any of the first LNPs described herein) and a second LNP (e.g., any of the second LNPs described herein) at a ratio of first LNP to second LNP of 1:10, 1:9, 1:8, 1:7, 1:6, 1:5, 1:4, 1:3, 1:2, 1:1, 1:0.75, 0.75:1, 2:1, 3:1, 4:1, 5:1, 6:1, 7:1, 8:1, 9:1, or 10:1. In some embodiments, the system comprise the first LNP and the second LNP at a ratio of 1 : 1.
[0231] In some embodiments, the system comprises a first LNP having a ratio of a first gene editor polynucleotide construct to a first attachment site-containing guide RNA (atgRNAl) of 1:10, 1:9, 1:8, 1:7, 1:6, 1:5, 1:4, 1:3, 1:2, 1:1, 1:0.75, 0.75:1, 2:1, 3:1, 4:1, 5:1, 6:1, 7:1, 8:1, 9:1, or 10:1. In some embodiments, the system includes a first LNP having a ratio of mRNA (i.e., mRNA encoding the gene editor protein) to atgRNAl of 2:1.
[0232] In some embodiments, the system comprise a second LNP having a ratio of a second gene editor polynucleotide construct to a second attachment site-containing guide RNA (atgRNA2) of 1:10, 1:9, 1:8, 1:7, 1:6, 1:5, 1:4, 1:3, 1:2, 1:1, 1:0.75, 0.75:1, 2:1, 3:1, 4:1, 5:1, 6:1, 7:1, 8:1, 9:1, or 10:1. In some embodiments, the system includes a second LNP having a ratio of mRNA (i.e., mRNA encoding the gene editor protein) to atgRNA2 of 2:1.
[0233] In some embodiments, the system comprises a ratio of gene editor polynucleotide construct (e.g., mRNA encoding the gene editor protein) to first atgRNA (atgRNAl) to second atgRNA (atgRNA2) of 1:0.25:0.25, l:0.5:0.5, 1:0.75:0.75, or 1:1:1. [0234] In some embodiments, the system comprises a mixture of LNPs comprising three or more LNPs, four or more LNPs, five or more LNPs, six or more LNPs, seven or more LNPs, eight or more LNPs, nine or more LNPs, or ten or more LNPs.
[0235] In some embodiments, where a split LNP (e.g., a mixture of two LNPs packaged with different cargo) is being used to site-specifically integrate the at least first integration recognition site into the genome, a vector comprising a template polynucleotide and a sequence that is an integration cognate (i.e., cognate to an integration recognition site site-specifically incorporated into the genome of a cell) can be delivered to the cell concurrently with the split LNPs or after delivery of the split LNPs. For example, after delivering the split LNPs to the cell, a vector that includes a template polynucleotide and a second integration recognition site that is a cognate pair with the first integration recognition site is delivered to the cell. In such embodiments, the sequence that is an integration cognate (e.g., a second integration recognition site) enables integration of the template polynucleotide or portion thereof when contacted with an integrase and the site- specifically incorporated first integration recognition site.
5.9.5. Vector Delivery of a template polynucleotide
[0236] In certain aspects the disclosure involves vectors, e.g. for delivering or introducing in a cell, but also for propagating these components (e.g. in prokaryotic cells). A used herein, a "vector" is a tool that allows or facilitates the transfer of an entity from one environment to another. It is a replicon, such as a plasmid, phage, or cosmid, into which another DNA segment may be inserted so as to bring about the replication of the inserted segment. Generally, a vector is capable of replication when associated with the proper control elements. In general, the term "vector" refers to a nucleic acid molecule capable of transporting another nucleic acid to which it has been linked. Vectors include, but are not limited to, nucleic acid molecules that are single- stranded, doublestranded, or partially double-stranded; nucleic acid molecules that comprise one or more free ends, no free ends (e.g. circular); nucleic acid molecules that comprise DNA, RNA, or both; and other varieties of polynucleotides known in the art. One type of vector is a ’’plasmid,” which refers to a circular double stranded DNA loop into which additional DNA segments can be inserted, such as by standard molecular cloning techniques. Another type of vector is a viral vector, wherein virally- derived DNA or RNA sequences are present in the vector for packaging into a virus (e.g. retroviruses, replication defective retroviruses, adenoviruses, replication defective adenoviruses, and adeno-associated viruses (AAVs)). Viral vectors also include polynucleotides carried by a virus for transfection into a host cell. Certain vectors are capable of autonomous replication in a host cell into which they are introduced (e.g. bacterial vectors having a bacterial origin of replication and episomal mammalian vectors). Other vectors (e.g., non-episomal mammalian vectors) are integrated into the genome of a host cell upon introduction into the host cell, and thereby are replicated along with the host genome. Moreover, certain vectors are capable of directing the expression of genes to which they are operatively-linked. Such vectors are referred to herein as "expression vectors." Vectors for and that result in expression in a eukaryotic cell can be referred to herein as "eukaryotic expression vectors." Common expression vectors of utility in recombinant DNA techniques are often in the form of plasmids.
[0237] Recombinant expression vectors can comprise a nucleic acid of the disclosure in a form suitable for expression of the nucleic acid in a host cell, which means that the recombinant expression vectors include one or more regulatory elements, which may be selected on the basis of the host cells to be used for expression, that is operatively-linked to the nucleic acid sequence to be expressed. Within a recombinant expression vector, "operably linked" is intended to mean that the nucleotide sequence of interest is linked to the regulatory element(s) in a manner that allows for expression of the nucleotide sequence (e.g. in an in vitro transcription/translation system or in a host cell when the vector is introduced into the host cell). With regards to recombination and cloning methods, mention is made of U.S. patent application Ser. No. 10/815,730, published Sep. 2, 2004 as US 2004-0171156 Al, the contents of which are herein incorporated by reference in their entirety.
[0238] Vector delivery, e.g., plasmid, viral delivery: The CRISPR enzyme, for instance a Type V protein such as C2cl or C2c3, and/or any of the present RNAs, for instance a guide RNA, can be delivered using any suitable vector, e.g., plasmid or viral vectors, such as adeno associated virus (AAV), lentivirus, adenovirus or other viral vector types, or combinations thereof. Effector proteins and one or more guide RNAs can be packaged into one or more vectors, e.g., plasmid or viral vectors. In some embodiments, the vector, e.g., plasmid or viral vector is delivered to the tissue of interest by, for example, an intramuscular injection, while other times the delivery is via intravenous, transdermal, intranasal, oral, mucosal, or other delivery methods. Such delivery may be either via a single dose, or multiple doses. One skilled in the art understands that the actual dosage to be delivered herein may vary greatly depending upon a variety of factors, such as the vector choice, the target cell, organism, or tissue, the general condition of the subject to be treated, the degree of transformation/modification sought, the administration route, the administration mode, the type of transformation/modification sought, etc. [0239] Such a dosage may further contain, for example, a carrier (water, saline, ethanol, glycerol, lactose, sucrose, calcium phosphate, gelatin, dextran, agar, pectin, peanut oil, sesame oil, etc.), a diluent, a pharmaceutically-acceptable carrier (e.g., phosphate -buffered saline), a pharmaceutically-acceptable excipient, and/or other compounds known in the art. The dosage may further contain one or more pharmaceutically acceptable salts such as, for example, a mineral acid salt such as a hydrochloride, a hydrobromide, a phosphate, a sulfate, etc.; and the salts of organic acids such as acetates, propionates, malonates, benzoates, etc. Additionally, auxiliary substances, such as wetting or emulsifying agents, pH buffering substances, gels or gelling materials, flavorings, colorants, microspheres, polymers, suspension agents, etc. may also be present herein. In addition, one or more other conventional pharmaceutical ingredients, such as preservatives, humectants, suspending agents, surfactants, antioxidants, anticaking agents, fillers, chelating agents, coating agents, chemical stabilizers, etc. may also be present, especially if the dosage form is a reconstitutable form. Suitable exemplary ingredients include microcrystalline cellulose, carboxymethylcellulose sodium, polysorbate 80, phenylethyl alcohol, chlorobutanol, potassium sorbate, sorbic acid, sulfur dioxide, propyl gallate, the parabens, ethyl vanillin, glycerin, phenol, parachlorophenol, gelatin, albumin and a combination thereof. A thorough discussion of pharmaceutically acceptable excipients is available in REMINGTON'S PHARMACEUTICAL SCIENCES (Mack Pub. Co., N.J. 1991) which is incorporated by reference herein.
[0240] In an embodiment herein the delivery is via an adenovirus, which may be at a single booster dose containing at least 1 x 105 particles (also referred to as particle units, pu) of adenoviral vector. In an embodiment herein, the dose preferably is at least about 1 x 106 particles (for example, about 1 x 106- 1 x 1011 particles), more preferably at least about 1 x 107 particles, more preferably at least about 1 x 108 particles (e.g., about 1 x 108- 1 x 1011 particles or about 1 x 109- 1 x 1012 particles), and most preferably at least about 1 x IO10 particles (e.g., about 1 x 109- 1 x IO10 particles or about 1 x 109-l x 1012 particles), or even at least about 1 x IO10 particles (e.g., about 1 x 1010-l x 1012 particles) of the adenoviral vector. Alternatively, the dose comprises no more than about 1 x 1014 particles, preferably no more than about 1 x 1013 particles, even more preferably no more than about 1 x 1012 particles, even more preferably no more than about 1 x 1011 particles, and most preferably no more than about 1 x IO10 particles (e.g., no more than about 1 x 109 particles). Thus, the dose may contain a single dose of adenoviral vector with, for example, about 1 x 106 particle units (pu), about 2 x 106 pu, about 4 x 106 pu, about 1 x 107 pu, about 2 x 107 pu, about 4 x 107 pu, about 1 x 108 pu, about 2 x 108 pu, about 4 x 108 pu, about 1 x 109 pu, about 2 x 109 pu, about 4 x 109 pu, about 1 x IO10 pu, about 2 x IO10 pu, about 4 x IO10 pu, about 1 x 1011 pu, about 2 x 1011 pu, about 4 x 1011 pu, about 1 x 1012 pu, about 2 x 1012 pu, or about 4 x 1012 pu of adenoviral vector. See, for example, the adenoviral vectors in U.S. Pat. No. 8,454,972 B2 to Nabel, et. al., granted on Jun. 4, 2013; incorporated by reference herein, and the dosages at col 29, lines 36-58 thereof. In an embodiment herein, the adenovirus is delivered via multiple doses.
[0241] In an embodiment herein, the delivery is via an AAV. A therapeutically effective dosage for in vivo delivery of the AAV to a human is believed to be in the range of from about 20 to about 50 ml of saline solution containing from about 1 x 1010 to about 1 x IO50 functional AAV/ml solution. The dosage may be adjusted to balance the therapeutic benefit against any side effects. In an embodiment herein, the AAV dose is generally in the range of concentrations of from about 1 x 105 to 1 x IO50 genomes AAV (sometimes referred to herein as “vector genomes” or “vg”), from about 1 x 108 to 1 x IO20 genomes AAV, from about 1 x 1010 to about 1 x 1016 genomes, or about 1 x 1011 to about 1 x 1016 genomes AAV. A human dosage may be about 1 x 1013 genomes AAV. Such concentrations may be delivered in from about 0.001 ml to about 100 ml, about 0.05 to about 50 ml, or about 10 to about 25 ml of a carrier solution. Other effective dosages can be readily established by one of ordinary skill in the art through routine trials establishing dose response curves. See, for example, U.S. Pat. No. 8,404,658 B2 to Hajjar, et al., granted on Mar. 26, 2013, at col. 27, lines 45-60.
[0242] The promoter used to drive nucleic acid-targeting effector protein coding nucleic acid molecule expression can include: AAV ITR can serve as a promoter: this is advantageous for eliminating the need for an additional promoter element (which can take up space in the vector). The additional space freed up can be used to drive the expression of additional elements (gRNA, etc.). Also, ITR activity is relatively weaker, so can be used to reduce potential toxicity due to over expression of nucleic acid-targeting effector protein. For ubiquitous expression, can use promoters: CMV, CAG, CBh, PGK, SV40, Ferritin heavy or light chains, etc. For brain or other CNS expression, can use promoters: SynapsinI for all neurons, CaMKIIalpha for excitatory neurons, GAD67 or GAD65 or VGAT for GABAergic neurons, etc. For liver expression, can use Albumin promoter. For lung expression, can use SP-B. For endothelial cells, can use ICAM. For hematopoietic cells can use IFNbeta or CD45. For Osteoblasts can use OG-2.
[0243] The promoter used to drive guide RNA can include: Pol III promoters such as U6 or Hl Use of Pol II promoter and intronic cassettes to express guide RNA Adeno Associated Virus (AAV).
[0244] Nucleic acid-targeting effector protein and one or more guide RNA can be delivered using adeno associated virus (AAV), lentivirus, adenovirus or other plasmid or viral vector types, in particular, using formulations and doses from, for example, U.S. Pat. No. 8,454,972 (formulations, doses for adenovirus), U.S. Pat. No. 8,404,658 (formulations, doses for AAV) and U.S. Pat. No. 5,846,946 (formulations, doses for DNA plasmids) and from clinical trials and publications regarding the clinical trials involving lentivirus, AAV and adenovirus. For examples, for AAV, the route of administration, formulation and dose can be as in U.S. Pat. No. 8,454,972 and as in clinical trials involving AAV. For Adenovirus, the route of administration, formulation and dose can be as in U.S. Pat. No. 8,404,658 and as in clinical trials involving adenovirus. For plasmid delivery, the route of administration, formulation and dose can be as in U.S. Pat. No. 5,846,946 and as in clinical studies involving plasmids. Doses may be based on or extrapolated to an average 70 kg individual (e.g., a male adult human), and can be adjusted for patients, subjects, mammals of different weight and species. Frequency of administration is within the ambit of the medical or veterinary practitioner (e.g., physician, veterinarian), depending on usual factors including the age, sex, general health, other conditions of the patient or subject and the particular condition or symptoms being addressed. The viral vectors can be injected into the tissue of interest. For celltype specific genome modification, the expression of nucleic acid-targeting effector can be driven by a cell-type specific promoter. For example, liver- specific expression might use the Albumin promoter and neuron- specific expression (e.g., for targeting CNS disorders) might use the Synapsin I promoter.
[0245] In terms of in vivo delivery, AAV is advantageous over other viral vectors for a couple of reasons: Low toxicity (this may be due to the purification method not requiring ultra centrifugation of cell particles that can activate the immune response) and Low probability of causing insertional mutagenesis because it doesn't integrate into the host genome.
[0246] AAV has a packaging limit of 4.5 or 4.75 Kb. This means that nucleic acid-targeting effector protein (such as a Type V protein such as C2cl or C2c3) as well as a promoter and transcription terminator have to be all fit into the same viral vector. Therefore embodiments of the disclosure include utilizing homologs of nucleic acid-targeting effector protein (such as a Type V protein such as C2cl or C2c3) that are shorter.
[0247] As to AAV, the AAV can be AAV1, AAV2, AAV5 or any combination thereof. One can select the AAV of the AAV with regard to the cells to be targeted; e.g., one can select AAV serotypes 1, 2, 5 or a hybrid capsid AAV1, AAV2, AAV5 or any combination thereof for targeting brain or neuronal cells; and one can select AAV4 for targeting cardiac tissue. AAV8 is useful for delivery to the liver. The herein promoters and vectors are preferred individually. [0248] Packaging cells are typically used to form virus particles that are capable of infecting a host cell. Such cells include 293 cells, which package adenovirus, and psi2 cells or PA317 cells, which package retrovirus. Viral vectors used in gene therapy are usually generated by producing a cell line that packages a nucleic acid vector into a viral particle. The vectors typically contain the minimal viral sequences required for packaging and subsequent integration into a host, other viral sequences being replaced by an expression cassette for the polynucleotide(s) to be expressed. The missing viral functions are typically supplied in trans by the packaging cell line. For example, AAV vectors used in gene therapy typically only possess ITR sequences from the AAV genome which are required for packaging and integration into the host genome. Viral DNA is packaged in a cell line, which contains a helper plasmid encoding the other AAV genes, namely rep and cap, but lacking ITR sequences. The cell line may also be infected with adenovirus as a helper. The helper virus promotes replication of the AAV vector and expression of AAV genes from the helper plasmid. The helper plasmid is not packaged in significant amounts due to a lack of ITR sequences. Contamination with adenovirus can be reduced by, e.g., heat treatment to which adenovirus is more sensitive than AAV. Additional methods for the delivery of nucleic acids to cells are known to those skilled in the art. See, for example, US20030087817, incorporated herein by reference.
[0249] Millington- Ward et al. (Molecular Therapy, vol. 19 no. 4, 642-649 April 2011) describes adeno-associated virus (AAV) vectors to deliver an RNA interference (RNAi)-based rhodopsin suppressor and a codon-modified rhodopsin replacement gene resistant to suppression due to nucleotide alterations at degenerate positions over the RNAi target site. An injection of either 6.0 x 108 vp or 1.8 x IO10 vp AAV were subretinally injected into the eyes by Millington- Ward et al. The AAV vectors of Millington- Ward et al. may be applied to the system of the present disclosure, contemplating a dose of about 2 x 1011 to about 6 x 1011 vp administered to a human.
[0250] Dalkara et al. (Sci Transl Med 5, 189ra76 (2013)) also relates to in vivo directed evolution to fashion an AAV vector that delivers wild-type versions of defective genes throughout the retina after noninjurious injection into the eyes' vitreous humor. Dalkara describes a 7 mer peptide display library and an AAV library constructed by DNA shuffling of cap genes from AAV1, 2, 4, 5, 6, 8, and 9. The rcAAV libraries and rAAV vectors expressing GFP under a CAG or Rho promoter were packaged and deoxyribonuclease-resistant genomic titers were obtained through quantitative PCR. The libraries were pooled, and two rounds of evolution were performed, each consisting of initial library diversification followed by three in vivo selection steps. In each such step, P30 rho-GFP mice were intravitreally injected with 2 ml of iodixanol-purified, phosphate- buffered saline (PBS) -dialyzed library with a genomic titer of about 1. times.10. sup.12 vg/ml. The AAV vectors of Dalkara et al. may be applied to the nucleic acid-targeting system of the present disclosure, contemplating a dose of about 1 x 1015 to about 1 x 1016 vg/ml administered to a human.
[0251] The tropism of a retrovirus can be altered by incorporating foreign envelope proteins, expanding the potential target population of target cells. Lentiviral vectors are retroviral vectors that are able to transduce or infect non-dividing cells and typically produce high viral titers. Selection of a retroviral gene transfer system would therefore depend on the target tissue. Retroviral vectors are comprised of cis-acting long terminal repeats with packaging capacity for up to 6-10 kb of foreign sequence. The minimum cis-acting LTRs are sufficient for replication and packaging of the vectors, which are then used to integrate the therapeutic gene into the target cell to provide permanent transgene expression. Widely used retroviral vectors include those based upon murine leukemia virus (MuLV), gibbon ape leukemia virus (GaLV), Simian Immuno deficiency virus (SW), human immuno deficiency virus (HIV), and combinations thereof (see, e.g., Buchscher et al., J. Virol. 66:2731-2739 (1992); Johann et al., J. Virol. 66:1635-1640 (1992); Sommnerfelt et al., Virol. 176:58-59 (1990); Wilson et al., J. Virol. 63:2374-2378 (1989); Miller et al., J. Virol. 65:2220-2224 (1991); PCT/US94/05700). In applications where transient expression is preferred, adenoviral based systems may be used. Adenoviral based vectors are capable of very high transduction efficiency in many cell types and do not require cell division. With such vectors, high titer and levels of expression have been obtained. This vector can be produced in large quantities in a relatively simple system. Adeno-associated virus ("AAV") vectors may also be used to transduce cells with target nucleic acids, e.g., in the in vitro production of nucleic acids and peptides, and for in vivo and ex vivo gene therapy procedures (see, e.g., West et al., Virology 160:38-47 (1987); U.S. Pat. No. 4,797,368; WO 93/24641; Kotin, Human Gene Therapy 5:793-801 (1994); Muzyczka, J. Clin. Invest. 94:1351 (1994). Construction of recombinant AAV vectors are described in a number of publications, including U.S. Pat. No. 5,173,414; Tratschin et al., Mol. Cell. Biol. 5:3251-3260 (1985); Tratschin, et al., Mol. Cell. Biol. 4:2072-2081 (1984); Hermonat & Muzyczka, PNAS 81:6466-6470 (1984); and Samulski et al., J. Virol. 63:03822-3828 (1989).
[0252] Packaging cells are typically used to form virus particles that are capable of infecting a host cell. Such cells include 293 cells, which package adenovirus, and yr2 cells or PA317 cells, which package retrovirus. Viral vectors used in gene therapy are usually generated by producing a cell line that packages a nucleic acid vector into a viral particle. The vectors typically contain the minimal viral sequences required for packaging and subsequent integration into a host, other viral sequences being replaced by an expression cassette for the polynucleotide(s) to be expressed. The missing viral functions are typically supplied in trans by the packaging cell line. For example, AAV vectors used in gene therapy typically only possess ITR sequences from the AAV genome which are required for packaging and integration into the host genome. Viral DNA is packaged in a cell line, which contains a helper plasmid encoding the other AAV genes, namely rep and cap, but lacking ITR sequences. The cell line may also be infected with adenovirus as a helper. The helper virus promotes replication of the AAV vector and expression of AAV genes from the helper plasmid. The helper plasmid is not packaged in significant amounts due to a lack of ITR sequences. Contamination with adenovirus can be reduced by, e.g., heat treatment to which adenovirus is more sensitive than AAV. Additional methods for the delivery of nucleic acids to cells are known to those skilled in the art. See, for example, US20030087817, incorporated herein by reference.
[0253] In some embodiments, a host cell is transiently or non-transiently transfected with one or more vectors described herein. In some embodiments, a cell is transfected as it naturally occurs in a subject. In some embodiments, a cell that is transfected is taken from a subject. Cells taken from a subject include, but are not limited to, hepatocytes or cells isolated from muscle, the CNS, eye or lung. Immunological cells are also contemplated, such as but not limited to T cells, HSCs, B-cells and NK cells.
[0254] Another useful method to deliver proteins, enzymes, and guides comprises transfection of messenger RNA (mRNA). Examples of mRNA delivery methods and compositions that may be utilized in the present disclosure including, for example, PCT/US2014/028330, US8822663B2, NZ700688A, ES2740248T3, EP2755693A4, EP2755986A4, WO2014152940A1, EP3450553B1, BR112016030852A2, and EP3362461A1. Expression of CRISPR systems in particular is described by W02020014577. Each of these publications are incorporated herein by reference in their entireties. Additional disclosure hereby incorporated by reference can be found in Kowalski et al., “Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery,” Mol Therap., 2019; 27(4): 710-728.
[0255] In some embodiments, the cell is derived from cells taken from a subject, such as a cell line. A wide variety of cell lines for tissue culture are known in the art. Examples of cell lines include, but are not limited to, C8161, CCRF-CEM, MOLT, mIMCD-3, NHDF, HeLa-S3, Huhl, Huh4, Huh7, HUVEC, HASMC, HEKn, HEKa, MiaPaCell, Panel, PC-3, TF1, CTLL-2, CIR, Rat6, CV1, RPTE, A10, T24, J82, A375, ARH-77, Calul, SW480, SW620, SKOV3, SK-UT, CaCo2, P388D1, SEM-K2, WEHI-231, HB56, TIB55, Jurkat, J45.01, LRMB, Bcl-1, BC-3, IC21, DLD2, Raw264.7, NRK, NRK-52E, MRC5, MEF, Hep G2, HeLa B, HeLa T4, COS, COS-1, COS-6, COS-M6A, BS-C-1 monkey kidney epithelial, BALB/3T3 mouse embryo fibroblast, 3T3 Swiss, 3T3-L1, 132-d5 human fetal fibroblasts; 10.1 mouse fibroblasts, 293-T, 3T3, 721, 9L, A2780, A2780ADR, A2780cis, A172, A20, A253, A431, A-549, ALC, B16, B35, BCP-1 cells, BEAS-2B, bEnd.3, BHK-21, BR 293, BxPC3, C3H-10T1/2, C6/36, Cal-27, CHO, CHO-7, CHOIR, CHO-K1, CHO-K2, CHO-T, CHO Dhfr-/-, COR-L23, COR-L23/CPR, COR-L23/5010, COR- L23/R23, COS-7, COV-434, CML Tl, CMT, CT26, D17, DH82, DU145, DuCaP, EL4, EM2, EM3, EMT6/AR1, EMT6/AR10.0, FM3, H1299, H69, HB54, HB55, HCA2, HEK-293, HeLa, Hepalclc7, HL-60, HMEC, HT-29, Jurkat, JY cells, K562 cells, Ku812, KCL22, KG1, KYO1, LNCap, Ma-Mel 1-48, MC-38, MCF-7, MCF-10A, MDA-MB-231, MDA-MB-468, MDA-MB- 435, MDCK II, MDCK II, MOR/0.2R, MONO-MAC 6, MTD-1A, MyEnd, NCI-H69/CPR, NCI- H69/LX10, NCI-H69/LX20, NCI-H69/LX4, NIH-3T3, NALM-1, NW- 145, OPCN/OPCT cell lines, Peer, PNT-1A/PNT 2, RenCa, RIN-5F, RMA/RMAS, Saos-2 cells, Sf-9, SkBr3, T2, T-47D, T84, THP1 cell line, U373, U87, U937, VCaP, Vero cells, WM39, WT-49, X63, YAC-1, YAR, and transgenic varieties thereof. Cell lines are available from a variety of sources known to those with skill in the art (see, e.g., the American Type Culture Collection (ATCC) (Manassas, Va.)). In some embodiments, a cell transfected with one or more vectors described herein is used to establish a new cell line comprising one or more vector-derived sequences.
[0256] In some embodiments, one or more vectors described herein are used to produce a nonhuman transgenic animal or transgenic plant. In some embodiments, the transgenic animal is a mammal, such as a mouse, rat, or rabbit. In certain embodiments, the organism or subject is a plant. In certain embodiments, the organism or subject or plant is algae. Methods for producing transgenic plants and animals are known in the art, and generally begin with a method of cell transfection, such as described herein.
[0257] In one aspect, the disclosure provides for methods of modifying a target polynucleotide in a prokaryotic or eukaryotic cell, which may be in vivo, ex vivo or in vitro. In some embodiments, the method comprises sampling a cell or population of cells from a human or non-human animal or plant (including micro-algae) and modifying the cell or cells. Culturing may occur at any stage ex vivo. The cell or cells may even be re-introduced into the non-human animal or plant (including micro-algae).
[0258] In plants, pathogens are often host-specific. For example, Fusariumn oxysporumf. sp. lycopersici causes tomato wilt but attacks only tomato, and F. oxysporum f. dianthii Puccinia graminis f. sp. tritici attacks only wheat. Plants have existing and induced defenses to resist most pathogens. Mutations and recombination events across plant generations lead to genetic variability that gives rise to susceptibility, especially as pathogens reproduce with more frequency than plants. In plants there can be non-host resistance, e.g., the host and pathogen are incompatible. There can also be Horizontal Resistance, e.g., partial resistance against all races of a pathogen, typically controlled by many genes and Vertical Resistance, e.g., complete resistance to some races of a pathogen but not to other races, typically controlled by a few genes. In a Gene-for-Gene level, plants and pathogens evolve together, and the genetic changes in one balance changes in other. Accordingly, using Natural Variability, breeders combine most useful genes for Yield. Quality, Uniformity, Hardiness, Resistance. The sources of resistance genes include native or foreign Varieties, Heirloom Varieties, Wild Plant Relatives, and Induced Mutations, e.g., treating plant material with mutagenic agents. Using the present disclosure, plant breeders are provided with a new tool to induce mutations. Accordingly, one skilled in the art can analyze the genome of sources of resistance genes, and in Varieties having desired characteristics or traits employ the present disclosure to induce the rise of resistance genes, with more precision than previous mutagenic agents and hence accelerate and improve plant breeding programs.
[0259] Examples of target polynucleotides include a sequence associated with a signaling biochemical pathway, e.g., a signaling biochemical pathway-associated gene or polynucleotide. Examples of target polynucleotides include a disease associated gene or polynucleotide. A “disease-associated” gene or polynucleotide refers to any gene or polynucleotide which is yielding transcription or translation products at an abnormal level or in an abnormal form in cells derived from a disease-affected tissues compared with tissues or cells of a non disease control. It may be a gene that becomes expressed at an abnormally high level; it may be a gene that becomes expressed at an abnormally low level, where the altered expression correlates with the occurrence and/or progression of the disease. A disease-associated gene also refers to a gene possessing mutation(s) or genetic variation that is directly responsible or is in linkage disequilibrium with a gene(s) that is responsible for the etiology of a disease. The transcribed or translated products may be known or unknown, and may be at a normal or abnormal level.
5.10. Lipid Nanoparticle Formulations, Compositions, and Delivery
[0260] In one aspect, this disclosure features compositions comprising LNP formulations capable of delivering a cargo into a liver sinusoidal endothelial cell (LSEC).
[0261] In one embodiment, a composition comprises: a lipid nanoparticle (LNP) formulation comprising a plurality of LNPs, wherein each LNP comprises: (i) an ionizable lipid; (ii) a cholesterol; (iii) a polyethyleneglycol (PEG)-modified lipid; (iv) optionally a neutral lipid, or (v) a combination of (i) - (iv); each LNP encapsulates a cargo; wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (LSEC) the encapsulated cargo. In some embodiments, the cargo is selected from: an mRNA, an siRNA, an antisense oligonucleotide, a DNA, a guide RNA (gRNA), a protein, a peptide, a chemotherapeutic agent, a small molecule drug, or a combination thereof. In some embodiments, the cargo is an mRNA and a gRNA. In some embodiments, the mRNA is a gene editor polynucleotide encoding a gene editor polypeptide and the gRNA is an attachment site-containing guide RNA (atgRNA).
[0262] In some embodiments, the composition includes: a lipid nanoparticle (LNP) formulation comprising a plurality of LNPs, wherein each LNP comprises: (i) a polyethyleneglycol (PEG)- modified lipid; (ii) a neutral lipid, or (iii) both (i) and (ii); wherein each LNP encapsulates: (i) a gene editor polynucleotide; (ii) at least a first attachment-site containing guide RNA (atgRNA); (iii) or both (i) and (ii), wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (LSEC) the (i) a gene editor polynucleotide; (ii) at least a first attachment- site containing guide RNA (atgRNA); (iii) or both (i) and (ii).
[0263] In another aspect, this disclosure features LNP formulations capable of delivering gene editing components into a liver sinusoidal endothelial cell (LSEC). In some embodiments, the lipid nanoparticle formulation includes: (a) a gene editor polynucleotide; (b) at least a first attachment- site containing guide RNA (atgRNA); (c) an ionizable lipid; (d) a cholesterol; (e) a polyethyleneglycol (PEG) -modified lipid; and (f) optionally a neutral lipid, wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (a), (b), (c), (d), (e), (f), or a combination thereof.
[0264] In some embodiments, the ionizable lipid is an ionizable cationic lipid. In some embodiments, the ionizable cationic lipid is selected from N-(2,3dioleyloxy)propyl)- N,N,Ntrimethylammonium chloride (DOTMA); N-(2,3dioleoyloxy)propyl)-N,N,N- trimethylamntonium chloride (DODAP); N,N-dioleyl-N,N-dimethylammonium chloride (DODAC); 1,2 bis (oleoyloxy)-3-(trimethylammonio) propane (DOTAP); N,NdistearylN,N- dimethylammonium bromide (DDAB); 3-(N — (N,N-dimethylaminoethane)-carbam- oyl)cholesterol (DC-Choi); diheptadecylamidoglycylspermidine (DHGS) and N-(l,2- dimyristyloxyprop-3-yl)-N,N-dimethyl-N-hydoxyethyl ammonium bromide (DMRIE), or a combination thereof [0265] In some embodiments, the composition or the LNP composition further comprises a anionic lipid. In some embodiments, wherein the ionizable lipid is replaced with an anionic lipid. In some embodiments, the anionic lipid is selected from: a phospholipid, phospholipid derivative, a sterol, a fatty acid, or a combination thereof.
[0266] In some embodiments, the PEG-modified lipid is select from 1,2-Dimyristoyl-sn-glycerol methoxypolyethylene glycol (PEG-DMG), PEG-disteryl glycerol (PEG-DSG), PEG- dipalmetoleyl, PEG-dioleyl, PEG-distearyl, PEG-diacylglycamide (PEG-DAG), PEG-dipalmitoyl phosphatidylethanolamine (PEG-DPPE), and PEG-l,2-dimyristyloxlpropyl-3-amine (PEG-c- DMA), or a combination thereof.
[0267] In some embodiments, the PEG-modified lipid comprises a moiety capable binding to a receptor on a liver sinusoidal endothelial cell (LSEC). In some embodiments, the moiety is capable of binding to a mannose receptor, a stabilin receptor, an Fc receptor, or a combination thereof.
[0268] In some embodiments, the moiety is capable of binding to a mannose receptor, a stabilin receptor, an Fc receptor, or a combination thereof. In some embodiments, the moiety is a mannose. In some embodiments, the PEG-modified lipid is a mannose-PEG lipid.
[0269] In some embodiments, the neutral lipid is a phospholipid.
[0270] In some embodiments, the phospholipid is a natural phospholipid selected from: phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylglycerol (PG), phosphatidylserine (PS), phosphatidylinositol (PI), Phosphatidic acid (phosphatidate) (PA), dipalmitoylphosphatidylcholine, monoacyl-phosphatidylcholine (lyso PC), l-palmitoyl-2-oleoyl- sn-glycero-3-phosphocholine (POPC), N-Acyl-PE, phosphoinositides, and phospho sphingolipids, or a combination thereof.
[0271] In some embodiments, the phospholipid is a phospholipid derivative selected from: phosphatidic acid (DMPA, DPPA, DSPA), phosphatidylcholine (DDPC, DLPC, DMPC, DPPC, DSPC, DOPC, POPC, DEPC), phosphatidylglycerol (DMPG, DPPG, DSPG, DOPG), phosphatidylethanolamine (DMPE, DPPE, DSPE DOPE), and phosphatidylserine (DOPS), or a combination thereof.
[0272] In some embodiments, the phospholipid is selected from: l,2-dioleoyl-sn-glycero-3- phospho-rac-(l-glycerol) sodium salt (DOPG); l,2-dilinoleoyl-sn-glycero-3-phosphocholine (DLPC); 1,2-dimyristoyl-sn-glycero-phosphocholine (DMPC); 1 ,2-dioleoyl-sn-glycero-3- phosphocholine (DOPC); l,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC); 1,2-distearoyl- sn-glycero-3-phosphocholine (DSPC); 1,2-diundecanoyl-sn-glycero-phosphocholine (DUPC); 1- palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC); l,2-di-0-octadecenyl-£-glycero-3- phosphocholine (18:0 Diether PC); l-oleoyl-2-cholesterylhemisuccinoyl-glycero-3- phosphocholine (OChemsPC); l-hexadecyl-sn-glycero-3-phosphocholine (C16 Lyso PC); 1,2- dilinolenoyl-sn-glycero-3-phosphocholine; l,2-diarachidonoyl-sn-glycero-3-phosphocholine; 1,2- didocosahexaenoyl-sn-glycero-3-phosphocholine; l,2-dioleoyl-sn-glycero-3- phosphoethanolamine (DOPE); l,2-diphytanoyl-sn-glycero-3-phosphoethanolamine (ME 16.0 PE); l,2-distearoyl-sn-glycero-3-phosphoethanolamine; l,2-dilinoleoyl-sn-glycero-3- phosphoethanolamine; 1 ,2-dilinolenoyl-sn-glycero-3-phosphoethanolamine; 1 ,2-diarachidonoyl- sn-glycero-3-phosphoethanolamine; and 1 ,2-didocosahexaenoyl-sn-glycero-3- phosphoethanolamine; and mixtures thereof.
[0273] In some embodiments, the phospholipid is DOPG.
[0274] In some embodiments, each LNP comprises: (i) modified PEGylated lipids; and (ii) neutral lipids.
[0275] In some embodiments, each LNP comprises (i) PEG-modified lipids comprising a moiety capable binding to a receptor on a liver sinusoidal endothelial cell (LSEC); and (ii) phospholipids.
[0276] In some embodiments, each LNP comprises mannose-PEG lipids and DOPGs.
[0277] In some embodiments, the LNP further comprises: cholesterol and an ionizable lipid.
[0278] In some embodiments, each LNP in the LNP formulation ranges from about 100 nm to about 200 nm.
[0279] In some embodiments, each LNP in the LNP formulation ranges from about 85 nm to about 120 nm.
[0280] In some embodiments, each LNP in the LNP formulation ranges from about 45 nm to about 65 nm.
[0281] In some embodiments, each LNP in the LNP formulation ranges from about 40 nm to about 50 nm.
[0282] In some embodiments, the encapsulation efficiency is about 80% to about 100%. [0283] In some embodiments, the encapsulation efficiency is no less than 84%.
[0284] In some embodiments, the encapsulation efficiency is no less than 90%.
[0285] In some embodiments, the polydispersity index of the composition ranges from about 0.005 to 0.5.
[0286] In some embodiments, the polydispersity index of the composition ranges from about 0.025 to 0.4.
[0287] In some embodiments, the LNP components are provided in the following ratios (e.g., molar ratios) 15-60% ionizable lipid, 4-40% phospholipid, 10-60% cholesterol, and 0-5% PEG- lipid.
[0288] In some embodiments, the LNP components are provided in the following ratios: 15-60% cKKE12, 4-40% DOPE or DOPG, 10-60% cholesterol, and 0-5% PEG2K-DMG. In some embodiments, the LNP components are provided in the following ratios: 40% cKKE12, 30% DOPE or DOPG, 28% Cholesterol, and 2% PEG2K-DMG.
[0289] In some embodiments, the LNP components are provided in the following ratios: 15-60% cKKE12, 4-40% DOPE or DOPG, 10-60% cholesterol, and 0-5% PEG2K-DMG/ Mannose- PEG2K-DSPE. In some embodiments, the LNP components are provided in the following ratios: 40% cKKE12, 30% DOPE or DOPG, 27% cholesterol, 0.5% PEG2K-DMG, and 2.5% Mannose- PEG2K-DSPE.
[0290] In some embodiments, the LNP components are provided in the following ratios: 15-60% ALC0315, 4-40% DSPC or DSPG, 10-60% Cholesterol, and 0-5% PEG2K-DMG. In some embodiments, the LNP components are provided in the following ratios: 46.3% ALC0315, 9.4% DSPC or DSPG, 42.7% Cholesterol, and 1.6% PEG2K-DMG.
[0291] In some embodiments, the LNP components are provided in the following ratios: 15-60% ALC0315, 4-40% DSPC or DSPG, 10-60% Cholesterol, and 0-5% PEG2K-DMG/Mannose- PEG2K-DSPE. In some embodiments, the LNP components are provided in the following ratios: 46.3% ALC0315, 9.4% DSPC or DSPG, 41.3% Cholesterol, 0.5% PEG2K-DMG, and 2.5% Mannose-PEG2K-DSPE.
[0292] In some embodiments, the LNP components are provided in the following ratios: 15- 60% 246C10, 4-40% DOPE or DOPG, 10-60% Cholesterol, and 0-5% C16-PEG2K-ceramide. In some embodiments, the LNP components are provided in the following ratios: 26.5% 246C10, 20% DOPE or DOPG, 50.5% Cholesterol, and 3% C16-PEG2K-ceramide.
[0293] In some embodiments, the LNP components are provided in the following ratios: 15- 60% 246C10, 4-40% DOPE or DOPG, 10-60% Cholesterol, and 0-5% C16-PEG2K- ceramide/Mannose-PEG2K-DSPE. In some embodiments, the LNP components are provided in the following ratios: 26.5% 246C10, 20% DOPE or DOPG, 50.5% Cholesterol, 0.5% C16- PEG2K-ceramide, and 2.5% Mannose-PEG2K-DSPE
[0294] In some embodiments, the gene editor polypeptide or polynucleotide encoding the gene editor polypeptide is packaged in a LNP and administered intravenously. In some embodiments, the gene editor polypeptide or polynucleotide encoding the gene editor polypeptide is packaged in a LNP and administered intrathecally. In some embodiments, the gene editor polypeptide or polynucleotide encoding the gene editor polypeptide is packaged in a LNP and administered by intracerebral ventricular injection. In some embodiments, the gene editor polypeptide or polynucleotide encoding the gene editor polypeptide is packaged in a LNP and administered by intracisternal magna administration. In some embodiments, the gene editor polypeptide or polynucleotide encoding the gene editor polypeptide is packaged in a LNP and administered by intravitreal injection.
[0295] The preparation of lipidmucleic acid complexes, including targeted liposomes such as immunolipid complexes, is well known to one of skill in the art (see, e.g., Crystal, Science 270:404-410 (1995); Blaese et al., Cancer Gene Ther. 2:291-297 (1995); Behr et al., Bioconjugate Chem. 5:382-389 (1994); Remy et al., Bioconjugate Chem. 5:647-654 (1994); Gao et al., Gene Therapy 2:710-722 (1995); Ahmad et al., Cancer Res. 52:4817-4820 (1992); U.S. Pat. Nos. 4,186,183, 4,217,344, 4,235,871, 4,261,975, 4,485,054, 4,501,728, 4,774,085, 4,837,028, and 4,946,787).
[0296] In another embodiment, LNP doses of about 0.01 to about 1 mg per kg of body weight administered intravenously are contemplated. Medications to reduce the risk of infusion-related reactions are contemplated, such as dexamethasone, acetampinophen, diphenhydramine or cetirizine, and ranitidine are contemplated. Multiple doses of about 0.3 mg per kilogram every 4 weeks for five doses are also contemplated.
[0297] The charge of the LNP must be taken into consideration. As cationic lipids combined with negatively charged lipids to induce nonbilayer structures that facilitate intracellular delivery. Because charged LNPs are rapidly cleared from circulation following intravenous injection, ionizable cationic lipids with pKa values below 7 were developed (see, e.g., Rosin et al, Molecular Therapy, vol. 19, no. 12, pages 1286-2200, December 2011). Negatively charged polymers such as RNA may be loaded into LNPs at low pH values (e.g., pH 4) where the ionizable lipids display a positive charge. However, at physiological pH values, the LNPs exhibit a low surface charge compatible with longer circulation times. Four species of ionizable cationic lipids have been focused upon, namely l,2-dilineoyl-3-dimethylammonium-propane (DLinDAP), 1,2- dilinoleyloxy-3-N,N-dimethylaminopropane (DLinDMA), 1 ,2-dilinoleyloxy-keto-N,N-dimethyl- 3-aminopropane (DLinKDMA), and l,2-dilinoleyl-4-(2-dimethylaminoethyl)-[l,3]-dioxolane (DlinKC2-DMA). It has been shown that LNP siRNA systems containing these lipids exhibit remarkably different gene silencing properties in hepatocytes in vivo, with potencies varying according to the series DlinKC2-DMA>DLinKDMA>DLinDMA»DLinDAP employing a Factor VII gene silencing model (see, e.g., Rosin et al, Molecular Therapy, vol. 19, no. 12, pages 1286-2200, December 2011). A dosage of 1 pg/ml of LNP in or associated with the LNP may be contemplated, especially for a formulation containing DlinKC2-DMA.
[0298] In some embodiments, the LNP composition comprises one or more one or more ionizable lipids. As used herein, the term “ionizable lipid” has its ordinary meaning in the art and may refer to a lipid comprising one or more charged moieties. In some embodiments, an ionizable lipid may be positively charged or negatively charged. In principle, there are no specific limitations concerning the ionizable lipids of the LNP compositions disclosed herein. In some embodiments, the one or more ionizable lipids are selected from the group consisting of 3-(didodecylamino)- Nl,Nl,4-tridodecyl-l-piperazineethanamine (KL10), Nl-[2-(didodecylamino)ethyl]-Nl,N4,N4- tridodecyl-l,4-piperazinediethanami- ne (KL22), 14,25-ditridecyl-15,18,21,24-tetraaza- octatriacontane (KL25), l,2-dilinoleyloxy-N,N-dimethylaminopropane (Dlin-DMA), 2,2- dilinoleyl-4-dimethylaminomethyl-[ 1 ,3]-dioxolane (Dlin-K-DMA), heptatriaconta-6,9,28,31 - tetraen-19-yl 4-(dimethylamino)butanoate (Dlin-MC3-DMA), 2,2-dilinoleyl-4-(2- dimethylaminoethyl)-[ 1 ,3]-dioxolane (Dlin-KC2-DMA), 1 ,2-dioleyloxy-N,N- dimethylaminopropane (DODMA), 2-({8-[(3)-cholest-5-en-3-yloxy]octyl}oxy)-N,N-dimethyl-3- [(9Z,12Z)-octad- eca-9,12-dien-l-yloxy]propan-l-amine (Octyl-CLinDMA), (2R)-2-({8-[(3)- cholest-5-en-3-yloxy]octyl}oxy)-N,N-dimethyl-3-[(9Z,12Z) — octadeca-9,12-dien-l- yloxy]propan-l -amine (Octyl-CLinDMA (2R)), and (2S)-2-({8-[(3)-cholest-5-en-3- yloxy]octyl}oxy)-N,N-dimethyl-3-[(9Z,12Z) — octadeca-9,12-dien-l-y loxy]propan- 1 -amine
(Octyl-CLinDMA (2S)). In one embodiment, the ionizable lipid may be selected from, but not limited to, an ionizable lipid described in International Publication Nos. WO2013086354 and WO2013116126.
[0299] In some embodiments, the lipid nanoparticle may include one or more (e.g., 1, 2, 3, 4, 5, 6, 7, or 8) cationic and/or ionizable lipids. Such cationic and/or ionizable lipids include, but are not limited to, 3-(didodecylamino)-Nl,Nl,4-tridodecyl-l-piperazineethanamine (KL10), Nl-[2- (didodecylamino)ethyl]-Nl,N4,N4-tridodecyl-l,4-piperazinediethanami- ne (KL22), 14,25- ditridecyl- 15, 18,21 ,24-tetraaza-octatriacontane (KL25), 1 ,2-dilinoleyloxy-N,N- dimethylaminopropane (Dlin-DMA) , 2,2-dilinoleyl-4-dimethylaminomethyl- [ 1 ,3] -dioxolane (Dlin-K-DMA), heptatriaconta-6,9,28,31-tetraen-19-yl 4-(dimethylamino)butanoate (Dlin-MC3- DMA), 2, 2-dilinoleyl-4-(2-dimethylaminoethyl)-[ 1,3] -dioxolane (Dlin-KC2-DMA), 2-({8- [(3.beta.)-cholest-5-en-3-yloxy]octyl]oxy)-N,N-dimethyl-3-[(9Z,12Z)- -octadeca-9,12-dien-l- yloxy]propan-l -amine (Octyl-CLinDMA), (2R)-2-({8-[(3.beta.)-cholest-5-en-3- yloxy]octyl]oxy)-N,N-dimethyl-3-[(9Z- ,12Z)-octadeca-9,12-dien-l-yl oxy]propan-l -amine (Octyl-CLinDMA (2R)), (2S)-2-({8-[(3Pcholest-5-en-3-yloxy]octyl]oxy)-N,N-dimethyl-3-[(9Z- ,12Z)-octadeca-9,12-dien-l-yl oxy]propan-l -amine (Octyl-CLinDMA (2S)).N,N-dioleyl-N,N- dimethylammonium chloride (“DODAC”); N-(2,3-dioleyloxy)propyl-N,N — N- triethylammonium chloride (“DOTMA”); N,N-distearyl-N,N-dimethylammonium bromide (“DDAB”); N-(2,3-dioleoyloxy)propyl)-N,N,N-trimethylammonium chloride (“DOTAP”); 1,2- Dioleyloxy-3-trimethylaminopropane chloride salt (“DOTAP. Cl”); 3-.beta.-(N — (N’,N’- dimethylaminoethane)-carbamoyl)cholesterol (“DC-Chol”), N-(l-(2,3-dioleyloxy)propyl)-N-2- (sperminecarboxamido)ethyl)-N,N-dimethyl- -ammonium trifluoracetate (“DOSPA”), dioctadecylamidoglycyl carboxy spermine (“DOGS”), l,2-dioleoyl-3-dimethylammonium propane (“DODAP”), N,N-dimethyl-2,3-dioleyloxy)propylamine (“DODMA”), and N-(l,2- dimyristyloxyprop-3-yl)-N,N-dimethyl-N-hydroxyethyl ammonium bromide (“DMRIE”). Additionally, a number of commercial preparations of cationic and/or ionizable lipids can be used, such as, e.g., LIPOFECTIN.RTM. (including DOTMA and DOPE, available from GIBCO/BRL), and LIPOFECT AMINE. RTM. (including DOSPA and DOPE, available from GIBCO/BRL). KL10, KL22, and KL25 are described, for example, in U.S. Pat. No. 8,691,750.
[0300] In some embodiments, the LNP composition comprises one or more amino lipids. The terms “amino lipid” and “cationic lipid” are used interchangeably herein to include those lipids and salts thereof having one, two, three, or more fatty acid or fatty alkyl chains and a pH-titratable amino head group (e.g., an alkylamino or dialkylamino head group). In principle, there are no specific limitations concerning the amino lipids of the LNP compositions disclosed herein. The cationic lipid is typically protonated (i.e., positively charged) at a pH below the pKa of the cationic lipid and is substantially neutral at a pH above the pKa. The cationic lipids can also be termed titratable cationic lipids. In some embodiments, the one or more cationic lipids include: a protonatable tertiary amine (e.g., pH-titratable) head group; alkyl chains, wherein each alkyl chain independently has 0 to 3 (e.g., 0, 1, 2, or 3) double bonds; and ether, ester, or ketal linkages between the head group and alkyl chains. Such cationic lipids include, but are not limited to, DSDMA, DODMA, DOTMA, DLinDMA, DLenDMA, .gamma.-DLenDMA, Dlin-K-DMA, Dlin-K-C2- DMA (also known as Dlin-C2K-DMA, XTC2, and C2K), Dlin-K-C3-DMA, Dlin-K-C4-DMA, Dlen-C2K-DMA, y-Dlen-C2-DMA, C12-200, cKK-E12, cKK-A12, cKK-012, Dlin-MC2-DMA (also known as MC2), and Dlin-MC3-DMA (also known as MC3).
[0301] Anionic lipids suitable for use in lipid nanoparticles include, but are not limited to, phosphatidylglycerol, cardiolipin, diacylphosphatidylserine, diacylphosphatidic acid, N- dodecanoyl phosphatidylethanoloamine, N-succinyl phosphatidylethanolamine, N-glutaryl phosphatidylethanolamine, lysylphosphatidylglycerol, and other anionic modifying groups joined to neutral lipids.
[0302] Neutral lipids (including both uncharged and zwitterionic lipids) suitable for use in lipid nanoparticles include, but are not limited to, diacylphosphatidylcholine, diacylphosphatidylethanolamine, ceramide, sphingomyelin, dihydrosphingomyelin, cephalin, sterols (e.g., cholesterol) and cerebrosides. In some embodiments, the lipid nanoparticle comprises cholesterol. Lipids having a variety of acyl chain groups of varying chain length and degree of saturation are available or may be isolated or synthesized by well-known techniques. Additionally, lipids having mixtures of saturated and unsaturated fatty acid chains and cyclic regions can be used. In some embodiments, the neutral lipids used in the disclosure are DOPE, DSPC, DPPC, POPC, or any related phosphatidylcholine. In some embodiments, the neutral lipid may be composed of sphingomyelin, dihydrosphingomyeline, or phospholipids with other head groups, such as serine and inositol.
[0303] In some embodiments, amphipathic lipids are included in nanoparticles. Exemplary amphipathic lipids suitable for use in nanoparticles include, but are not limited to, sphingolipids, phospholipids, fatty acids, and amino lipids.
[0304] The lipid composition of the pharmaceutical composition may comprise one or more phospholipids, for example, one or more saturated or (poly)unsaturated phospholipids or a combination thereof. In general, phospholipids comprise a phospholipid moiety and one or more fatty acid moieties.
[0305] A phospholipid moiety can be selected, for example, from the non-limiting group consisting of phosphatidyl choline, phosphatidyl ethanolamine, phosphatidyl glycerol, phosphatidyl serine, phosphatidic acid, 2-lysophosphatidyl choline, and a sphingomyelin.
[0306] A fatty acid moiety can be selected, for example, from the non-limiting group consisting of lauric acid, myristic acid, myristoleic acid, palmitic acid, palmitoleic acid, stearic acid, oleic acid, linoleic acid, alpha-linolenic acid, erucic acid, phytanoic acid, arachidic acid, arachidonic acid, eicosapentaenoic acid, behenic acid, docosapentaenoic acid, and docosahexaenoic acid.
[0307] Particular amphipathic lipids can facilitate fusion to a membrane. For example, a cationic phospholipid can interact with one or more negatively charged phospholipids of a membrane (e.g., a cellular or intracellular membrane). Fusion of a phospholipid to a membrane can allow one or more elements (e.g., a therapeutic agent) of a lipid-containing composition (e.g., LNPs) to pass through the membrane permitting, e.g., delivery of the one or more elements to a target tissue.
[0308] Non-natural amphipathic lipid species including natural species with modifications and substitutions including branching, oxidation, cyclization, and alkynes are also contemplated. For example, a phospholipid can be functionalized with or cross-linked to one or more alkynes (e.g., an alkenyl group in which one or more double bonds is replaced with a triple bond). Under appropriate reaction conditions, an alkyne group can undergo a copper-catalyzed cycloaddition upon exposure to an azide. Such reactions can be useful in functionalizing a lipid bilayer of a nanoparticle composition to facilitate membrane permeation or cellular recognition or in conjugating a nanoparticle composition to a useful component such as a targeting or imaging moiety (e.g., a dye).
[0309] Phospholipids include, but are not limited to, glycerophospholipids such as phosphatidylcholines, phosphatidylethanolamines, phosphatidylserines, phosphatidylinositols, phosphatidy glycerols, and phosphatidic acids. Phospholipids also include phospho sphingolipid, such as sphingomyelin.
[0310] In some embodiments, the LNP composition comprises one or more phospholipids. In some embodiments, the phospholipid is selected from the group consisting of 1,2-dilinoleoyl-sn- glycero-3-phosphocholine (DLPC), 1,2-dimyristoyl-sn-glycero-phosphocholine (DMPC), 1,2- dioleoyl-sn-glycero-3-phosphocholine (DOPC), l,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), l,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), 1,2-diundecanoyl-sn-glycero- phosphocholine (DUPC), l-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1,2-di-O- octadecenyl-sn-glycero-3-phosphocholine (18:0 Diether PC), l-oleoyl-2- cholesterylhemisuccinoyl-sn-glycero-3-phosphocholine (OchemsPC), l-hexadecyl-sn-glycero-3- phosphocholine (C16 Lyso PC), l,2-dilinolenoyl-sn-glycero-3-phosphocholine, 1,2- diarachidonoyl-sn-glycero-3-phosphocholine, l,2-didocosahexaenoyl-sn-glycero-3- phosphocholine, l,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-diphytanoyl-sn- glycero-3-phosphoethanolamine (ME 16:0 PE), l,2-distearoyl-sn-glycero-3- phosphoethanolamine, 1 ,2-dilinoleoyl-sn-glycero-3-phosphoethanolamine, 1 ,2-dilinolenoyl-sn- glycero-3-phosphoethanolamine, l,2-diarachidonoyl-sn-glycero-3-phosphoethanolaminel,2- didocosahexaenoyl — sn-glycero-3-phosphoethanolamine, l,2-dioleoyl-sn-glycero-3-phospho- rac-(l -glycerol) sodium salt (DOPG), sphingomyelin, and any mixtures thereof.
[0311] Other phosphorus-lacking compounds, such as sphingolipids, glyco sphingolipid families, diacylglycerols, and .beta. -acyloxyacids, may also be used. Additionally, such amphipathic lipids can be readily mixed with other lipids, such as triglycerides and sterols.
[0312] In some embodiments, the LNP composition comprises one or more helper lipids. The term “helper lipid” as used herein refers to lipids that enhance transfection (e.g., transfection of an LNP comprising an mRNA that encodes a site-directed endonuclease, such as a SpCas9 polypeptide). In principle, there are no specific limitations concerning the helper lipids of the LNP compositions disclosed herein. Without being bound to any particular theory, it is believed that the mechanism by which the helper lipid enhances transfection includes enhancing particle stability. In some embodiments, the helper lipid enhances membrane fusogenicity. Generally, the helper lipid of the LNP compositions disclosure herein can be any helper lipid known in the art. Non-limiting examples of helper lipids suitable for the compositions and methods include steroids, sterols, and alkyl resorcinols. Particularly helper lipids suitable for use in the present disclosure include, but are not limited to, saturated phosphatidylcholine (PC) such as distearoyl-PC (DSPC) and dipalymitoyl-PC (DPPC), dioleoylphosphatidylethanolamine (DOPE), 1,2-dilinoleoyl-sn- glycero-3-phosphocholine (DLPC), cholesterol, 5-heptadecylresorcinol, and cholesterol hemisuccinate. In some embodiments, the helper lipid of the LNP composition includes cholesterol.
[0313] In some embodiments, the LNP composition comprises one or more structural lipids. As used herein, the term “structural lipid” refers to sterols and also to lipids containing sterol moieties. Without being bound to any particular theory, it is believed that the incorporation of structural lipids into the LNPs mitigates aggregation of other lipids in the particle. Structural lipids can be selected from the group including but not limited to, cholesterol, fecosterol, sitosterol, ergosterol, campesterol, stigmasterol, brassicasterol, tomatidine, tomatine, ursolic acid, alpha-tocopherol, hopanoids, phytosterols, steroids, and mixtures thereof. In some embodiments, the structural lipid is a sterol. As defined herein, "sterols" are a subgroup of steroids consisting of steroid alcohols. In certain embodiments, the structural lipid is a steroid. In some embodiments, the structural lipid is cholesterol. In certain embodiments, the structural lipid is an analog of cholesterol.
[0314] The lipid component of a lipid nanoparticle composition may include one or more molecules comprising polyethylene glycol, such as PEG or PEG-modified lipids. In some embodiments, the LNP composition disclosed herein comprise one or more polyethylene glycol (PEG) lipid. The term “PEG-lipid” refers to polyethylene glycol (PEG)-modified lipids. Such lipids are also referred to as PEGylated lipids. Non-limiting examples of PEG-lipids include PEG- modified phosphatidylethanolamine and phosphatidic acid, PEG-ceramide conjugates (e.g., PEG- CerC14 or PEG-CerC20), PEG-modified dialkylamines and PEG-modified 1,2-diacyloxypropan- 3-amines For example, a PEG lipid can be PEG-c-DOMG, PEG-DMG, PEG-DLPE, PEG-DMPE, PEG-DPPC, or a PEG-DSPE lipid. In some embodiments, the PEG-lipid includes, but not limited to 1,2-dimyristoyl-sn-glycerol methoxypolyethylene glycol (PEG-DMG), 1,2-distearoyl-sn- glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)] (PEG-DSPE), PEG-disteryl glycerol (PEG-DSG), PEG-dipalmetoleyl, PEG-dioleyl, PEG-distearyl, PEG-diacylglycamide (PEG-DAG), PEG-dipalmitoyl phosphatidylethanolamine (PEG-DPPE), or PEG- 1,2- dimyristyloxlpropyl-3-amine (PEG-c-DMA). In some embodiments, the PEG-lipid is selected from the group consisting of a PEG-modified phosphatidylethanolamine, a PEG-modified phosphatidic acid, a PEG-modified ceramide, a PEG-modified dialkylamine, a PEG-modified diacylglycerol, a PEG-modified dialkylglycerol, and mixtures thereof. In some embodiments, the lipid moiety of the PEG-lipids includes those having lengths of from about C. sub.14 to about C.sub.22, preferably from about C. sub.14 to about C. sub.16. In some embodiments, a PEG moiety, for example a mPEG-NH.sub.2, has a size of about 1000, 2000, 5000, 10,000, 15,000 or 20,000 daltons. In some embodiment, the PEG-lipid is PEG2k-DMG. In some embodiments, the one or more PEG lipids of the LNP composition comprises PEG-DMPE. In some embodiments, the one or more PEG lipids of the LNP composition comprises PEG-DMG.
[0315] In some embodiments, the ratio between the lipid components and the nucleic acid molecules of the LNP composition, e.g., the weight ratio, is sufficient for (i) formation of LNPs with desired characteristics, e.g., size, charge, and (ii) delivery of a sufficient dose of nucleic acid at a dose of the lipid component(s) that is tolerable for in vivo administration as readily ascertained by one of skill in the art.
[0316] In certain embodiments, it is desirable to target a nanoparticle, e.g., a lipid nanoparticle, using a targeting moiety that is specific to a cell type and/or tissue type. In some embodiments, a nanoparticle may be targeted to a particular cell, tissue, and/or organ using a targeting moiety. In particular embodiments, a nanoparticle comprises a targeting moiety. Exemplary non-limiting targeting moieties include ligands, cell surface receptors, glycoproteins, vitamins (e.g., riboflavin) and antibodies (e.g., full-length antibodies, antibody fragments (e.g., Fv fragments, single chain Fv (scFv) fragments, Fab’ fragments, or F(ab’)2 fragments), single domain antibodies, camelid antibodies and fragments thereof, human antibodies and fragments thereof, monoclonal antibodies, and multispecific antibodies (e.g., bispecific antibodies)). In some embodiments, the targeting moiety may be a polypeptide. The targeting moiety may include the entire polypeptide (e.g., peptide or protein) or fragments thereof. A targeting moiety is typically positioned on the outer surface of the nanoparticle in such a manner that the targeting moiety is available for interaction with the target, for example, a cell surface receptor. A variety of different targeting moieties and methods are known and available in the art, including those described, e.g., in Sapra et al., Prog. Eipid Res. 42(5):439-62, 2003 and Abra et al., J. Eiposome Res. 12:1-3, 2002.
[0317] In some embodiments, a lipid nanoparticle (e.g., a liposome) may include a surface coating of hydrophilic polymer chains, such as polyethylene glycol (PEG) chains (see, e.g., Allen et al., Biochimica et Biophysica Acta 1237: 99-108, 1995; DeFrees et al., Journal of the American Chemistry Society 118: 6101-6104, 1996; Blume et al., Biochimica et Biophysica Acta 1149: ISO- 184, 1993; Klibanov et al., Journal of Eiposome Research 2: 321-334, 1992; U.S. Pat. No. 5,013,556; Zalipsky, Bioconjugate Chemistry 4: 296-299, 1993; Zalipsky, FEBS Fetters 353: 71- 74, 1994; Zalipsky, in Stealth Eiposomes Chapter 9 (Easic and Martin, Eds) CRC Press, Boca Raton Fla., 1995). In one approach, a targeting moiety for targeting the lipid nanoparticle is linked to the polar head group of lipids forming the nanoparticle. In another approach, the targeting moiety is attached to the distal ends of the PEG chains forming the hydrophilic polymer coating (see, e.g., Klibanov et al., Journal of Eiposome Research 2: 321-334, 1992; Kirpotin et al., FEBS Letters 388: 115-118, 1996).
[0318] Standard methods for coupling the targeting moiety or moieties may be used. For example, phosphatidylethanolamine, which can be activated for attachment of targeting moieties, or derivatized lipophilic compounds, such as lipid-derivatized bleomycin, can be used. Antibody- targeted liposomes can be constructed using, for instance, liposomes that incorporate protein A (see, e.g., Renneisen et al., J. Bio. Chem., 265:16337-16342, 1990 and Leonetti et al., Proc. Natl. Acad. Sci. (USA), 87:2448-2451, 1990). Other examples of antibody conjugation are disclosed in U.S. Pat. No. 6,027,726. Examples of targeting moieties can also include other polypeptides that are specific to cellular components, including antigens associated with neoplasms or tumors. Polypeptides used as targeting moieties can be attached to the liposomes via covalent bonds (see, for example Heath, Covalent Attachment of Proteins to Liposomes, 149 Methods in Enzymology 111-119 (Academic Press, Inc. 1987)). Other targeting methods include the biotin-avidin system.
[0319] In some embodiments, a lipid nanoparticle includes a targeting moiety that targets the lipid nanoparticle to a cell including, but not limited to, hepatocytes, colon cells, epithelial cells, hematopoietic cells, epithelial cells, endothelial cells, lung cells, bone cells, stem cells, mesenchymal cells, neural cells, cardiac cells, adipocytes, vascular smooth muscle cells, cardiomyocytes, skeletal muscle cells, beta cells, pituitary cells, synovial lining cells, ovarian cells, testicular cells, fibroblasts, B cells, T cells, reticulocytes, leukocytes, granulocytes, and tumor cells (including primary tumor cells and metastatic tumor cells). In particular embodiments, the targeting moiety targets the lipid nanoparticle to a hepatocyte.
[0320] The lipid nanoparticles described herein may be lipidoid-based. The synthesis of lipidoids has been extensively described and formulations containing these compounds are particularly suited for delivery of polynucleotides (see Mahon et al., Bioconjug Chem. 2010 21:1448-1454; Schroeder et al., J Intern Med. 2010 267:9-21; Akinc et al., Nat. Biotechnol. 2008 26:561-569; Love et al., Proc Natl Acad Sci USA. 2010 107: 1864-1869; Siegwart et al., Proc Natl Acad Sci USA. 2011 108:12996-3001).
[0321] The characteristics of optimized lipidoid formulations for intramuscular or subcutaneous routes may vary significantly depending on the target cell type and the ability of formulations to diffuse through the extracellular matrix into the blood stream. While a particle size of less than 150 nm may be desired for effective hepatocyte delivery due to the size of the endothelial fenestrae (see e.g., Akinc et al., Mol Ther. 2009 17:872-879), use of lipidoid oligonucleotides to deliver the formulation to other cells types including, but not limited to, endothelial cells, myeloid cells, and muscle cells may not be similarly size-limited.
[0322] In one aspect, effective delivery to myeloid cells, such as monocytes, lipidoid formulations may have a similar component molar ratio. Different ratios of lipidoids and other components including, but not limited to, a neutral lipid (e.g., diacylphosphatidylcholine), cholesterol, a PEGylated lipid (e.g., PEG-DMPE), and a fatty acid (e.g., an omega-3 fatty acid) may be used to optimize the formulation of the mRNA or system for delivery to different cell types including, but not limited to, hepatocytes, myeloid cells, muscle cells, etc. Exemplary lipidoids include, but are not limited to, Dlin-DMA, Dlin-K-DMA, Dlin-KC2-DMA, 98N12-5, C 12-200 (including variants and derivatives), Dlin-MC3-DMA and analogs thereof. The use of lipidoid formulations for the localized delivery of nucleic acids to cells (such as, but not limited to, adipose cells and muscle cells) via either subcutaneous or intramuscular delivery, may also not require all of the formulation components which may be required for systemic delivery, and as such may comprise the lipidoid and the mRNA or system.
[0323] According to the present disclosure, a system described herein may be formulated by mixing the mRNA or system, or individual components of the system, with the lipidoid at a set ratio prior to addition to cells. In vivo formulations may require the addition of extra ingredients to facilitate circulation throughout the body. After formation of the particle, a system or individual components of a system is added and allowed to integrate with the complex. The encapsulation efficiency is determined using a standard dye exclusion assays.
[0324] In vivo delivery of systems may be affected by many parameters, including, but not limited to, the formulation composition, nature of particle PEGylation, degree of loading, oligonucleotide to lipid ratio, and biophysical parameters such as particle size (Akinc et al., Mol Ther. 2009 17:872-879; herein incorporated by reference in its entirety). As an example, small changes in the anchor chain length of poly(ethylene glycol) (PEG) lipids may result in significant effects on in vivo efficacy. Formulations with the different lipidoids, including, but not limited to penta[3-(l- laurylaminopropionyl)]-triethylenetetramine hydrochloride (TETA-5LAP; aka 98N12-5, see Murugaiah et al., Analytical Biochemistry, 401:61 (2010)), C12-200 (including derivatives and variants), MD1, Dlin-DMA, Dlin-K-DMA, Dlin-KC2-DMA and Dlin-MC3-DMA can be tested for in vivo activity. The lipidoid referred to herein as “98N12-5” is disclosed by Akinc et al., Mol Ther. 2009 17:872-879). The lipidoid referred to herein as “C12-200” is disclosed by Love et al., Proc Natl Acad Sci USA. 2010 107:1864-1869 and Liu and Huang, Molecular Therapy. 2010669- 670.
[0325] LNPs in which a nucleic acid is entrapped within the lipid portion of the particle and is protected from degradation, can be formed by any method known in the art including, but not limited to, a continuous mixing method, a direct dilution process, and an in-line dilution process. Additional techniques and methods suitable for the preparation of the LNPs described herein include coacervation, microemulsions, supercritical fluid technologies, phase-inversion temperature (PIT) techniques.
[0326] In some embodiments, the LNPs used herein are produced via a continuous mixing method, e.g., a process that includes providing an aqueous solution a nucleic acid described herein in a first reservoir, providing an organic lipid solution in a second reservoir (wherein the lipids present in the organic lipid solution are solubilized in an organic solvent, e.g., a lower alkanol such as ethanol), and mixing the aqueous solution with the organic lipid solution such that the organic lipid solution mixes with the aqueous solution so as to substantially instantaneously produce a lipid vesicle (e.g., liposome) encapsulating the nucleic acid molecule within the lipid vesicle. This process and the apparatus for carrying out this process are known in the art. More information in this regard can be found in, for example, U.S. Patent Publication No. 20040142025. The action of continuously introducing lipid and buffer solutions into a mixing environment, such as in a mixing chamber, causes a continuous dilution of the lipid solution with the buffer solution, thereby producing a lipid vesicle substantially instantaneously upon mixing. By mixing the aqueous solution comprising a nucleic acid molecule with the organic lipid solution, the organic lipid solution undergoes a continuous stepwise dilution in the presence of the buffer solution (e.g., aqueous solution) to produce a nucleic acid-lipid particle.
[0327] In some embodiments, the LNPs used herein are produced via a direct dilution process that includes forming a lipid vesicle (e.g., liposome) solution and immediately and directly introducing the lipid vesicle solution into a collection vessel containing a controlled amount of dilution buffer. In some embodiments, the collection vessel includes one or more elements configured to stir the contents of the collection vessel to facilitate dilution. In some embodiments, the amount of dilution buffer present in the collection vessel is substantially equal to the volume of lipid vesicle solution introduced thereto.
[0328] In some embodiments, the LNPs are produced via an in-line dilution process in which a third reservoir containing dilution buffer is fluidly coupled to a second mixing region. In these embodiments, the lipid vesicle (e.g., liposome) solution formed in a first mixing region is immediately and directly mixed with dilution buffer in the second mixing region. These processes and the apparatuses for carrying out direct dilution and in-line dilution processes are known in the art. More information in this regard can be found in, for example, U.S. Patent Publication No. 20070042031. 5.11. Additional delivery modalities
[0329] This disclosure is not limited to systems and methods described herein. Any delivery method that is capable of delivering the systems described herein can be used as long as it is capable of site-specifically integrating a template polynucleotide into the genome of a cell.
5.12. Genes and Targets
[0330] This disclosure provides compositions, systems and methods for correcting or replacing genes or gene fragments (including introns or exons) or inserting genes in new locations. In certain embodiments, such a method comprises recombination or integration into a safe harbor site (SHS). A frequently used human SHS is the AAVS1 site on chromosome 19q, initially identified as a site for recurrent adeno-associated virus insertion. Another locus comprises the human homolog of the murine Rosa26 locus. Yet another SHS comprises the human Hl l locus on chromosome 22. In some cases, a complete gene may be prohibitively large and replacement of an entire gene impractical. In certain embodiments, a method of the disclosure comprises recombining corrective gene fragments into a defective locus.
[0331] The methods and compositions can be used to target, without limitation, stem cells for example induced pluripotent stem cells (iPSCs), HSCs, HSPCs, mesenchymal stem cells, or neuronal stem cells and cells at various stages of differentiation. In certain embodiments, methods and compositions of the disclosure are adapted to target organoids, including patient derived organoids.
[0332] In certain embodiments, methods and compositions of the disclosure are adapted to treat muscle cells, not limited to cardiomyocytes for Duchene Muscular Dystrophy (DMD). The dystrophin gene is the largest gene in the human genome, spanning ~2.3 Mb of DNA. DMD is composed of 79 exons resulting in a 14-kb full-length mRNA. Common mutations include mutations that disrupt the reading frame of generate a premature stop codon. An aspect of DMD that lends it to gene editing as a therapeutic approach is the modular structure of the dystrophin protein. Redundancy in the central rod domain permits the deletion of internal segments of the gene that may harbor loss-of-function mutations, thereby restoring the open reading frame (ORFs). In some embodiments, the methods and systems described herein are used to treat DMD by site- specifically integrating in the genome a polynucleotide template that repairs or replaces all or a portion of the defective DMD gene. [0333] The following are non-limiting diseases that may be treated utilizing the methods and compositions of the present disclosure:
Inherited Retinal Diseases:
• Stargardt Disease (ABCA4)
• Leber congenital amaurosis 10 (CEP290)
• X linked Retinitis Pigmentosa (RPGR)
• Autosomal Dominant Retinitis Pigmentosa (RHO)
Liver Diseases:
• Wilson’s disease (ATP7B)
• Alpha- 1 antitrypsin (SERPINA1)
Intellectual Disabilities:
• Rett Syndrome (MECP2)
• SYNGAP1-ID (SYNGAP1)
• CDKL5 deficiency disorder (CDKL5)
Peripheral Neuropathies:
• Charcot-Marie-Tooth 2A (MFN2)
Lung Diseases:
• Cystic Fibrosis (CFTR)
• Alpha- 1 Antitrypsin (SERPINA1)
Autoimmune diseases:
• IgA Nephropathy (Berger’s disease)
• Anti-Neutrophil Cytoplasmic Antibody (ANCA) Vasculitis
• Systemic Lupus Erythematosus (SLE) / Lupus Nephritis (LN)
• Membranous Nephropathy (MN)
• C3 glomerulonephritis (C3GN)
Blood disorders:
• Sickle Cell
• Hemophilia
• Factor VIII or
• Factor IX Ornithine transcarbamylase deficiency (OTCD)
Homocystinuria (HCU)
Phenylketonuria (PKU)
Cancer
• Prostate cancer
• Renal cell cancer
• Thyroid cancer
[0334] CFTR (cystic fibrosis transmembrane conductance regulator). The most common cystic fibrosis (CF) mutation F508del removes a single amino acid. In some embodiments, recombining human CFTR into an SHS of a cell that expresses CFTR F508del is a corrective treatment path. In some embodiments, the methods and systems described herein are used to CF by site-specifically integrating in the genome a polynucleotide template that corrects the mutation causing CF. Proposed validation is detection of persistent CFTR mRNA and protein expression in transduced cells.
[0335] Sickle cell disease (SCD) is caused by mutation of a specific amino acid — valine to glutamic acid at amino acid position 6. In some embodiments, SCD is corrected by recombination of the HBB gene into a safe harbor site (SHS) and by demonstrating correction in a proportion of target cells that is high enough to produce a substantial benefit. In some embodiments, the methods and systems described herein are used to sickle cell disease by site-specifically integrating in the genome a polynucleotide template that corrects the mutation causing the disease. In some embodiments, validation is detection of persistent HBB mRNA and protein expression in transduced cells.
[0336] DMD — Duchenne Muscular Dystrophy. The dystrophin gene is the largest gene in the human genome, spanning ~2.3 Mb of DNA. DMD is composed of 79 exons resulting in a 14- kb full-length mRNA. Common mutations include mutations that disrupt the reading frame of generate a premature stop codon. An aspect of DMD that lends it to gene editing as a therapeutic approach is the modular structure of the dystrophin protein. Redundancy in the central rod domain permits the deletion of internal segments of the gene that may harbor loss-of-function mutations, thereby restoring the open reading frame (ORFs).
[0337] In some embodiments, recombination will be into safe harbor sites (SHS). A frequently used human SHS is the AAVS1 site on chromosome 19q, initially identified as a site for recurrent adeno-associated virus insertion. In some embodiments, the site is the human homolog of the e murine Rosa26 locus (pubmed.ncbi.nlm.nih.gov/18037879). In some embodiments, the site is the human Hl l locus on chromosome 22. Proposed target cells for recombination include stem cells for example induced pluripotent stem cells (iPSCs) and cells at various stages of differentiation. In some cases, a complete gene may be prohibitively large and replacement of an entire gene impractical. In such instances, rescuing mutants by recombining in corrected gene fragments with the methods and systems described herein is a corrective option.
[0338] In some embodiments, correcting mutations in exon 44 (or 51) by recombining in a corrective coding sequence downstream of exon 43 (or 50), using the methods and systems described herein is a corrective option. Proposed validation is detection of persistent DMD mRNA and protein expression in transduced cells.
[0339] F8 (Factor VIII). A large proportion of severe hemophilia A patients harbor one of two types of chromosomal inversions in the FVIII gene. The recombinase technology and methods described herein are well suited to correcting such inversions (and other mutations) by recombining of the FVIII gene into a SHS.
[0340] In some embodiments, correcting factor VIII deficiency by recombining the FVIII gene into an SHS is a corrective path. In some embodiments, the methods and systems described herein are used to correct factor VIII deficiency by site- specifically integrating in the genome a polynucleotide template that corrects the mutation causing the FIX deficiency. Proposed validation is detection of persistent FVIII mRNA and protein expression in transduced cells.
[0341] Factor 9 (Factor IX) Hemophilia B, also called factor IX (FIX) deficiency is a genetic disorder caused by missing or defective factor IX, a clotting protein.
[0342] In some embodiments, the methods and systems described herein are used to correct factor IX deficiency by site-specifically integrating in the genome a polynucleotide template that corrects the mutation causing the FIX deficiency. Proposed validation is detection of persistent FiX mRNA and protein expression in transduced cells.
[0343] Ornithine transcarbamylase deficiency (OTCD). Ornithine transcarbamylase deficiency is a rare genetic condition that causes ammonia to build up in the blood. The condition - more commonly called OTC deficiency - is more common in boys than girls and tends to be more severe when symptoms emerge shortly after birth. [0344] In some embodiments, the methods and systems described herein are used to correct OTC deficiency by site- specifically integrating in the genome a polynucleotide template that corrects the mutation causing the OTC deficiency or integrates a polynucleotide encoding a functional ornithine transcarbamylase enzyme. Proposed validation is detection of persistent OTC mRNA and protein expression in transduced cells.
[0345] Phenylketonuria, also called PKU, is a rare inherited disorder that causes an amino acid called phenylalanine to build up in the body. PKU is caused by a change in the phenylalanine hydroxylase (PAH) gene. This gene helps create the enzyme needed to break down phenylalanine.
[0346] In some embodiments, the methods and systems described herein are used to correct PKU by site-specifically integrating in the genome a polynucleotide template that corrects the mutation causing the PKU deficiency or integrates a polynucleotide encoding a functional phenylalanine hydroxylase (PAH) gene. Proposed validation is detection of persistent PAH mRNA and protein expression in transduced cells.
[0347] Homocystinuria (HCU). Homocystinuria is elevation of the amino acid, homocysteine (protein building block coming from our diet) in the urine or blood. Common causes of HCU include: problems with the enzyme cystathionine beta synthase (CBS), which converts homocysteine to the amino acid cystathionine (which then becomes cysteine) and needs the vitamin B6 (pyridoxine); and roblems with converting homocysteine to the amino acid methionine.
[0348] In some embodiments, the methods and systems described herein are used to correct HCU by site-specifically integrating in the genome a polynucleotide template that corrects the mutation causing the HCU or integrates a polynucleotide encoding a functional copy of a gene (e.g., CBS) able to reduce or prevent buildup of homocysteine in the urine. Proposed validation is detection of persistent CBS mRNA and protein expression in transduced cells.
[0349] IgA Nephropathy (Berger’s disease). IgA nephropathy, also known as Berger's disease, is a kidney /autoimmune disease that occurs when an antibody called immunoglobulin A (IgA) builds up in the kidneys.
[0350] In some embodiments, the methods and systems described herein are used to treat Berger’s disease by administering to a patient an iPSC-derived Natural Killer cell that includes a polynucleotide site-specifically integrated in the genome of the cell using the methods described herein. Upon administering the iPSC-NK cell to the patient, the iPSC-NK cell is capable of removing native cells (e.g., B cells) that are responsible, at least in part, for the symptoms of Berger’s disease.
[0351] Anti-Neutrophil Cytoplasmic Antibody (ANCA) Vasculitis. ANCA vasculitis is an autoimmune disease affecting small blood vessels in the body. It is caused by autoantibodies called ANCAs, or Anti-Neutrophilic Cytoplasmic Autoantibodies. ANCAs target and attack a certain kind of white blood cells called neutrophils.
[0352] In some embodiments, the methods and systems described herein are used to treat ANCA vasculitis by administering to a patient an iPSC-derived Natural Killer cell that includes a polynucleotide site- specifically integrated in the genome of the cell using the methods described herein. Upon administering the iPSC-NK cell to the patient, the iPSC-NK cell is capable of removing native cells (e.g., B cells) that are responsible, at least in part, for the symptoms of ANCA vasculitis.
[0353] Systemic Lupus Erythematosus (SLE) / Lupus Nephritis (LN). Lupus is an autoimmune — a disorder in which the body’s immune system attacks the body’s own cells and organs.
[0354] In some embodiments, the methods and systems described herein are used to treat SLE/LN by administering to a patient an iPSC-derived Natural Killer cell that includes a polynucleotide site- specifically integrated in the genome of the cell using the methods described herein. Upon administering the iPSC-NK cell to the patient, the iPSC-NK cell is capable of removing native cells (e.g., B cells) that are responsible, at least in part, for the symptoms of SLE/LN.
[0355] Membranous Nephropathy (MN). MN is a kidney disease that affects the filters (glomeruli) of the kidney and can cause protein in the urine, as well as decreased kidney function and swelling. It can sometimes be called membranous glomerulopathy as well (these terms can be used interchangeably and mean the same thing).
[0356] In some embodiments, the methods and systems described herein are used to treat MN by administering to a patient an iPSC-derived Natural Killer cell that includes a polynucleotide site- specific ally integrated in the genome of the cell using the methods described herein. Upon administering the iPSC-NK cell to the patient, the iPSC-NK cell is capable of removing native cells (e.g., B cells) that are responsible, at least in part, for the symptoms of MN. [0357] C3 glomerulonephritis (C3GN). C3 glomerulopathy is a group of related conditions that cause the kidneys to malfunction. The major features of C3 glomerulopathy include high levels of protein in the urine (proteinuria), blood in the urine (hematuria), reduced amounts of urine, low levels of protein in the blood, and swelling in many areas of the body. Affected individuals may have particularly low levels of a protein called complement component 3 (or C3) in the blood.
[0358] In some embodiments, the methods and systems described herein are used to treat C3 glomerulopathy by administering to a patient an iPSC-derived Natural Killer cell that includes a polynucleotide site- specifically integrated in the genome of the cell using the methods described herein. Upon administering the iPSC-NK cell to the patient, the iPSC-NK cell is capable of removing native cells (e.g., B cells) that are responsible, at least in part, for the symptoms of C3 glomerulopathy.
5.13. Methods of treatment
[0359] In another aspect, methods of treatment are presented. The method comprises administering an effective amount of the pharmaceutical composition comprising the nucleic acid construct or vectorized nucleic acid construct described above to a patient in need thereof. In some embodiments, the system (e.g., any of the systems described herein) are delivered to a cell ex vivo and the cell is then administered to the subject. In some embodiments, the systems (e.g., any of the systems described herein) are delivered to a patient, thereby delivering to a cell in vivo.
[0360] DNA or RNA viral vectors can be administered directly to patients (in vivo) or they can be used to treat cells in vitro, and the modified cells may optionally be administered to patients (ex vivo). Conventional viral based systems to be used herein could include retroviral, lentivirus, adenoviral, adeno-associated and herpes simplex virus vectors for gene transfer. Integration in the host genome is possible with the retrovirus, lentivirus, and adeno-associated virus gene transfer methods, often resulting in long term expression of the inserted transgene. Additionally, high transduction efficiencies have been observed in many different cell types and target tissues.
[0361] In some embodiments, the co-delivery system described herein (e.g., a gene editor construct packaged in a LNP and a donor template packaged in a vector) is administered intravenously. In some embodiments, the co-delivery system described herein (e.g., a gene editor construct packaged in a LNP and a donor template packaged in a vector) is administered intrathecally. In some embodiments, the co-delivery system described herein (e.g., a gene editor construct packaged in a LNP and a donor template packaged in a vector) is administered by intracerebral ventricular injection. In some embodiments, the co-delivery system described herein (e.g., a gene editor construct packaged in a LNP and a donor template packaged in a vector) is administered by intracisternal magna administration. In some embodiments, the co-delivery system described herein (e.g., a gene editor construct packaged in a LNP and a donor template packaged in a vector) is administered by intravitreal injection.
[0362] Methods of non-viral delivery of the donor DNA template described herein include lipofection, nucleofection, microinjection, biolistics, virosomes, liposomes, immunoliposomes, polycation or lipidmucleic acid conjugates, naked DNA, artificial virions, and agent-enhanced uptake of DNA. Lipofection is described in e.g., U.S. Pat. Nos. 5,049,386, 4,946,787; and 4,897,355) and lipofection reagents are sold commercially (e.g., Transfectam™ and Lipofectin™). Cationic and neutral lipids that are suitable for efficient receptor-recognition lipofection of polynucleotides include those of Feigner, WO 91/17424; WO 91/16024. Delivery can be to cells (e.g. in vitro or ex vivo administration) or target tissues (e.g. in vivo administration).
5.13.1. mRNA delivery
[0363] Another useful method to deliver proteins, enzymes, and guides comprises transfection of messenger RNA (mRNA). Examples of mRNA delivery methods and compositions that may be utilized in the present disclosure including, for example, PCT/US2014/028330, US8822663B2, NZ700688A, ES2740248T3, EP2755693A4, EP2755986A4, WO2014152940A1, EP3450553B1, BR112016030852A2, and EP3362461A1. Expression of CRISPR systems in particular is described by W02020014577. Each of these publications are incorporated herein by reference in their entireties. Additional disclosure hereby incorporated by reference can be found in Kowalski et al., “Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery,” Mol Therap., 2019; 27(4): 710-728.
6. EXAMPLES
6.1. Example 1: Delivery of gene editor polynucleotide sequence packaged in LNP and donor template packaged in AAV
[0364] A gene editor polynucleotide construct is packaged into a LNP (FIG. 1), wherein the gene editor polynucleotide sequence comprises a polynucleotide sequence encoding a prime editor protein linked to an integrase via peptide linker a polynucleotide sequence encoding an attachment site-containing guide RNA (atgRNA); a polynucleotide sequence encoding a nickase guide RNA (ngRNA).
[0365] A donor template polynucleotide construct is packaged in an AAV vector (FIG. 2). [0366] Co-administration of the gene editor construct packaged LNP and the donor template packaged AAV co-delivers the gene editor construct to a cell cytoplasm and the donor template to a cell nucleus. By use of programmable genome editing to place integrase landing site at a desired location in the genome, the direct activity of the associated integrase to the specific genomic site is guided. Gene editor construct expression, with template co-delivery, results in integration of template “cargo” at a precisely defined target location.
6.2. Example 2: Delivery of gene editor polynucleotide sequence packaged in LNP and donor template capable of self-circularization packaged in AAV
[0367] A gene editor polynucleotide construct is packaged into a LNP (FIG. 1), wherein the gene editor polynucleotide sequence comprises a polynucleotide sequence encoding a prime editor protein linked to an integrase via peptide linker a polynucleotide sequence encoding an attachment site-containing guide RNA (atgRNA); a polynucleotide sequence encoding a nickase guide RNA (ngRNA).
[0368] A donor template polynucleotide construct is packaged in an AAV vector (FIG. 2).
[0369] Co-administration of the gene editor construct packaged LNP and the donor template packaged AAV co-delivers the gene editor construct to a cell cytoplasm and the donor template to a cell nucleus. Integrase-mediated self-circularization of donor template occurs at integration target recognition sites within the AAV genome (FIG. 3). By use of programmable genome editing to place an orthogonal integrase landing site (i.e., distinct att site from att sites used for selfcircularization) at a desired location in the genome, the direct activity of the associated integrase to the specific genomic site is guided. Gene editor construct expression, with template co-delivery and integrase-mediated circularization of template, results in integration of template “cargo” at a precisely defined target location.
6.3. Example 3: Delivery of gene editor polynucleotide sequence packaged in LNP and atgRNA, ngRNA, and donor template co-packaged in AAV
[0370] A gene editor polynucleotide construct is packaged into a LNP (FIG. 4), wherein the gene editor polynucleotide sequence comprises a polynucleotide sequence encoding a prime editor protein linked to an integrase via peptide linker.
[0371] A polynucleotide sequence encoding an attachment site-containing guide RNA (atgRNA), a polynucleotide sequence encoding a nicking guide RNA (ngRNA), and donor template are packaged in an AAV vector (FIG. 4). [0372] Co-administration of the gene editor construct packaged LNP and the atgRNA, ngRNA, donor template packaged AAV co-delivers the gene editor construct to a cell. Integrase- mediated self-circularization of donor template occurs at integration target recognition sites within the AAV genome (FIG. 3). By use of programmable genome editing to place an orthogonal integrase landing site (i.e., distinct att site from att sites used for self-circularization) at a desired location in the genome, the direct activity of the associated integrase to the specific genomic site is guided. Gene editor construct expression, with atgRNA, ngRNA, and template co-delivery and integrase-mediated circularization of template, results in integration of template “cargo” at a precisely defined target location.
6.4. Example 4: Delivery of gene editor polynucleotide sequence and ngRNA packaged in LNP and atgRNA and donor template co-packaged in AAV
[0373] A gene editor polynucleotide construct and a nicking guide RNA (ngRNA) are packaged into a LNP (FIG. 5), wherein the gene editor polynucleotide sequence comprises a polynucleotide sequence encoding a prime editor protein linked to an integrase via peptide linker.
[0374] A polynucleotide sequence encoding an attachment site-containing guide RNA (atgRNA) and donor template are packaged in an AAV vector (FIG. 5).
[0375] Co-administration of the gene editor construct and ngRNA packaged LNP and the atgRNA, donor template packaged AAV co-delivers the gene editor construct to a cell. Integrase- mediated self-circularization of donor template occurs at integration target recognition sites within the AAV genome (FIG. 3). By use of programmable genome editing to place an orthogonal integrase landing site (i.e., distinct att site from att sites used for self-circularization) at a desired location in the genome, the direct activity of the associated integrase to the specific genomic site is guided. Gene editor construct expression, with atgRNA, ngRNA, and template co-delivery and integrase-mediated circularization of template, results in integration of template “cargo” at a precisely defined target location.
6.5. Example 5: Intramolecular circularization of plasmid and packaged AAV genomes
[0376] Three self-complementary AAV (scAAV) genomes were designed and generated to verify recombinase/integrase-mediated intramolecular circularization of a DNA cargo from within a linear AAV genome (FIGs. 6A-6B). Circularization of a scAAV genome is mediated by one of Cre, FLPe (thermostable mutant), or Bxbl. Further, the scAAV genomes are comprised of a DNA cargo of interest (“payload”) and an attP site (GT central dinucleotide for circularization orthogonality) for gene insertion into a genome placed attB beacon site. Expected recombinase/integrase-mediated intramolecular circularization products are illustrated in FIG. 7. A universal ddPCR probe capable of binding any linear or circularized AAV genome was designed, wherein the universal ddPCR probe is designed to only give signal upon cognate recombinase/integrase mediated circularization (FIGs. 8A-8B). Circularization products are amplified by use of a circle junction PCR primer set that is designed to amplify only circular products due to primer direction constraints. To confirm Bxbl mediated circularization specifically, an attR scar quencher-fluorophore probe was designed. In addition, a template reference primer set was designed and generated to quantify total template DNA (linear or circular confirmation) (FIGs. 8A-8B).
[0377] Intracellular circularization of either plasmid or packaged AAV genomes were screened in HEK293 cells (35K cells per well) (FIG. 9). Plasmids (25 fmol pDNA = IX or 50 fmol pDNA = 2X) encoding one of Cre, FEPe, or Bxbl were transfected by Eipofectamine 3000. Plasmid genome substrates were transfected at a dose of 1E10 copies per well using Eipofectamine 3000 (FIG. 9). Additionally, AAV genomes were packaged in AAV-DJ capsids and delivered at a dose of 3E5 genomes per cell or 1E10 genomes per well. Circularization ddPCR analysis was conducted three days post transfection.
[0378] FIG. 10 demonstrates circularization of AAV pDNA and packaged AAV genomic DNA for both IX Bxbl and 2X Bxbl conditions (confirmed by use of attR ddPCR primer set). Further, replicates that lacked either Bxbl or AAV pDNA substrate demonstrated insignificant circularization. All three of the Cre-, FEPe-, and Bxbl -targeted AAV pDNA substrates demonstrated circularization upon cognate recombinase/integrase introduction, as confirmed by using the universal ddPCR probe (FIG. 11). Moreover, Cre-, FEPe-, and Bxbl -mediated circularization of packaged AAV DJ genomes substrates were demonstrated and confirmed using the universal ddPCR probe (FIG. 12).
[0379] As shown in FIG. 13, the Bxbl-mediated attR scar probe provided similar percent circularization quantification compared to the universal probe.
6.6. Example 6: In vitro beacon placement in primary mouse hepatocytes and primary human hepatocytes using mRNA and AAV for co-delivery
[0380] This example assessed the efficiency of in vitro beacon placement in primary human hepatocytes using mRNA delivering of a polynucleotide encoding a gene editor polynucleotide construct and AAV to deliver the first and second atgRNA. See FIG. 15 for a non-limiting example of a dual atgRNA-mediated insertion of an integration recognition site.
[0381] In the mouse experiments, the mRNA and AAV were delivered into the primary mouse hepatocytes (PMH) using (i) concurrent delivery (“co-dose”), (ii) AAV delivery followed by a “1- day delay” before delivery of the mRNA, or (iii) AAV delivery followed by a “2-day delay” before delivery of the mRNA. Beacon placement was then assessed using next-generation sequencing of DNA isolated from cells subjected to the delivery conditions mentioned above. The mRNA encoding the gene editor polynucleotide construct was delivered in various amounts per well: 2000 ng, 1000 ng, 500 ng, 250 ng, 125 ng, 62.5 ng, and 31.25 ng. AAV encoding the first and second atgRNA (see Table 12). The primary mouse hepatocyte data is shown in FIG. 16 and the human primary hepatocyte data is shown in FIG. 17. [0382] As shown in FIG. 16, in primary mouse hepatocytes (PMH) delivering the first atgRNAs (SEQ ID NO: 543) and the second atgRNA (SEQ ID NO: 544) using AAV at day 0 and then delivering the mRNA encoding the gene editing polynucleotide construct at day 2 (“2 day delay”) resulted in greater than 10% beacon placement for each amount of mRNA tested. Surprisingly, a 2 day delay resulted in greater beacon placement than either no delay (“co-dose) or a 1 day delay.
[0383] As shown in FIG. 17, in primary human hepatocytes (PHH), using AAV to deliver the first atgRNA (SEQ ID NO: 545) and the second atgRNA (SEQ ID NO: 546) and mRNA to deliver the gene editing polynucleotide construct resulted in about 17% beacon placement.
[0384] Taken together, this data showed robust ex vivo beacon placement in primary mouse and primary human hepatocytes.
6.7. Example 7: In vivo beacon placement with mRNA + AAV guide
[0385] In vivo beacon placement in mice was assessed using AAV to deliver the first and second atgRNAs and mRNA to delivery the gene editing polynucleotide construct.
[0386] In these experiments, mice were administered AAV containing the first atgRNA (SEQ ID NO: 543; Table 12) and the second atgRNA (SEQ ID NO: 544) targeting the Nolcl locus at 3E11 to 1E12 vector genomes (vg) per animal two 2 weeks prior to administration of the mRNA containing the gene editing polynucleotide construct (see FIG. 18). mRNA was delivered using various LNP formulations (e.g., LNP #F1 , LNP #F2, and LNP #F3) at concentrations ranging from 5 mg/kg to 0.5 mg/kg via intravenous injection (see FIG. 18). After delivery of the mRNA, liver tissue was harvested, genomic DNA was isolated, and beacon efficiency was assessed by NGS. As shown in FIG. 18, three conditions resulted in vivo beacon placement efficiency greater than 10%.
[0387] Taken together, this data provided proof-of-concept for successful in vivo beacon placement using AAV to deliver the first and second atgRNA and LNPs to deliver the mRNA encoding the gene editor polynucleotide construct.
6.8. Example 8: In vivo integration in mice using AAV to deliver the template polynucleotide and Adenovirus to deliver BxBl
[0388] In vivo integration efficiency in AttP mice was assessed using adenovirus to deliver an integrase (e.g., Bxbl) and an AAV to deliver the template polynucleotide. [0389] For these experiments, the adenovirus (i.e., adenovirus containing polynucleotide encoding the integrase) and the AAV (i.e., AAV containing the template polynucleotide and an attB site) were administered to mice containing dual AttP sites integrated in to the Rosa26 locus (B6.RosaBxb-GT/GA; female, Strain# 036152). The Rosa26 locus included a first AttP site comprising a GT dinucleotide and a second AttP site comprising a GA dinucleotide. The AAV was a scAAV8 containing a vector having a template polynucleotide and a 38 bp GT AttB site. The Adenovirus was an adenovirus-type 5 (Ad5) containing a polynucleotide encoding Bxbl (“Bxbl AdV”) (SEQ ID NO: 563; Table 14). Mice were administered the adenovirus and AAV according to the experimental details in Table 13.
[0390] Ten days after administration of the AdV and AAV viruses, liver punches were collected and genomic DNA was isolated. ddPCR of the genomic DNA was used to assess integration efficiency.
[0391] As shown in FIG. 19, administering the AAV and AdV resulted in in vivo integration of the donor polynucleotide template into the AttP mice. In particular, 3E10 vg/animal BxB 1 AdV resulted in about 7% in vivo integration efficiency (see FIG. 19). Administering increased amounts of BxB 1 AdV, 1E11 vg/animal, resulted in higher integration efficiency, about 11%, in AttP mice than with lower amount of 3E10 vg/animal (see FIG. 19).
[0392] Overall, this data establishes proof-of-concept for in vivo integration using an adenovirus to deliver and drive expression of Bxbl and an AAV to deliver the template polynucleotide to be integrated into a mammalian genome, in this case, the mouse genome.
6.9. Example 9: In vivo beacon placement in neonatal mice using split LNP
[0393] In vivo beacon placement was assessed in neonatal mice following administration of a single dose of a mixture of two LNPs. The first LNP contained mRNA encoding a prime editing system and a first synthetic atgRNA (atgRNAl). The mRNA and atgRNAl were included at 1:1 ratio in the first LNP. The second LNP contained mRNA encoding a prime editing system and a second synthetic atgRNA (atgRNA2). The mRNA and atgRNA2 were included at a 1:1 ratio in the second LNP. Each of the first and second atgRNAs targeted the mouse Nolcl locus and each encoded a portion of an integration recognition site (a “beacon”). AtgRNAl and atgRNA2 together included a 6bp overlap. The first and second LNPs were combined 1:1 as mixture prior to administration. The first atgRNA and second atgRNA are provide in Table 15, where the atgRNA include one or more 2’O-methyl modifications and one or more phosphorothioate linkages.
[0394] The LNP mixture was administered to the neonatal mice (2-5 day old CD-I mice) according to the experimental details in Table 16.
[0395] Eight days after administration of the LNP mixture in vivo beacon placement was assessed. In particular, at day 8 post administration, liver samples (either whole liver for groups 1-3 or liver punches from each lobe for groups 4-6 (see Table 13)) were collected and genomic DNA was isolated. Beacon placement was detected using ddPCR and NGS.
[0396] As shown in FIG. 20A, ddPCR revealed about 1% beacon placement (in Nolcl alleles) following administration of a 3mg/kg dose of the LNP mixture. Confirmation of beacon placement using NGS showed about 7% beacon placement (in Nolcl alleles) following administration of a 3 mg/kg dose of the LNP mixture (see FIG. 20B). In order to determine what percentage of the integrated beacons included the expected integration recognition site (“perfect beacon”), an NGS- based assay was used to make this assessment. As shown in FIG. 20C, about 1% of the integrated beacons contained the expected integration recognition site.
[0397] Neonates were also assessed at six weeks after administration of the LNP mixture. Beacon placement was detected using ddPCR and NGS. As shown in FIG. 21A., at six weeks post administration, ddPCR revealed about 4% beacon placement (in Nolcl alleles) for a 3mg/kg dose of the LNP mixture. Confirmation of beacon placement using NGS showed about 15% beacon placement (in Nolcl alleles) for a 3 mg/kg dose of the LNP mixture (see FIG. 21B). Assessment of the percent of integrated beacons that included the expected integration recognition site (“perfect beacon”) revealed that about 3.5% of beacons were comprised of perfect beacons (see FIG. 21C).
[0398] Overall, this data demonstrated successful in vivo site-specific integration of an integration recognition site. In particular, this data showed that a split LNP approach can be used for site- specifically integrating an integration recognition site in vivo in a mammalian genome, in this case neonatal mice.
6.10. Example 10: In vivo beacon placement in mice using split LNP
[0399] In vivo beacon placement was assessed in adult mice using a single dose mixture of two LNPs. The first LNP contained mRNA encoding a prime editing system and a first synthetic atgRNA (atgRNAl). The mRNA and atgRNAl were included at different ratios (e.g., 1:0.5, 1:1, and 1 :2) ratio in the first LNP. The second LNP contained mRNA encoding a prime editing system and a second synthetic atgRNA (atgRNA2). The mRNA and atgRNA2 were included at different ratios (e.g., 1:0.5, 1:1, and 1:2) ratio in the second LNP. Here, the first and second atgRNAs targeted mouse Factor IX (“mF9”) locus and each encoded a portion of an integration recognition site (“beacon”). Similar to Example 9, atgRNAl and atgRNA2 together included a 6bp overlap and were combined 1:1 as mixture prior to administration. The first atgRNA and second atgRNA are provide in Table 17, where the atgRNA include one or more 2’0-methyl modifications and one or more phosphorothioate linkages.
[0400] In particular, the LNP mixture was administered to female CD-I mice 6-8 weeks old according to the experimental details in Table 18. Table 18. Experimental details for in vivo beacon placement in adult mice
*1:0.25:0.25 = mRNA:atgRNAl 1 :0.5; mRNA:atgRNA2 1 :0.5; LNPs mixed 1 : 1 **1:0.5:0.5 = mRNA:atgRNAl 1 : 1; mRNA:atgRNA2 1 : 1; LNPs mixed 1 : 1
***1 : 1 : 1= mRNA:atgRNAl 1 :2; mRNA:atgRNA2 1 :2; LNPs mixed 1 : 1
[0401] Eight days after administration of the LNP mixture in vivo beacon placement was assessed. In particular, at day 8 post administration, liver samples (i.e., liver punches of each lobe (see Table 14)) were collected and genomic DNA was isolated. Beacon placement was detected using ddPCR and NGS.
[0402] As shown in FIG. 22A, ddPCR revealed about 0.8% beacon placement (in mF9 alleles) following administration of a 1:0.25:0.25 ratio of mRNA:atgRNAl:atgRNA2. Confirmation of beacon placement using NGS showed about 14% beacon placement (in mF9 alleles) following administration of the 1:0.25:0.25 ratio of mRNA:atgRNAl:atgRNA2 (see FIG. 22B). Similar to Example 9, an NGS-based assay was used to determined what percentages of the integrated beacons included the expected integration recognition site (“perfect beacon”). As shown in FIG. 22C, about 0.02% of the beacons placed in the mF9 locus were “perfect” beacons.
[0403] Overall, this data showed successful in vivo site-specific integration of an integration recognition site in adult mice. In particular, this data showed that the ratio of mRNA to atgRNA is an important consideration in determining efficacy of in vivo site- specific integration of an integration recognition site. 6.11. Example 11: Assessment of LNP formulations capable of targeting LSECs
[0404] A set of experiments were performed to identify LNP formulations capable of targeting LSECs. Methods of preparing LNPs are known in the art. Table 19 show the LNP formulations and the modifications tested. Table 19 also shows the size, polydispersity (PDI), and encapsulation efficiency (EE% as determined by a Ribogreen assay) for the LNP formulations. LNP were diluted 100-fold in lx PBS in cuvettes and analyzed using Dynamic Light Scattering (DLS) with a Malvern Zetasizer instrument and software to determine size and PDI. Encapsulation efficiency was calculated by analyzing the free and encapsulated RNA using the Ribogreen dye (ThermoFisher) according to manufacturer’s protocol. Control LNPs include the unmodified four lipid components while the DOPG, Mannose, and DOPG/Mannose groups indicate a phospholipid, PEG-lipid, or both substitutions in the LNP.
[0405] LNP compositions for these experiments included components at the following ratios: included: “cKKE12”: 40% cKKE12, 30% DOPE or DOPG, 28% Cholesterol, 2% PEG2K-DMG; “cKKE12-Mannose”: 40% cKKE12, 30% DOPE or DOPG, 27% cholesterol, 0.5% PEG2K-DMG, 2.5% Mannose-PEG2K-DSPE; “ALC0315”: 46.3% ALC0315, 9.4% DSPC or DSPG, 42.7% Cholesterol, 1.6% PEG2K-DMG; “ALC-Mannose”: 46.3% ALC0315, 9.4% DSPC or DSPG, 41.3% Cholesterol, 0.5% PEG2K-DMG, 2.5% Mannose-PEG2K-DSPE; “246C10”: 26.5% 246C10, 20% DOPE or DOPG, 50.5% Cholesterol, 3% C16-PEG2K-ceramide; and “246C10 Mannose”: 26.5% 246C10, 20% DOPE or DOPG, 50.5% Cholesterol, 0.5% C16-PEG2K- ceramide, 2.5% Mannose-PEG2K-DSPE.
[0406] Experimental conditions were carried out as described in FIG. 23 A and FIG. 23B.
[0407] FIGs. 24A-24B show representative images of liver sinusoidal endothelial cells (LSECs) contacted with the indicated LNP formulations: cKK (500 ng GFP); cKK-DOPG (500 ng GFP). Out of the four formulations, GFP expression was most robust when encapsulated and delivered using a cKK formulation. No GFP expression was observed in ALC (consistent with other in-vitro results in different cell types).
[0408] FIG. 25 shows representative images of LSECs contacted with the indicated LNP formulations: cKK-Mannose (500 ng GFP); or cKK-DOPG/Mannose (500 ng GFP). FIG. 26 shows representative images of the controls (i.e., LSECs transfected using with GFP lipofectamine or untreated LSECs).
[0409] Overall, this Data showed LNPs having the indicated formulations can be used to successfully target mouse LSECs in-vitro.
6.12. Example 12: Assessment of guide RNAs for targeting Factor VIII in LSECs
[0410] A set of experiments was performed in Hepa 1-6 cells to screen for guide RNA (gRNA) that target human factor VIII (“F8”). This experiment served, in part, as a proof of concept for using the indicated LNP formulations to deliver guide RNAs and polynucleotides encoding gene editor polynucleotides to LSECs. As described herein, F8 was targeted because it is expressed exclusively expressed in LSECs and when missing or defective, thereby causing Hemophilia A. Therefore, targeting F8 provided an opportunity to target a disease relevant gene in a disease relevant tissue using the optimized LNP formulations to do so.
[0411] In particular, Hepa 1-6 cells were plated at 15,000 cells per well and transfected one day post plating with five different F8 knockout (KO) guide RNAs (gRNAs) (see FIG. 27). F8 gRNAs were transfected at a concentration of 60 nM per guide. Cas9 was transfected at a concentration of 200 ng. gRNAs used are shown in Table 20.
[0412] FIG. 28 shows results of initial knockout guides screened in Hepa 1-6 cells. Successful F8 knockout was observed with all guides tested (see FIG. 28). Similar efficiencies were observed in guides 2, 3, and 4 (see FIG. 28). Guides 2, 3, and 4 were selected for further formulations studies (see Example 13).
6.13. Example 13: Assessment of LNP formulations to deliver gene editing reagents to LSECs in vitro
[0413] A proof-of-concept study was performed to assess use of various LNP formulations for delivering gRNA 2 (“G2”), gRNA 3 (“G3”) (data not shown) and gRNA 4 (“G4”) to LSECs.
[0414] In particular, LNPs encapsulating G2, G3, or G4 were added to the LSEC culture at four different concentrations (500 ng, 250 ng, 125 ng, and 62.5 ng). The cells were harvested 3 days after introduction of the LNPs and assessed for % indel formation.
[0415] The data showed that guide 4 (G4) had the strongest knockout among the 3 guides encapsulated in LNPs (see PIG. 29 and PIG. 30; data for guide 3 not shown). Notably, G4 was also had the highest % indel in the hepa 1-6 screen (see PIG. 28). PIG. 29 and PIG. 30 also showed that a cKK LNP formulation comprising a DOPG modification produced the most robust %indel among the different LNP modifications for each of the different concentrations tested. These results are consistent with the data in Example 11, which showed that LNP formulations having a DOPG modification (e.g., a cKK LNP formulation comprising a DOPG modification or a ALC LNP formulation comprising a DOPG modification) produced the most robust GPP expression. [0416] As shown in FIG. 31, LNPs with a modification (e.g., a PEG-modified lipid or a phospholipid (e.g., one of the indicated phospholipids)) are more efficient than unmodified LNPs at producing indels in LSEC. When comparing the ratio LSEC targeting to hepatocyte targeting, LNPs with a modification (e.g., a PEG-modified lipid or a phospholipid (e.g., one of the indicated phospholipids)) are clearly more efficient than unmodified LNPs at targeting LSECs.
[0417] FIG. 32 shows that the LNP modification (mod 1), a complete substitution/replacement of a neutrally charged phospholipid (DSPC) with a negatively charged phospholipid (DOPG), resulted much higher LSEC targeting efficiency compared to the hepatocyte targeting with the modified LNP.
[0418] Overall, this data showed successful delivery of gene editing components to LSECs using LNP formulations comprising DOPG modifications.
7. EQUIVALENTS AND INCORPORATION BY REFERENCE
[0419] All references cited herein are incorporated by reference to the same extent as if each individual publication, database entry (e.g. Genbank sequences or GenelD entries), patent application, or patent, was specifically and individually indicated incorporated by reference in its entirety, for all purposes. This statement of incorporation by reference is intended by Applicants, pursuant to 37 C.F.R. § 1.57(b)(1), to relate to each and every individual publication, database entry (e.g. Genbank sequences or GenelD entries), patent application, or patent, each of which is clearly identified in compliance with 37 C.F.R. § 1.57(b)(2), even if such citation is not immediately adjacent to a dedicated statement of incorporation by reference. The inclusion of dedicated statements of incorporation by reference, if any, within the specification does not in any way weaken this general statement of incorporation by reference. Citation of the references herein is not intended as an admission that the reference is pertinent prior art, nor does it constitute any admission as to the contents or date of these publications or documents.
[0420] It is an object of the invention not to encompass within the invention any previously known product, process of making the product, or method of using the product such that Applicant reserves the right and hereby disclose a disclaimer of any previously known product, process, or method. It is further noted that the invention does not intend to encompass within the scope of the invention any product, process, or making of the product or method of using the product, which does not meet the written description and enablement requirements of the USPTO (35 U.S.C. 112(a)) or the EPO (Article 83 of the EPC), such that Applicant reserves the right and hereby disclose a disclaimer of any previously described product, process of making the product, or method of using the product. It may be advantageous in the practice of the invention to be in compliance with Art. 53(c) EPC and Rule 28(b) and (c) EPC. Nothing herein is to be construed as a promise. It is noted that in this disclosure and particularly in the claims and/or paragraphs, terms such as "comprises", "comprised", "comprising" and the like can have the meaning attributed to it in U.S. Patent law; e.g., they can mean "includes", "included", "including", and the like; and that terms such as "consisting essentially of" and "consists essentially of" have the meaning ascribed to them in U.S. Patent law, e.g., they allow for elements not explicitly recited, but exclude elements that are found in the prior art or that affect a basic or novel characteristic of the invention.
[0421] While the invention has been particularly shown and described with reference to a preferred embodiment and various alternate embodiments, it is understood by persons skilled in the relevant art that various changes in form and details can be made therein without departing from the spirit and scope of the invention.
Claims
1. A composition comprising: a lipid nanoparticle (LNP) formulation comprising a plurality of LNPs, wherein each LNP comprises:
(i) an ionizable lipid;
(ii) a cholesterol; and
(iii) a polyethyleneglycol (PEG)-modified lipid; and
(iv) optionally a neutral lipid, each LNP encapsulates a cargo; wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (LSEC) the encapsulated cargo.
2. The composition of claim 1, wherein the cargo is selected from: an mRNA, an siRNA, an antisense oligonucleotide, a DNA, a guide RNA (gRNA), a protein, a peptide, a chemotherapeutic agent, a small molecule drug, or a combination thereof.
3. The composition of claim 2, wherein the cargo is an mRNA and a gRNA.
4. The composition of claim 3, wherein the mRNA is a gene editor polynucleotide encoding a gene editor polypeptide and the gRNA is an attachment site-containing guide RNA (atgRNA).
5. A composition comprising: a lipid nanoparticle (LNP) formulation comprising a plurality of LNPs, wherein each LNP comprises:
(i) a polyethylene glycol (PEG) -modified lipid;
(ii) a neutral lipid, or
(iii) both (i) and (ii); wherein each LNP encapsulates:
(i) a gene editor polynucleotide;
(ii) at least a first attachment- site containing guide RNA (atgRNA);
(iii) or both (i) and (ii), wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (LSEC) (i) the gene editor polynucleotide; (ii) the at least a first attachment-site containing guide RNA (atgRNA); or (iii) both (i) and (ii).
6. A lipid nanoparticle (LNP) composition comprising:
(a) a gene editor polynucleotide;
(b) at least a first attachment- site containing guide RNA (atgRNA);
(c) an ionizable lipid;
(d) a cholesterol; and
(e) a polyethylene glycol (PEG)-modified lipid; and
(f) optionally a neutral lipid, wherein the LNP formulation is capable of delivering into a liver sinusoidal endothelial cell (LSEC) the gene editor polynucleotide and the at least first attachment-site containing guide RNA (atgRNA).
7. The composition or LNP composition of any one of claims 1-6, wherein the ionizable lipid is an ionizable cationic lipid.
8. The composition or LNP composition of claim 7, wherein the ionizable cationic lipid is selected from N-(2,3dioleyloxy)propyl)-N,N,Ntrimethylammonium chloride (DOTMA); N-(2,3dioleoyloxy)propyl)-N,N,N-trimethylamntonium chloride (DODAP); N,N-dioleyl-N,N-dimethylammonium chloride (DODAC); 1,2 bis (oleoyloxy)-3-(trimethylammonio) propane (DOTAP); N,NdistearylN,N- dimethylammonium bromide (DDAB); 3-(N — (N,N-dimethylaminoethane)-carbam- oyl)cholesterol (DC-Choi); diheptadecylamidoglycylspermidine (DHGS) and N-(l,2- dimyristyloxyprop-3-yl)-N,N-dimethyl-N-hydoxyethyl ammonium bromide (DMRIE), or a combination thereof
9. The composition or LNP composition of any one of claims 1-8, further comprising a anionic lipid.
10. The composition or LNP composition of any one of claims 1-8, wherein the ionizable lipid is replaced with an anionic lipid.
11. The composition of LNP composition of claim 9 or 10, wherein the anionic lipid is selected from: a phospholipid, phospholipid derivative, a sterol, a fatty acid, or a combination thereof.
12. The composition or LNP composition of any one of claims 1-11, wherein the PEG- modified lipid is select from 1,2-Dimyristoyl-sn-glycerol methoxypolyethylene glycol (PEG-DMG), PEG-disteryl glycerol (PEG-DSG), PEG-dipalmetoleyl, PEG-dioleyl, PEG-distearyl, PEG-diacylglycamide (PEG-DAG), PEG-dipalmitoyl phosphatidylethanolamine (PEG-DPPE), and PEG-l,2-dimyristyloxlpropyl-3-amine (PEG-c-DMA), or a combination thereof.
13. The composition of LNP composition of any one of claims 1-12, wherein the PEG- modified lipid comprises a moiety capable binding to a receptor on a liver sinusoidal endothelial cell.
14. The composition or LNP composition of claim 13, wherein the moiety is capable of binding to a mannose receptor, a stabilin receptor, an Fc receptor, or a combination thereof.
15. The composition or LNP composition of claim 1-14, wherein the moiety is a mannose.
16. The composition or LNP composition of claim 15, wherein the PEG-modified lipid is a mannose-PEG lipid.
17. The composition or LNP composition of any one of claims 1-16, wherein the neutral lipid is a phospholipid.
18. The composition or LNP composition of claim 17, wherein the phospholipid is a natural phospholipid selected from: phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylglycerol (PG), phosphatidylserine (PS), phosphatidylinositol (PI), Phosphatidic acid (phosphatidate) (PA), dipalmitoylphosphatidylcholine, monoacyl-phosphatidylcholine (lyso PC), 1- palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), N-Acyl-PE, phosphoinositides, and phospho sphingolipids, or a combination thereof.
19. The composition or LNP composition of claim 16, wherein the phospholipid is a phospholipid derivative selected from: phosphatidic acid (DMPA, DPPA, DSPA), phosphatidylcholine (DDPC, DLPC, DMPC, DPPC, DSPC, DOPC, POPC, DEPC), phosphatidylglycerol (DMPG, DPPG, DSPG, DOPG), phosphatidylethanolamine (DMPE, DPPE, DSPE DOPE), and phosphatidylserine (DOPS), or a combination thereof.
20. The composition or LNP composition of claim 17, wherein the phospholipid is selected from: l,2-dioleoyl-sn-glycero-3-phospho-rac-(l-glycerol) sodium salt (DOPG); l,2-dilinoleoyl-sn-glycero-3-phosphocholine (DLPC); 1,2-dimyristoyl-sn- glycero-phosphocholine (DMPC); 1 ,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC); l,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC); 1,2-distearoyl-sn- glycero-3-phosphocholine (DSPC); 1 ,2-diundecanoyl-sn-glycero-phosphocholine (DUPC); l-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC); l,2-di-0- octadecenyl-£-glycero-3-phosphocholine (18:0 Diether PC); l-oleoyl-2- cholesterylhemisuccinoyl-glycero-3-phosphocholine (OChemsPC); 1-hexadecyl-sn- glycero-3-phosphocholine (Cl 6 Lyso PC); l,2-dilinolenoyl-sn-glycero-3- phosphocholine; l,2-diarachidonoyl-sn-glycero-3-phosphocholine; 1,2- didocosahexaenoyl-sn-glycero-3-phosphocholine; l,2-dioleoyl-sn-glycero-3- phosphoethanolamine (DOPE); l,2-diphytanoyl-sn-glycero-3-phosphoethanolamine (ME 16.0 PE); l,2-distearoyl-sn-glycero-3-phosphoethanolamine; 1,2-dilinoleoyl-sn- glycero-3-phosphoethanolamine; 1 ,2-dilinolenoyl-sn-glycero-3- phosphoethanolamine; 1 ,2-diarachidonoyl-sn-glycero-3-phosphoethanolamine; and l,2-didocosahexaenoyl-sn-glycero-3-phosphoethanolamine; and mixtures thereof.
21. The composition or LNP composition of any one of claims 18-20, wherein the phospholipid is DOPG.
22. The composition or LNP composition of any one of claims 1-21, wherein each LNP in the LNP formulation comprises: (i) modified PEGylated lipids; and (ii) neutral lipids.
23. The composition or LNP composition of claim 22, wherein each LNP in the LNP formulation comprises (i) PEG-modified lipids comprising a moiety capable binding to a receptor on a liver sinusoidal endothelial cell (LSEC); and (ii) phospholipids.
24. The composition or LNP composition of claim 22 or 23, wherein each LNP in the LNP formulation comprises mannose-PEG lipids and DOPGs.
25. The composition or LNP composition of any one of claims 1-24, wherein the LNP further comprises: cholesterol and an ionizable lipid.
26. The composition or LNP composition of any one of claims 1-25, wherein each LNP in the LNP formulation ranges from about 100 nm to about 200 nm.
27. The composition or LNP composition of any one of claims 1-25, wherein each LNP in the LNP formulation ranges from about 85 nm to about 120 nm.
28. The composition or LNP composition of any one of claims 1-25, wherein each LNP in the LNP formulation ranges from about 45 nm to about 65 nm.
29. The composition or LNP composition of any one of claims 1-25, wherein each LNP in the LNP formulation ranges from about 40 nm to about 50 nm.
30. The composition or LNP composition of any one of claims 1-29, wherein the encapsulation efficiency is about 80% to about 100%.
31. The composition or LNP composition of any one of claims 1-30, wherein the encapsulation efficiency is no less than 84%.
32. The composition or LNP composition of any one of claims 1-30, wherein the encapsulation efficiency is no less than 90%.
33. The composition or LNP composition of any one of claims 1-32, wherein the polydispersity index of the composition ranges from about 0.005 to 0.5.
34. The composition or LNP composition of any one of claims 1-33, wherein the polydispersity index of the composition ranges from about 0.025 to 0.4.
35. The composition or LNP composition of any one of claims 1-34, wherein the gene editor polynucleotide comprises a nucleotide sequence encoding a nickase and a nucleotide sequence encoding a reverse transcriptase.
36. The method of claim 35, wherein the nickase is linked to the reverse transcriptase by a linker.
37. The method of claim 36, wherein the first atgRNA comprises:
(i) a domain that is capable of guiding the nickase to a target sequence; and
(ii) a reverse transcriptase (RT) template that comprises at least a portion of an at least first integration recognition site, whereby the at least portion of the at least first integration recognition site is integrated into the genome of the cell at the target sequence.
38. The composition or LNP composition of any one of claims 4-37, wherein the LNPs further comprises a nicking gRNA.
39. The composition or LNP composition of any one of claims 4-37, wherein the LNPs further comprise a second atgRNA.
40. The composition or LNP composition of claim 39, wherein the first atgRNA and the second atgRNA are an at least first pair of atgRNAs, wherein the at least first pair of atgRNAs have domains that are capable of guiding the prime editor system to a target sequence; the first atgRNA further includes a first RT template that comprises at least a portion of the first integration recognition site; the second atgRNA further includes a second RT template that comprises at least a portion of the first integration recognition site, and the first atgRNA and the second atgRNAs collectively encode the entirety of the first integration recognition site, whereby the first integration recognition site is integrated into the genome of the cell at the target sequence.
41. The composition or LNP composition of any one of claims 4-40, further comprising an integration enzyme or a polynucleotide encoding an integration enzyme.
42. The composition or LNP composition of claim 41, wherein the integration enzyme or the polynucleotide encoding the integration enzyme is encapsulated in an LNP along with (i) a gene editor polynucleotide; (ii) at least a first attachment- site containing guide RNA (atgRNA); or (iii) both (i) and (ii),
43. The composition or LNP composition of any one of claims 4-42, further comprising a donor polynucleotide template.
44. A liver sinusoidal endothelial cell (LSEC) comprising the composition or LNP composition of any one of claims 1-43.
45. A genetically engineered liver sinusoidal endothelia cell (LSEC) made with the composition or LNP composition of any one of claims 1-43.
46. A method for site-specifically incorporating an integration recognition site into a liver sinusoidal endothelial cell (LSEC) genome at a target DNA sequence, comprising: incorporating an integration target recognition site into the genome by delivering into the cell: the composition or LNP composition of any one of claims 1-43.
47. A method for site- specifically integrating a donor polynucleotide template into a liver sinusoidal endothelial cell (LSEC) genome at a target DNA sequence, comprising incorporating an integration recognition site into the genome by delivering into the cell: the composition or LNP composition of any one of claims 1-43; and integrating the donor polynucleotide template into the genome by delivering into the cell: an integration enzyme; and a donor polynucleotide template, wherein the donor polynucleotide template is linked to a sequence that is an integration cognate of the integration recognition site present in the atgRNA, and wherein the donor polynucleotide template is integrated into the genome at the incorporated genomic integration recognition site by the integration enzyme.
48. A genetically engineered liver sinusoidal endothelia cell (LSEC) made using the composition or LNP composition of any one of claims 1-43 or the methods of claims 46 or 47.
Applications Claiming Priority (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| US202363501633P | 2023-05-11 | 2023-05-11 | |
| US63/501,633 | 2023-05-11 |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| WO2024234006A1true WO2024234006A1 (en) | 2024-11-14 |
Family
ID=91621254
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/US2024/029153PendingWO2024234006A1 (en) | 2023-05-11 | 2024-05-13 | Systems, compositions, and methods for targeting liver sinusodial endothelial cells (lsecs) |
Country Status (1)
| Country | Link |
|---|---|
| WO (1) | WO2024234006A1 (en) |
Citations (105)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US4186183A (en) | 1978-03-29 | 1980-01-29 | The United States Of America As Represented By The Secretary Of The Army | Liposome carriers in chemotherapy of leishmaniasis |
| US4217344A (en) | 1976-06-23 | 1980-08-12 | L'oreal | Compositions containing aqueous dispersions of lipid spheres |
| US4235871A (en) | 1978-02-24 | 1980-11-25 | Papahadjopoulos Demetrios P | Method of encapsulating biologically active materials in lipid vesicles |
| US4261975A (en) | 1979-09-19 | 1981-04-14 | Merck & Co., Inc. | Viral liposome particle |
| US4485054A (en) | 1982-10-04 | 1984-11-27 | Lipoderm Pharmaceuticals Limited | Method of encapsulating biologically active materials in multilamellar lipid vesicles (MLV) |
| US4501728A (en) | 1983-01-06 | 1985-02-26 | Technology Unlimited, Inc. | Masking of liposomes from RES recognition |
| US4774085A (en) | 1985-07-09 | 1988-09-27 | 501 Board of Regents, Univ. of Texas | Pharmaceutical administration systems containing a mixture of immunomodulators |
| US4797368A (en) | 1985-03-15 | 1989-01-10 | The United States Of America As Represented By The Department Of Health And Human Services | Adeno-associated virus as eukaryotic expression vector |
| US4837028A (en) | 1986-12-24 | 1989-06-06 | Liposome Technology, Inc. | Liposomes with enhanced circulation time |
| US4897355A (en) | 1985-01-07 | 1990-01-30 | Syntex (U.S.A.) Inc. | N[ω,(ω-1)-dialkyloxy]- and N-[ω,(ω-1)-dialkenyloxy]-alk-1-yl-N,N,N-tetrasubstituted ammonium lipids and uses therefor |
| US4946787A (en) | 1985-01-07 | 1990-08-07 | Syntex (U.S.A.) Inc. | N-(ω,(ω-1)-dialkyloxy)- and N-(ω,(ω-1)-dialkenyloxy)-alk-1-yl-N,N,N-tetrasubstituted ammonium lipids and uses therefor |
| US5013556A (en) | 1989-10-20 | 1991-05-07 | Liposome Technology, Inc. | Liposomes with enhanced circulation time |
| US5049386A (en) | 1985-01-07 | 1991-09-17 | Syntex (U.S.A.) Inc. | N-ω,(ω-1)-dialkyloxy)- and N-(ω,(ω-1)-dialkenyloxy)Alk-1-YL-N,N,N-tetrasubstituted ammonium lipids and uses therefor |
| WO1991016024A1 (en) | 1990-04-19 | 1991-10-31 | Vical, Inc. | Cationic lipids for intracellular delivery of biologically active molecules |
| WO1991017424A1 (en) | 1990-05-03 | 1991-11-14 | Vical, Inc. | Intracellular delivery of biologically active substances by means of self-assembling lipid complexes |
| US5173414A (en) | 1990-10-30 | 1992-12-22 | Applied Immune Sciences, Inc. | Production of recombinant adeno-associated virus vectors |
| WO1993024641A2 (en) | 1992-06-02 | 1993-12-09 | The United States Of America, As Represented By The Secretary, Department Of Health & Human Services | Adeno-associated virus with inverted terminal repeat sequences as promoter |
| US5846946A (en) | 1996-06-14 | 1998-12-08 | Pasteur Merieux Serums Et Vaccins | Compositions and methods for administering Borrelia DNA |
| US6027726A (en) | 1994-09-30 | 2000-02-22 | Inex Phamaceuticals Corp. | Glycosylated protein-liposome conjugates and methods for their preparation |
| US20030087817A1 (en) | 1999-01-12 | 2003-05-08 | Sangamo Biosciences, Inc. | Regulation of endogenous gene expression in cells using zinc finger proteins |
| US20040142025A1 (en) | 2002-06-28 | 2004-07-22 | Protiva Biotherapeutics Ltd. | Liposomal apparatus and manufacturing methods |
| US20040171156A1 (en) | 1995-06-07 | 2004-09-02 | Invitrogen Corporation | Recombinational cloning using nucleic acids having recombination sites |
| US20070042031A1 (en) | 2005-07-27 | 2007-02-22 | Protiva Biotherapeutics, Inc. | Systems and methods for manufacturing liposomes |
| US8404658B2 (en) | 2007-12-31 | 2013-03-26 | Nanocor Therapeutics, Inc. | RNA interference for the treatment of heart failure |
| US8454972B2 (en) | 2004-07-16 | 2013-06-04 | The United States Of America, As Represented By The Secretary, Department Of Health And Human Services | Method for inducing a multiclade immune response against HIV utilizing a multigene and multiclade immunogen |
| WO2013086354A1 (en) | 2011-12-07 | 2013-06-13 | Alnylam Pharmaceuticals, Inc. | Biodegradable lipids for the delivery of active agents |
| WO2013116126A1 (en) | 2012-02-01 | 2013-08-08 | Merck Sharp & Dohme Corp. | Novel low molecular weight, biodegradable cationic lipids for oligonucleotide delivery |
| US8691750B2 (en) | 2011-05-17 | 2014-04-08 | Axolabs Gmbh | Lipids and compositions for intracellular delivery of biologically active compounds |
| US8697359B1 (en) | 2012-12-12 | 2014-04-15 | The Broad Institute, Inc. | CRISPR-Cas systems and methods for altering expression of gene products |
| EP2755986A1 (en) | 2011-09-12 | 2014-07-23 | Moderna Therapeutics, Inc. | Engineered nucleic acids and methods of use thereof |
| EP2755693A2 (en) | 2011-09-12 | 2014-07-23 | Moderna Therapeutics, Inc. | Engineered nucleic acids and methods of use thereof |
| US8795965B2 (en) | 2012-12-12 | 2014-08-05 | The Broad Institute, Inc. | CRISPR-Cas component systems, methods and compositions for sequence manipulation |
| US8822663B2 (en) | 2010-08-06 | 2014-09-02 | Moderna Therapeutics, Inc. | Engineered nucleic acids and methods of use thereof |
| WO2014152940A1 (en) | 2013-03-14 | 2014-09-25 | Shire Human Genetic Therapies, Inc. | Mrna therapeutic compositions and use to treat diseases and disorders |
| US8865406B2 (en) | 2012-12-12 | 2014-10-21 | The Broad Institute Inc. | Engineering and optimization of improved systems, methods and enzyme compositions for sequence manipulation |
| US8889356B2 (en) | 2012-12-12 | 2014-11-18 | The Broad Institute Inc. | CRISPR-Cas nickase systems, methods and compositions for sequence manipulation in eukaryotes |
| US8906616B2 (en) | 2012-12-12 | 2014-12-09 | The Broad Institute Inc. | Engineering of systems, methods and optimized guide compositions for sequence manipulation |
| US8993233B2 (en) | 2012-12-12 | 2015-03-31 | The Broad Institute Inc. | Engineering and optimization of systems, methods and compositions for sequence manipulation with functional domains |
| WO2015056756A1 (en) | 2013-10-18 | 2015-04-23 | 国立大学法人熊本大学 | Method of inducing kidney from pluripotent stem cells |
| US9023649B2 (en) | 2012-12-17 | 2015-05-05 | President And Fellows Of Harvard College | RNA-guided human genome engineering |
| US9068179B1 (en) | 2013-12-12 | 2015-06-30 | President And Fellows Of Harvard College | Methods for correcting presenilin point mutations |
| US9074199B1 (en) | 2013-11-19 | 2015-07-07 | President And Fellows Of Harvard College | Mutant Cas9 proteins |
| US9163284B2 (en) | 2013-08-09 | 2015-10-20 | President And Fellows Of Harvard College | Methods for identifying a target site of a Cas9 nuclease |
| US9228207B2 (en) | 2013-09-06 | 2016-01-05 | President And Fellows Of Harvard College | Switchable gRNAs comprising aptamers |
| US9267135B2 (en) | 2013-06-04 | 2016-02-23 | President And Fellows Of Harvard College | RNA-guided transcriptional regulation |
| NZ700688A (en) | 2009-12-01 | 2016-02-26 | Shire Human Genetic Therapies | Delivery of mrna for the augmentation of proteins and enzymes in human genetic diseases |
| US9322006B2 (en) | 2011-07-22 | 2016-04-26 | President And Fellows Of Harvard College | Evaluation and improvement of nuclease cleavage specificity |
| US9322037B2 (en) | 2013-09-06 | 2016-04-26 | President And Fellows Of Harvard College | Cas9-FokI fusion proteins and uses thereof |
| US9359599B2 (en) | 2013-08-22 | 2016-06-07 | President And Fellows Of Harvard College | Engineered transcription activator-like effector (TALE) domains and uses thereof |
| US9405700B2 (en) | 2010-11-04 | 2016-08-02 | Sonics, Inc. | Methods and apparatus for virtualization in an integrated circuit |
| US9526784B2 (en) | 2013-09-06 | 2016-12-27 | President And Fellows Of Harvard College | Delivery system for functional nucleases |
| US9587252B2 (en) | 2013-07-10 | 2017-03-07 | President And Fellows Of Harvard College | Orthogonal Cas9 proteins for RNA-guided gene regulation and editing |
| US9777262B2 (en) | 2013-03-13 | 2017-10-03 | President And Fellows Of Harvard College | Mutants of Cre recombinase |
| US9790490B2 (en) | 2015-06-18 | 2017-10-17 | The Broad Institute Inc. | CRISPR enzymes and systems |
| BR112016030852A2 (en) | 2014-07-02 | 2018-01-16 | Shire Human Genetic Therapies | rna messenger encapsulation |
| US9914939B2 (en) | 2013-07-26 | 2018-03-13 | President And Fellows Of Harvard College | Genome engineering |
| US10000772B2 (en) | 2012-05-25 | 2018-06-19 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| EP3362461A1 (en) | 2015-10-16 | 2018-08-22 | Modernatx, Inc. | Mrna cap analogs with modified phosphate linkage |
| US10077453B2 (en) | 2014-07-30 | 2018-09-18 | President And Fellows Of Harvard College | CAS9 proteins including ligand-dependent inteins |
| US10113163B2 (en) | 2016-08-03 | 2018-10-30 | President And Fellows Of Harvard College | Adenosine nucleobase editors and uses thereof |
| US10167457B2 (en) | 2015-10-23 | 2019-01-01 | President And Fellows Of Harvard College | Nucleobase editors and uses thereof |
| US10190137B2 (en) | 2013-11-07 | 2019-01-29 | Editas Medicine, Inc. | CRISPR-related methods and compositions with governing gRNAS |
| US10266886B2 (en) | 2016-12-09 | 2019-04-23 | The Broad Institute, Inc. | CRISPR effector system based diagnostics |
| US10377998B2 (en) | 2013-12-12 | 2019-08-13 | The Broad Institute, Inc. | CRISPR-CAS systems and methods for altering expression of gene products, structural information and inducible modular CAS enzymes |
| US10375938B2 (en) | 2015-10-08 | 2019-08-13 | President And Fellows Of Harvard College | Multiplexed genome editing |
| US10385336B2 (en) | 2014-09-05 | 2019-08-20 | Vilnius University | Programmable RNA shredding by the type III-A CRISPR-Cas system of Streptococcus thermophilus |
| US10494621B2 (en) | 2015-06-18 | 2019-12-03 | The Broad Institute, Inc. | Crispr enzyme mutations reducing off-target effects |
| EP3450553B1 (en) | 2014-03-24 | 2019-12-25 | Translate Bio, Inc. | Mrna therapy for treatment of ocular diseases |
| US10519454B2 (en) | 2014-08-06 | 2019-12-31 | Toolgen Incorporated | Genome editing using Campylobacter jejuni CRISPR/CAS system-derived RGEN |
| WO2020014577A1 (en) | 2018-07-13 | 2020-01-16 | Allele Biotechnology And Pharmaceuticals, Inc. | Methods of achieving high specificity of genome editing |
| US10550372B2 (en) | 2013-12-12 | 2020-02-04 | The Broad Institute, Inc. | Systems, methods and compositions for sequence manipulation with optimized functional CRISPR-Cas systems |
| ES2740248T3 (en) | 2011-06-08 | 2020-02-05 | Translate Bio Inc | Lipid nanoparticle compositions and methods for mRNA administration |
| US10577630B2 (en) | 2013-06-17 | 2020-03-03 | The Broad Institute, Inc. | Delivery and use of the CRISPR-Cas systems, vectors and compositions for hepatic targeting and therapy |
| US10612011B2 (en) | 2015-07-30 | 2020-04-07 | President And Fellows Of Harvard College | Evolution of TALENs |
| US20200109398A1 (en) | 2018-08-28 | 2020-04-09 | Flagship Pioneering, Inc. | Methods and compositions for modulating a genome |
| US10648020B2 (en) | 2015-06-18 | 2020-05-12 | The Broad Institute, Inc. | CRISPR enzymes and systems |
| US10689691B2 (en) | 2014-12-19 | 2020-06-23 | The Broad Institute, Inc. | Unbiased identification of double-strand breaks and genomic rearrangement by genome-wide insert capture sequencing |
| US10711285B2 (en) | 2013-06-17 | 2020-07-14 | The Broad Institute, Inc. | Optimized CRISPR-Cas double nickase systems, methods and compositions for sequence manipulation |
| US10731181B2 (en) | 2012-12-06 | 2020-08-04 | Sigma, Aldrich Co. LLC | CRISPR-based genome modification and regulation |
| US10745677B2 (en) | 2016-12-23 | 2020-08-18 | President And Fellows Of Harvard College | Editing of CCR5 receptor gene to protect against HIV infection |
| US10781444B2 (en) | 2013-06-17 | 2020-09-22 | The Broad Institute, Inc. | Functional genomics using CRISPR-Cas systems, compositions, methods, screens and applications thereof |
| WO2020191153A2 (en) | 2019-03-19 | 2020-09-24 | The Broad Institute, Inc. | Methods and compositions for editing nucleotide sequences |
| US10787684B2 (en) | 2013-11-19 | 2020-09-29 | President And Fellows Of Harvard College | Large gene excision and insertion |
| US10851380B2 (en) | 2012-10-23 | 2020-12-01 | Toolgen Incorporated | Methods for cleaving a target DNA using a guide RNA specific for the target DNA and Cas protein-encoding nucleic acid or Cas protein |
| US10851357B2 (en) | 2013-12-12 | 2020-12-01 | The Broad Institute, Inc. | Compositions and methods of use of CRISPR-Cas systems in nucleotide repeat disorders |
| US10851369B2 (en) | 2016-06-21 | 2020-12-01 | President And Fellows Of Harvard College | Frequency-based modulation of diverse species in a nucleic acid library |
| US10930367B2 (en) | 2012-12-12 | 2021-02-23 | The Broad Institute, Inc. | Methods, models, systems, and apparatus for identifying target sequences for Cas enzymes or CRISPR-Cas systems for target sequences and conveying results thereof |
| US10946108B2 (en) | 2013-06-17 | 2021-03-16 | The Broad Institute, Inc. | Delivery, use and therapeutic applications of the CRISPR-Cas systems and compositions for targeting disorders and diseases using viral components |
| US10954514B2 (en) | 2014-12-12 | 2021-03-23 | The Broad Institute, Inc. | Escorted and functionalized guides for CRISPR-Cas systems |
| US10968257B2 (en) | 2018-04-03 | 2021-04-06 | The Broad Institute, Inc. | Target recognition motifs and uses thereof |
| US11001829B2 (en) | 2014-09-25 | 2021-05-11 | The Broad Institute, Inc. | Functional screening with optimized functional CRISPR-Cas systems |
| US11008588B2 (en) | 2013-06-17 | 2021-05-18 | The Broad Institute, Inc. | Delivery, engineering and optimization of tandem guide systems, methods and compositions for sequence manipulation |
| US11021740B2 (en) | 2017-03-15 | 2021-06-01 | The Broad Institute, Inc. | Devices for CRISPR effector system based diagnostics |
| US11041173B2 (en) | 2012-12-12 | 2021-06-22 | The Broad Institute, Inc. | Delivery, engineering and optimization of systems, methods and compositions for sequence manipulation and therapeutic applications |
| US11060115B2 (en) | 2015-06-18 | 2021-07-13 | The Broad Institute, Inc. | CRISPR enzymes and systems |
| US11071790B2 (en) | 2014-10-29 | 2021-07-27 | Massachusetts Eye And Ear Infirmary | Method for efficient delivery of therapeutic molecules in vitro and in vivo |
| US11085072B2 (en) | 2016-08-31 | 2021-08-10 | President And Fellows Of Harvard College | Methods of generating libraries of nucleic acid sequences for detection via fluorescent in situ sequencing |
| US11104937B2 (en) | 2017-03-15 | 2021-08-31 | The Broad Institute, Inc. | CRISPR effector system based diagnostics |
| US11104967B2 (en) | 2015-07-22 | 2021-08-31 | President And Fellows Of Harvard College | Evolution of site-specific recombinases |
| US11111521B2 (en) | 2011-12-22 | 2021-09-07 | President And Fellows Of Harvard College | Compositions and methods for analyte detection |
| US11111472B2 (en) | 2014-10-31 | 2021-09-07 | Massachusetts Institute Of Technology | Delivery of biomolecules to immune cells |
| WO2021226558A1 (en) | 2020-05-08 | 2021-11-11 | The Broad Institute, Inc. | Methods and compositions for simultaneous editing of both strands of a target double-stranded nucleotide sequence |
| WO2022087235A1 (en) | 2020-10-21 | 2022-04-28 | Massachusetts Institute Of Technology | Systems, methods, and compositions for site-specific genetic engineering using programmable addition via site-specific targeting elements (paste) |
| WO2022139528A1 (en)* | 2020-12-24 | 2022-06-30 | (주)인핸스드바이오 | Lipid nanoparticles comprising mannose or uses thereof |
| WO2022241040A1 (en)* | 2021-05-11 | 2022-11-17 | Seattle Children's Hospital D/B/A Seattle Children's Research Institute | Gene editing for expression of functional factor viii for the treatment of hemophilia |
- 2024
- 2024-05-13WOPCT/US2024/029153patent/WO2024234006A1/enactivePending
Patent Citations (209)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US4217344A (en) | 1976-06-23 | 1980-08-12 | L'oreal | Compositions containing aqueous dispersions of lipid spheres |
| US4235871A (en) | 1978-02-24 | 1980-11-25 | Papahadjopoulos Demetrios P | Method of encapsulating biologically active materials in lipid vesicles |
| US4186183A (en) | 1978-03-29 | 1980-01-29 | The United States Of America As Represented By The Secretary Of The Army | Liposome carriers in chemotherapy of leishmaniasis |
| US4261975A (en) | 1979-09-19 | 1981-04-14 | Merck & Co., Inc. | Viral liposome particle |
| US4485054A (en) | 1982-10-04 | 1984-11-27 | Lipoderm Pharmaceuticals Limited | Method of encapsulating biologically active materials in multilamellar lipid vesicles (MLV) |
| US4501728A (en) | 1983-01-06 | 1985-02-26 | Technology Unlimited, Inc. | Masking of liposomes from RES recognition |
| US5049386A (en) | 1985-01-07 | 1991-09-17 | Syntex (U.S.A.) Inc. | N-ω,(ω-1)-dialkyloxy)- and N-(ω,(ω-1)-dialkenyloxy)Alk-1-YL-N,N,N-tetrasubstituted ammonium lipids and uses therefor |
| US4897355A (en) | 1985-01-07 | 1990-01-30 | Syntex (U.S.A.) Inc. | N[ω,(ω-1)-dialkyloxy]- and N-[ω,(ω-1)-dialkenyloxy]-alk-1-yl-N,N,N-tetrasubstituted ammonium lipids and uses therefor |
| US4946787A (en) | 1985-01-07 | 1990-08-07 | Syntex (U.S.A.) Inc. | N-(ω,(ω-1)-dialkyloxy)- and N-(ω,(ω-1)-dialkenyloxy)-alk-1-yl-N,N,N-tetrasubstituted ammonium lipids and uses therefor |
| US4797368A (en) | 1985-03-15 | 1989-01-10 | The United States Of America As Represented By The Department Of Health And Human Services | Adeno-associated virus as eukaryotic expression vector |
| US4774085A (en) | 1985-07-09 | 1988-09-27 | 501 Board of Regents, Univ. of Texas | Pharmaceutical administration systems containing a mixture of immunomodulators |
| US4837028A (en) | 1986-12-24 | 1989-06-06 | Liposome Technology, Inc. | Liposomes with enhanced circulation time |
| US5013556A (en) | 1989-10-20 | 1991-05-07 | Liposome Technology, Inc. | Liposomes with enhanced circulation time |
| WO1991016024A1 (en) | 1990-04-19 | 1991-10-31 | Vical, Inc. | Cationic lipids for intracellular delivery of biologically active molecules |
| WO1991017424A1 (en) | 1990-05-03 | 1991-11-14 | Vical, Inc. | Intracellular delivery of biologically active substances by means of self-assembling lipid complexes |
| US5173414A (en) | 1990-10-30 | 1992-12-22 | Applied Immune Sciences, Inc. | Production of recombinant adeno-associated virus vectors |
| WO1993024641A2 (en) | 1992-06-02 | 1993-12-09 | The United States Of America, As Represented By The Secretary, Department Of Health & Human Services | Adeno-associated virus with inverted terminal repeat sequences as promoter |
| US6027726A (en) | 1994-09-30 | 2000-02-22 | Inex Phamaceuticals Corp. | Glycosylated protein-liposome conjugates and methods for their preparation |
| US20040171156A1 (en) | 1995-06-07 | 2004-09-02 | Invitrogen Corporation | Recombinational cloning using nucleic acids having recombination sites |
| US5846946A (en) | 1996-06-14 | 1998-12-08 | Pasteur Merieux Serums Et Vaccins | Compositions and methods for administering Borrelia DNA |
| US20030087817A1 (en) | 1999-01-12 | 2003-05-08 | Sangamo Biosciences, Inc. | Regulation of endogenous gene expression in cells using zinc finger proteins |
| US20040142025A1 (en) | 2002-06-28 | 2004-07-22 | Protiva Biotherapeutics Ltd. | Liposomal apparatus and manufacturing methods |
| US8454972B2 (en) | 2004-07-16 | 2013-06-04 | The United States Of America, As Represented By The Secretary, Department Of Health And Human Services | Method for inducing a multiclade immune response against HIV utilizing a multigene and multiclade immunogen |
| US20070042031A1 (en) | 2005-07-27 | 2007-02-22 | Protiva Biotherapeutics, Inc. | Systems and methods for manufacturing liposomes |
| US8404658B2 (en) | 2007-12-31 | 2013-03-26 | Nanocor Therapeutics, Inc. | RNA interference for the treatment of heart failure |
| NZ700688A (en) | 2009-12-01 | 2016-02-26 | Shire Human Genetic Therapies | Delivery of mrna for the augmentation of proteins and enzymes in human genetic diseases |
| US8822663B2 (en) | 2010-08-06 | 2014-09-02 | Moderna Therapeutics, Inc. | Engineered nucleic acids and methods of use thereof |
| US9405700B2 (en) | 2010-11-04 | 2016-08-02 | Sonics, Inc. | Methods and apparatus for virtualization in an integrated circuit |
| US8691750B2 (en) | 2011-05-17 | 2014-04-08 | Axolabs Gmbh | Lipids and compositions for intracellular delivery of biologically active compounds |
| ES2740248T3 (en) | 2011-06-08 | 2020-02-05 | Translate Bio Inc | Lipid nanoparticle compositions and methods for mRNA administration |
| US9322006B2 (en) | 2011-07-22 | 2016-04-26 | President And Fellows Of Harvard College | Evaluation and improvement of nuclease cleavage specificity |
| US10323236B2 (en) | 2011-07-22 | 2019-06-18 | President And Fellows Of Harvard College | Evaluation and improvement of nuclease cleavage specificity |
| EP2755986A1 (en) | 2011-09-12 | 2014-07-23 | Moderna Therapeutics, Inc. | Engineered nucleic acids and methods of use thereof |
| EP2755693A2 (en) | 2011-09-12 | 2014-07-23 | Moderna Therapeutics, Inc. | Engineered nucleic acids and methods of use thereof |
| WO2013086354A1 (en) | 2011-12-07 | 2013-06-13 | Alnylam Pharmaceuticals, Inc. | Biodegradable lipids for the delivery of active agents |
| US11111521B2 (en) | 2011-12-22 | 2021-09-07 | President And Fellows Of Harvard College | Compositions and methods for analyte detection |
| WO2013116126A1 (en) | 2012-02-01 | 2013-08-08 | Merck Sharp & Dohme Corp. | Novel low molecular weight, biodegradable cationic lipids for oligonucleotide delivery |
| US10988780B2 (en) | 2012-05-25 | 2021-04-27 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10513712B2 (en) | 2012-05-25 | 2019-12-24 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10400253B2 (en) | 2012-05-25 | 2019-09-03 | The Regents Of The University Of California | Methods and compositions or RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10385360B2 (en) | 2012-05-25 | 2019-08-20 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10415061B2 (en) | 2012-05-25 | 2019-09-17 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10421980B2 (en) | 2012-05-25 | 2019-09-24 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10428352B2 (en) | 2012-05-25 | 2019-10-01 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10358659B2 (en) | 2012-05-25 | 2019-07-23 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10358658B2 (en) | 2012-05-25 | 2019-07-23 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US11028412B2 (en) | 2012-05-25 | 2021-06-08 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US11008589B2 (en) | 2012-05-25 | 2021-05-18 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US11008590B2 (en) | 2012-05-25 | 2021-05-18 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US11001863B2 (en) | 2012-05-25 | 2021-05-11 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10407697B2 (en) | 2012-05-25 | 2019-09-10 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10988782B2 (en) | 2012-05-25 | 2021-04-27 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10982230B2 (en) | 2012-05-25 | 2021-04-20 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10351878B2 (en) | 2012-05-25 | 2019-07-16 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10337029B2 (en) | 2012-05-25 | 2019-07-02 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10982231B2 (en) | 2012-05-25 | 2021-04-20 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10308961B2 (en) | 2012-05-25 | 2019-06-04 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10900054B2 (en) | 2012-05-25 | 2021-01-26 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10301651B2 (en) | 2012-05-25 | 2019-05-28 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10793878B1 (en) | 2012-05-25 | 2020-10-06 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10443076B2 (en) | 2012-05-25 | 2019-10-15 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10774344B1 (en) | 2012-05-25 | 2020-09-15 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10752920B2 (en) | 2012-05-25 | 2020-08-25 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10487341B2 (en) | 2012-05-25 | 2019-11-26 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10676759B2 (en) | 2012-05-25 | 2020-06-09 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10669560B2 (en) | 2012-05-25 | 2020-06-02 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10266850B2 (en) | 2012-05-25 | 2019-04-23 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10519467B2 (en) | 2012-05-25 | 2019-12-31 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10640791B2 (en) | 2012-05-25 | 2020-05-05 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10626419B2 (en) | 2012-05-25 | 2020-04-21 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10612045B2 (en) | 2012-05-25 | 2020-04-07 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10597680B2 (en) | 2012-05-25 | 2020-03-24 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10577631B2 (en) | 2012-05-25 | 2020-03-03 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10000772B2 (en) | 2012-05-25 | 2018-06-19 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10570419B2 (en) | 2012-05-25 | 2020-02-25 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10563227B2 (en) | 2012-05-25 | 2020-02-18 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10227611B2 (en) | 2012-05-25 | 2019-03-12 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10113167B2 (en) | 2012-05-25 | 2018-10-30 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10550407B2 (en) | 2012-05-25 | 2020-02-04 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10533190B2 (en) | 2012-05-25 | 2020-01-14 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10526619B2 (en) | 2012-05-25 | 2020-01-07 | The Regents Of The University Of California | Methods and compositions for RNA-directed target DNA modification and for RNA-directed modulation of transcription |
| US10851380B2 (en) | 2012-10-23 | 2020-12-01 | Toolgen Incorporated | Methods for cleaving a target DNA using a guide RNA specific for the target DNA and Cas protein-encoding nucleic acid or Cas protein |
| US10731181B2 (en) | 2012-12-06 | 2020-08-04 | Sigma, Aldrich Co. LLC | CRISPR-based genome modification and regulation |
| US8771945B1 (en) | 2012-12-12 | 2014-07-08 | The Broad Institute, Inc. | CRISPR-Cas systems and methods for altering expression of gene products |
| US8889356B2 (en) | 2012-12-12 | 2014-11-18 | The Broad Institute Inc. | CRISPR-Cas nickase systems, methods and compositions for sequence manipulation in eukaryotes |
| US8889418B2 (en) | 2012-12-12 | 2014-11-18 | The Broad Institute Inc. | Engineering and optimization of improved systems, methods and enzyme compositions for sequence manipulation |
| US8865406B2 (en) | 2012-12-12 | 2014-10-21 | The Broad Institute Inc. | Engineering and optimization of improved systems, methods and enzyme compositions for sequence manipulation |
| US9840713B2 (en) | 2012-12-12 | 2017-12-12 | The Broad Institute Inc. | CRISPR-Cas component systems, methods and compositions for sequence manipulation |
| US10930367B2 (en) | 2012-12-12 | 2021-02-23 | The Broad Institute, Inc. | Methods, models, systems, and apparatus for identifying target sequences for Cas enzymes or CRISPR-Cas systems for target sequences and conveying results thereof |
| US8795965B2 (en) | 2012-12-12 | 2014-08-05 | The Broad Institute, Inc. | CRISPR-Cas component systems, methods and compositions for sequence manipulation |
| US8697359B1 (en) | 2012-12-12 | 2014-04-15 | The Broad Institute, Inc. | CRISPR-Cas systems and methods for altering expression of gene products |
| US8871445B2 (en) | 2012-12-12 | 2014-10-28 | The Broad Institute Inc. | CRISPR-Cas component systems, methods and compositions for sequence manipulation |
| US9822372B2 (en) | 2012-12-12 | 2017-11-21 | The Broad Institute Inc. | CRISPR-Cas component systems, methods and compositions for sequence manipulation |
| US11041173B2 (en) | 2012-12-12 | 2021-06-22 | The Broad Institute, Inc. | Delivery, engineering and optimization of systems, methods and compositions for sequence manipulation and therapeutic applications |
| US8999641B2 (en) | 2012-12-12 | 2015-04-07 | The Broad Institute Inc. | Engineering and optimization of systems, methods and compositions for sequence manipulation with functional domains |
| US8993233B2 (en) | 2012-12-12 | 2015-03-31 | The Broad Institute Inc. | Engineering and optimization of systems, methods and compositions for sequence manipulation with functional domains |
| US8945839B2 (en) | 2012-12-12 | 2015-02-03 | The Broad Institute Inc. | CRISPR-Cas systems and methods for altering expression of gene products |
| US8932814B2 (en) | 2012-12-12 | 2015-01-13 | The Broad Institute Inc. | CRISPR-Cas nickase systems, methods and compositions for sequence manipulation in eukaryotes |
| US8906616B2 (en) | 2012-12-12 | 2014-12-09 | The Broad Institute Inc. | Engineering of systems, methods and optimized guide compositions for sequence manipulation |
| US8895308B1 (en) | 2012-12-12 | 2014-11-25 | The Broad Institute Inc. | Engineering and optimization of improved systems, methods and enzyme compositions for sequence manipulation |
| US10435708B2 (en) | 2012-12-17 | 2019-10-08 | President And Fellows Of Harvard College | RNA-guided human genome engineering |
| US9023649B2 (en) | 2012-12-17 | 2015-05-05 | President And Fellows Of Harvard College | RNA-guided human genome engineering |
| US10273501B2 (en) | 2012-12-17 | 2019-04-30 | President And Fellows Of Harvard College | RNA-guided human genome engineering |
| US9970024B2 (en) | 2012-12-17 | 2018-05-15 | President And Fellows Of Harvard College | RNA-guided human genome engineering |
| US10717990B2 (en) | 2012-12-17 | 2020-07-21 | President And Fellows Of Harvard College | RNA-guided human genome engineering |
| US9260723B2 (en) | 2012-12-17 | 2016-02-16 | President And Fellows Of Harvard College | RNA-guided human genome engineering |
| US9777262B2 (en) | 2013-03-13 | 2017-10-03 | President And Fellows Of Harvard College | Mutants of Cre recombinase |
| WO2014152940A1 (en) | 2013-03-14 | 2014-09-25 | Shire Human Genetic Therapies, Inc. | Mrna therapeutic compositions and use to treat diseases and disorders |
| US9267135B2 (en) | 2013-06-04 | 2016-02-23 | President And Fellows Of Harvard College | RNA-guided transcriptional regulation |
| US10640789B2 (en) | 2013-06-04 | 2020-05-05 | President And Fellows Of Harvard College | RNA-guided transcriptional regulation |
| US10767194B2 (en) | 2013-06-04 | 2020-09-08 | President And Fellows Of Harvard College | RNA-guided transcriptional regulation |
| US11008588B2 (en) | 2013-06-17 | 2021-05-18 | The Broad Institute, Inc. | Delivery, engineering and optimization of tandem guide systems, methods and compositions for sequence manipulation |
| US10946108B2 (en) | 2013-06-17 | 2021-03-16 | The Broad Institute, Inc. | Delivery, use and therapeutic applications of the CRISPR-Cas systems and compositions for targeting disorders and diseases using viral components |
| US10577630B2 (en) | 2013-06-17 | 2020-03-03 | The Broad Institute, Inc. | Delivery and use of the CRISPR-Cas systems, vectors and compositions for hepatic targeting and therapy |
| US10781444B2 (en) | 2013-06-17 | 2020-09-22 | The Broad Institute, Inc. | Functional genomics using CRISPR-Cas systems, compositions, methods, screens and applications thereof |
| US10711285B2 (en) | 2013-06-17 | 2020-07-14 | The Broad Institute, Inc. | Optimized CRISPR-Cas double nickase systems, methods and compositions for sequence manipulation |
| US9587252B2 (en) | 2013-07-10 | 2017-03-07 | President And Fellows Of Harvard College | Orthogonal Cas9 proteins for RNA-guided gene regulation and editing |
| US10329587B2 (en) | 2013-07-10 | 2019-06-25 | President And Fellows Of Harvard College | Orthogonal Cas9 proteins for RNA-guided gene regulation and editing |
| US10563225B2 (en) | 2013-07-26 | 2020-02-18 | President And Fellows Of Harvard College | Genome engineering |
| US9914939B2 (en) | 2013-07-26 | 2018-03-13 | President And Fellows Of Harvard College | Genome engineering |
| US9163284B2 (en) | 2013-08-09 | 2015-10-20 | President And Fellows Of Harvard College | Methods for identifying a target site of a Cas9 nuclease |
| US10508298B2 (en) | 2013-08-09 | 2019-12-17 | President And Fellows Of Harvard College | Methods for identifying a target site of a CAS9 nuclease |
| US10954548B2 (en) | 2013-08-09 | 2021-03-23 | President And Fellows Of Harvard College | Nuclease profiling system |
| US9359599B2 (en) | 2013-08-22 | 2016-06-07 | President And Fellows Of Harvard College | Engineered transcription activator-like effector (TALE) domains and uses thereof |
| US11046948B2 (en) | 2013-08-22 | 2021-06-29 | President And Fellows Of Harvard College | Engineered transcription activator-like effector (TALE) domains and uses thereof |
| US10227581B2 (en) | 2013-08-22 | 2019-03-12 | President And Fellows Of Harvard College | Engineered transcription activator-like effector (TALE) domains and uses thereof |
| US10597679B2 (en) | 2013-09-06 | 2020-03-24 | President And Fellows Of Harvard College | Switchable Cas9 nucleases and uses thereof |
| US9340799B2 (en) | 2013-09-06 | 2016-05-17 | President And Fellows Of Harvard College | MRNA-sensing switchable gRNAs |
| US9999671B2 (en) | 2013-09-06 | 2018-06-19 | President And Fellows Of Harvard College | Delivery of negatively charged proteins using cationic lipids |
| US9388430B2 (en) | 2013-09-06 | 2016-07-12 | President And Fellows Of Harvard College | Cas9-recombinase fusion proteins and uses thereof |
| US10858639B2 (en) | 2013-09-06 | 2020-12-08 | President And Fellows Of Harvard College | CAS9 variants and uses thereof |
| US9340800B2 (en) | 2013-09-06 | 2016-05-17 | President And Fellows Of Harvard College | Extended DNA-sensing GRNAS |
| US10912833B2 (en) | 2013-09-06 | 2021-02-09 | President And Fellows Of Harvard College | Delivery of negatively charged proteins using cationic lipids |
| US9737604B2 (en) | 2013-09-06 | 2017-08-22 | President And Fellows Of Harvard College | Use of cationic lipids to deliver CAS9 |
| US9526784B2 (en) | 2013-09-06 | 2016-12-27 | President And Fellows Of Harvard College | Delivery system for functional nucleases |
| US9322037B2 (en) | 2013-09-06 | 2016-04-26 | President And Fellows Of Harvard College | Cas9-FokI fusion proteins and uses thereof |
| US10682410B2 (en) | 2013-09-06 | 2020-06-16 | President And Fellows Of Harvard College | Delivery system for functional nucleases |
| US9228207B2 (en) | 2013-09-06 | 2016-01-05 | President And Fellows Of Harvard College | Switchable gRNAs comprising aptamers |
| WO2015056756A1 (en) | 2013-10-18 | 2015-04-23 | 国立大学法人熊本大学 | Method of inducing kidney from pluripotent stem cells |
| US10640788B2 (en) | 2013-11-07 | 2020-05-05 | Editas Medicine, Inc. | CRISPR-related methods and compositions with governing gRNAs |
| US10190137B2 (en) | 2013-11-07 | 2019-01-29 | Editas Medicine, Inc. | CRISPR-related methods and compositions with governing gRNAS |
| US10435679B2 (en) | 2013-11-19 | 2019-10-08 | President And Fellows Of Harvard College | Mutant Cas9 proteins |
| US10683490B2 (en) | 2013-11-19 | 2020-06-16 | President And Fellows Of Harvard College | Mutant Cas9 proteins |
| US10787684B2 (en) | 2013-11-19 | 2020-09-29 | President And Fellows Of Harvard College | Large gene excision and insertion |
| US10100291B2 (en) | 2013-11-19 | 2018-10-16 | President And Fellows Of Harvard College | Mutant Cas9 proteins |
| US9074199B1 (en) | 2013-11-19 | 2015-07-07 | President And Fellows Of Harvard College | Mutant Cas9 proteins |
| US10377998B2 (en) | 2013-12-12 | 2019-08-13 | The Broad Institute, Inc. | CRISPR-CAS systems and methods for altering expression of gene products, structural information and inducible modular CAS enzymes |
| US11053481B2 (en) | 2013-12-12 | 2021-07-06 | President And Fellows Of Harvard College | Fusions of Cas9 domains and nucleic acid-editing domains |
| US10465176B2 (en) | 2013-12-12 | 2019-11-05 | President And Fellows Of Harvard College | Cas variants for gene editing |
| US10550372B2 (en) | 2013-12-12 | 2020-02-04 | The Broad Institute, Inc. | Systems, methods and compositions for sequence manipulation with optimized functional CRISPR-Cas systems |
| US10851357B2 (en) | 2013-12-12 | 2020-12-01 | The Broad Institute, Inc. | Compositions and methods of use of CRISPR-Cas systems in nucleotide repeat disorders |
| US9068179B1 (en) | 2013-12-12 | 2015-06-30 | President And Fellows Of Harvard College | Methods for correcting presenilin point mutations |
| US9840699B2 (en) | 2013-12-12 | 2017-12-12 | President And Fellows Of Harvard College | Methods for nucleic acid editing |
| EP3450553B1 (en) | 2014-03-24 | 2019-12-25 | Translate Bio, Inc. | Mrna therapy for treatment of ocular diseases |
| BR112016030852A2 (en) | 2014-07-02 | 2018-01-16 | Shire Human Genetic Therapies | rna messenger encapsulation |
| US10077453B2 (en) | 2014-07-30 | 2018-09-18 | President And Fellows Of Harvard College | CAS9 proteins including ligand-dependent inteins |
| US10704062B2 (en) | 2014-07-30 | 2020-07-07 | President And Fellows Of Harvard College | CAS9 proteins including ligand-dependent inteins |
| US10519454B2 (en) | 2014-08-06 | 2019-12-31 | Toolgen Incorporated | Genome editing using Campylobacter jejuni CRISPR/CAS system-derived RGEN |
| US10385336B2 (en) | 2014-09-05 | 2019-08-20 | Vilnius University | Programmable RNA shredding by the type III-A CRISPR-Cas system of Streptococcus thermophilus |
| US11001829B2 (en) | 2014-09-25 | 2021-05-11 | The Broad Institute, Inc. | Functional screening with optimized functional CRISPR-Cas systems |
| US11071790B2 (en) | 2014-10-29 | 2021-07-27 | Massachusetts Eye And Ear Infirmary | Method for efficient delivery of therapeutic molecules in vitro and in vivo |
| US11111472B2 (en) | 2014-10-31 | 2021-09-07 | Massachusetts Institute Of Technology | Delivery of biomolecules to immune cells |
| US10954514B2 (en) | 2014-12-12 | 2021-03-23 | The Broad Institute, Inc. | Escorted and functionalized guides for CRISPR-Cas systems |
| US10689691B2 (en) | 2014-12-19 | 2020-06-23 | The Broad Institute, Inc. | Unbiased identification of double-strand breaks and genomic rearrangement by genome-wide insert capture sequencing |
| US10876100B2 (en) | 2015-06-18 | 2020-12-29 | The Broad Institute, Inc. | Crispr enzyme mutations reducing off-target effects |
| US11060115B2 (en) | 2015-06-18 | 2021-07-13 | The Broad Institute, Inc. | CRISPR enzymes and systems |
| US9790490B2 (en) | 2015-06-18 | 2017-10-17 | The Broad Institute Inc. | CRISPR enzymes and systems |
| US10494621B2 (en) | 2015-06-18 | 2019-12-03 | The Broad Institute, Inc. | Crispr enzyme mutations reducing off-target effects |
| US11091798B2 (en) | 2015-06-18 | 2021-08-17 | The Broad Institute, Inc. | CRISPR enzymes and systems |
| US10648020B2 (en) | 2015-06-18 | 2020-05-12 | The Broad Institute, Inc. | CRISPR enzymes and systems |
| US11104967B2 (en) | 2015-07-22 | 2021-08-31 | President And Fellows Of Harvard College | Evolution of site-specific recombinases |
| US10612011B2 (en) | 2015-07-30 | 2020-04-07 | President And Fellows Of Harvard College | Evolution of TALENs |
| US11078469B2 (en) | 2015-07-30 | 2021-08-03 | President And Fellows Of Harvard College | Evolution of TALENs |
| US10375938B2 (en) | 2015-10-08 | 2019-08-13 | President And Fellows Of Harvard College | Multiplexed genome editing |
| US11064684B2 (en) | 2015-10-08 | 2021-07-20 | President And Fellows Of Harvard College | Multiplexed genome editing |
| US10925263B2 (en) | 2015-10-08 | 2021-02-23 | President And Fellows Of Harvard College | Multiplexed genome editing |
| US10959413B2 (en) | 2015-10-08 | 2021-03-30 | President And Fellows Of Harvard College | Multiplexed genome editing |
| EP3362461A1 (en) | 2015-10-16 | 2018-08-22 | Modernatx, Inc. | Mrna cap analogs with modified phosphate linkage |
| US10167457B2 (en) | 2015-10-23 | 2019-01-01 | President And Fellows Of Harvard College | Nucleobase editors and uses thereof |
| US10851369B2 (en) | 2016-06-21 | 2020-12-01 | President And Fellows Of Harvard College | Frequency-based modulation of diverse species in a nucleic acid library |
| US10113163B2 (en) | 2016-08-03 | 2018-10-30 | President And Fellows Of Harvard College | Adenosine nucleobase editors and uses thereof |
| US10947530B2 (en) | 2016-08-03 | 2021-03-16 | President And Fellows Of Harvard College | Adenosine nucleobase editors and uses thereof |
| US11085072B2 (en) | 2016-08-31 | 2021-08-10 | President And Fellows Of Harvard College | Methods of generating libraries of nucleic acid sequences for detection via fluorescent in situ sequencing |
| US10266887B2 (en) | 2016-12-09 | 2019-04-23 | The Broad Institute, Inc. | CRISPR effector system based diagnostics |
| US10266886B2 (en) | 2016-12-09 | 2019-04-23 | The Broad Institute, Inc. | CRISPR effector system based diagnostics |
| US10745677B2 (en) | 2016-12-23 | 2020-08-18 | President And Fellows Of Harvard College | Editing of CCR5 receptor gene to protect against HIV infection |
| US11104937B2 (en) | 2017-03-15 | 2021-08-31 | The Broad Institute, Inc. | CRISPR effector system based diagnostics |
| US11021740B2 (en) | 2017-03-15 | 2021-06-01 | The Broad Institute, Inc. | Devices for CRISPR effector system based diagnostics |
| US10968257B2 (en) | 2018-04-03 | 2021-04-06 | The Broad Institute, Inc. | Target recognition motifs and uses thereof |
| WO2020014577A1 (en) | 2018-07-13 | 2020-01-16 | Allele Biotechnology And Pharmaceuticals, Inc. | Methods of achieving high specificity of genome editing |
| US20200109398A1 (en) | 2018-08-28 | 2020-04-09 | Flagship Pioneering, Inc. | Methods and compositions for modulating a genome |
| WO2020191248A1 (en) | 2019-03-19 | 2020-09-24 | The Broad Institute, Inc. | Method and compositions for editing nucleotide sequences |
| WO2020191249A1 (en) | 2019-03-19 | 2020-09-24 | The Broad Institute, Inc. | Methods and compositions for editing nucleotide sequences |
| WO2020191239A1 (en) | 2019-03-19 | 2020-09-24 | The Broad Institute, Inc. | Methods and compositions for editing nucleotide sequences |
| WO2020191234A1 (en) | 2019-03-19 | 2020-09-24 | The Broad Institute, Inc. | Methods and compositions for editing nucleotide sequences |
| WO2020191241A1 (en) | 2019-03-19 | 2020-09-24 | The Broad Institute, Inc. | Methods and compositions for editing nucleotide sequences |
| WO2020191171A1 (en) | 2019-03-19 | 2020-09-24 | The Broad Institute, Inc. | Methods and compositions for editing nucleotide sequences |
| WO2020191153A2 (en) | 2019-03-19 | 2020-09-24 | The Broad Institute, Inc. | Methods and compositions for editing nucleotide sequences |
| WO2020191233A1 (en) | 2019-03-19 | 2020-09-24 | The Broad Institute, Inc. | Methods and compositions for editing nucleotide sequences |
| WO2020191242A1 (en) | 2019-03-19 | 2020-09-24 | The Broad Institute, Inc. | Methods and compositions for editing nucleotide sequences |
| WO2020191245A1 (en) | 2019-03-19 | 2020-09-24 | The Broad Institute, Inc. | Methods and compositions for editing nucleotide sequences |
| WO2020191246A1 (en) | 2019-03-19 | 2020-09-24 | The Broad Institute, Inc. | Methods and compositions for editing nucleotide sequences |
| WO2020191243A1 (en) | 2019-03-19 | 2020-09-24 | The Broad Institute, Inc. | Methods and compositions for editing nucleotide sequences |
| WO2021226558A1 (en) | 2020-05-08 | 2021-11-11 | The Broad Institute, Inc. | Methods and compositions for simultaneous editing of both strands of a target double-stranded nucleotide sequence |
| WO2022087235A1 (en) | 2020-10-21 | 2022-04-28 | Massachusetts Institute Of Technology | Systems, methods, and compositions for site-specific genetic engineering using programmable addition via site-specific targeting elements (paste) |
| US11572556B2 (en) | 2020-10-21 | 2023-02-07 | Massachusetts Institute Of Technology | Systems, methods, and compositions for site-specific genetic engineering using programmable addition via site-specific targeting elements (paste) |
| WO2022139528A1 (en)* | 2020-12-24 | 2022-06-30 | (주)인핸스드바이오 | Lipid nanoparticles comprising mannose or uses thereof |
| EP4268808A1 (en)* | 2020-12-24 | 2023-11-01 | EnhancedBio Inc. | Lipid nanoparticles comprising mannose or uses thereof |
| WO2022241040A1 (en)* | 2021-05-11 | 2022-11-17 | Seattle Children's Hospital D/B/A Seattle Children's Research Institute | Gene editing for expression of functional factor viii for the treatment of hemophilia |
Non-Patent Citations (66)
| Title |
|---|
| "Covalent Attachment of Proteins to Liposomes", vol. 111, 1987, ACADEMIC PRESS, INC, pages: 119 |
| ABRA ET AL., J. LIPOSOME RES., vol. 12, 2002, pages 1 - 3 |
| AHMAD ET AL., CANCER RES, vol. 52, 1992, pages 4817 - 4820 |
| AKINC ET AL., MOL THER, vol. 17, 2009, pages 872 - 879 |
| AKINC ET AL., NAT. BIOTECHNOL., vol. 26, 2008, pages 561 - 569 |
| ALLEN ET AL., BIOCHIMICA ET BIOPHYSICA ACTA, vol. 1237, 1995, pages 99 - 108 |
| ANZALONE ET AL., BIORXIV, 2 November 2021 (2021-11-02) |
| ANZALONE ET AL., NATURE, vol. 576, 2019, pages 149 |
| BLAESE ET AL., CANCER GENE THER, vol. 2, 1995, pages 291 - 297 |
| BLUME ET AL., BIOCHIMICA ET BIOPHYSICA ACTA, vol. 1149, 1993, pages 180 - 184 |
| BUCHSCHER ET AL., J. VIROL., vol. 66, 1992, pages 1635 - 1640 |
| CHEN ET AL., CELL, vol. 184, 28 October 2021 (2021-10-28), pages 1 - 18 |
| CRYSTAL, SCIENCE, vol. 270, 1995, pages 404 - 410 |
| DALKARA ET AL., SCI TRANSL MED, vol. 5, 2013, pages 89ra76 |
| DEFREES ET AL., JOURNAL OF THE AMERICAN CHEMISTRY SOCIETY, vol. 118, 1996, pages 6101 - 6104 |
| DURRANT ET AL., NAT. BIOTECHNOL., 2022 |
| EL-MAARRI ET AL., HAMOSTASEOLOGIE, vol. 40, no. S01, November 2020 (2020-11-01), pages S26 - S31 |
| GAO ET AL., GENE THERAPY, vol. 2, 1995, pages 710 - 722 |
| GASIUNAS ET AL.: "A catalogue of biochemically diverse CRISPR-Cas9 orthologs", NATURE COMMUNICATIONS, vol. 11, pages 5512 |
| HERMONATMUZYCZKA, PNAS, vol. 81, 1984, pages 6466 - 6470 |
| IOANNIDI ELEONORA I. ET AL: "Drag-and-drop genome insertion without DNA cleavage with CRISPR-directed integrases", BIORXIV, 1 November 2021 (2021-11-01), XP093015571, Retrieved from the Internet <URL:https://www.biorxiv.org/content/10.1101/2021.11.01.466786v1.full.pdf> [retrieved on 20230119], DOI: 10.1101/2021.11.01.466786* |
| IONNIDI ET AL.: "Drag-and-drop genome insertion without DNA cleavage with CRISPR directed integrases", BIORXIV 2021 11.01 466786 |
| JIANG ET AL., NAT. BIOTECHNOLOGY, 14 October 2021 (2021-10-14) |
| KARVELIS ET AL.: "PAM recognition by miniature CRISPR-Cas12f nucleases triggers programmable double-stranded DNA target cleavage", NUCLEIC ACIDS RESEARCH, vol. 48, no. 9, 21 May 2020 (2020-05-21), pages 5016 - 23, XP055920188, DOI: 10.1093/nar/gkaa208 |
| KIRPOTIN ET AL., FEBS LETTERS, vol. 388, 1996, pages 115 - 118 |
| KLIBANOV ET AL., JOURNAL OF LIPOSOME RESEARCH, vol. 2, 1992, pages 321 - 334 |
| KOTIN, HUMAN GENE THERAPY, vol. 5, 1994, pages 793 - 801 |
| KOWALSKI ET AL.: "Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery", MOL THERAP., vol. 27, no. 4, 2019, pages 710 - 728, XP055634628, DOI: 10.1016/j.ymthe.2019.02.012 |
| LEONETTI ET AL., PROC. NATL. ACAD. SCI. (USA, vol. 87, 1990, pages 2448 - 2451 |
| LIUHUANG, MOLECULAR THERAPY, 2010, pages 669 - 670 |
| LOVE ET AL., PROC NATL ACAD SCI USA, vol. 107, 2010, pages 1864 - 1869 |
| LOVE ET AL., PROC NATL ACAD SCI USA., vol. 107, 2010, pages 1864 - 1869 |
| MAHON ET AL., BIOCONJUG CHEM, vol. 21, 2010, pages 1448 - 1454 |
| MAKAROVA ET AL.: "C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector", SCIENCE, vol. 353, no. 6299, 2016, XP055407082, DOI: 10.1126/science.aaf5573 |
| MAKAROVA ET AL.: "Classification and Nomenclature of CRISPR-Cas Systems: Where from Here?", THE CRISPR JOURNAL, vol. 1, no. 5, 2018, XP055619311, DOI: 10.1089/crispr.2018.0033 |
| MILLER ET AL., J. VIROL., vol. 65, 1991, pages 2220 - 2224 |
| MILLINGTON-WARD ET AL., MOLECULAR THERAPY, vol. 19, no. 4, April 2011 (2011-04-01), pages 642 - 649 |
| MURUGAIAH ET AL., ANALYTICAL BIOCHEMISTRY, vol. 401, 2010, pages 61 |
| MUTHUSAMY JAYARAMAN ET AL: "Maximizing the Potency of siRNA Lipid Nanoparticles for Hepatic Gene Silencing In Vivo", ANGEWANDTE CHEMIE INTERNATIONAL EDITION, VERLAG CHEMIE, HOBOKEN, USA, vol. 51, no. 34, 10 July 2012 (2012-07-10), pages 8529 - 8533, XP072070912, ISSN: 1433-7851, DOI: 10.1002/ANIE.201203263* |
| MUZYCZKA, J. CLIN. INVEST., vol. 94, 1994, pages 1351 |
| NISHIMASU ET AL.: "Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA", CELL, vol. 156, 27 February 2014 (2014-02-27), pages 935 - 949, XP028667665, DOI: 10.1016/j.cell.2014.02.001 |
| OH, Y ET AL.: "Expansion of the prime editing modality with Cas9 from Francisella novicida", BIORXIV 2021.05.25.445577 |
| RAMAKRISHNAN ET AL.: "Nuclear export signal (NES) of transposases affects the transposition activity of mariner-like elements Ppmarl and Ppmar2 of moso bamboo", MOB DNA, vol. 10, 19 August 2019 (2019-08-19), pages 35 |
| REMY ET AL., BIOCONJUGATE CHEM, vol. 5, 1994, pages 647 - 654 |
| RENNEISEN ET AL., J. BIO. CHEM., vol. 265, 1990, pages 16337 - 16342 |
| ROSIN ET AL., MOLECULAR THERAPY, vol. 19, no. 12, December 2011 (2011-12-01), pages 1286 - 2200 |
| SAMULSKI ET AL., J. VIROL., vol. 63, 1989, pages 03822 - 3828 |
| SAPRA ET AL., PROG. LIPID RES., vol. 42, no. 5, 2003, pages 439 - 62 |
| SCHMIDT ET AL.: "Improved CRISPR genome editing using small highly active and specific engineered RNA-guided nucleases", NAT COMMUN, vol. 12, 2021, pages 4219 |
| SCHROEDER ET AL., J INTERN MED, vol. 267, 2010, pages 9 - 21 |
| SHAH ET AL.: "Protospacer recognition motifs: mixed identities and functional diversity", RNA BIOLOGY, vol. 10, no. 5, pages 891 - 899 |
| SIEGWART ET AL., PROC NATL ACAD SCI USA., vol. 108, 2011, pages 12996 - 3001 |
| SOMMNERFELT ET AL., VIROL, vol. 176, 1990, pages 58 - 59 |
| SWARTS ET AL.: "Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a", MOLECULAR CELL, vol. 66, 20 April 2017 (2017-04-20), pages 221 - 233, XP055569665, DOI: 10.1016/j.molcel.2017.03.016 |
| TRATSCHIN ET AL., MOL. CELL. BIOL, vol. 5, 1985, pages 3251 - 3260 |
| TRATSCHIN ET AL., MOL. CELL. BIOL., vol. 4, 1984, pages 2072 - 2081 |
| WEST ET AL., VIROLOGY, vol. 160, 1987, pages 38 - 47 |
| WILSON ET AL., J. VIROL, vol. 63, 1989, pages 2374 - 2378 |
| XU ET AL.: "Accuracy and efficiency define Bxb1 integrase as the best of fifteen candidate serine recombinases for the integration of DNA into the human genome", BMC BIOTECHNOL, vol. 13, 20 October 2013 (2013-10-20), pages 87 |
| XU ET AL.: "Comparison and optimization of ten phage encoded serine integrases for genome engineering in Saccharomyces cerevisiae", BMC BIOTECHNOL, vol. 16, 9 February 2016 (2016-02-09), pages 13 |
| XU, X ET AL.: "Engineered Miniature CRISPR-Cas System for Mammalian Genome Regulation and Editing", MOLECULAR CELL, vol. 81, no. 20, 21 October 2021 (2021-10-21), pages 4333 - 45 |
| YAMANO ET AL.: "Crystal structure of Cpf1 in complex with guide RNA and target DNA", CELL, vol. 165, 2016, pages 949 - 962, XP029530759, DOI: 10.1016/j.cell.2016.04.003 |
| YAMANO ET AL.: "Crystal structure of Cpfl in complex with guide RNA and target DNA", CELL, vol. 165, 5 May 2016 (2016-05-05), pages 949 - 962 |
| ZALIPSKY, BIOCONJUGATE CHEMISTRY, vol. 4, 1993, pages 296 - 299 |
| ZALIPSKY, FEBS LETTERS, vol. 353, 1994, pages 71 - 74 |
| ZALIPSKY: "Stealth Liposomes", 1995, CRC PRESS |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| US20250057980A1 (en) | Co-Delivery of a Gene Editor Construct and a Donor Template | |
| JP7581405B2 (en) | Lipid Nanoparticle Formulations for CRISPR/CAS Components | |
| US12263227B2 (en) | Optimized mRNA encoding CAS9 for use in LNPs | |
| JP2024003220A (en) | Gene editing using modified closed-ended DNA (CEDNA) | |
| AU2022375820A1 (en) | Single construct platform for simultaneous delivery of gene editing machinery and nucleic acid cargo | |
| WO2023039440A9 (en) | Hbb-modulating compositions and methods | |
| US12188060B2 (en) | Messenger RNA encoding Cas9 for use in genome-editing systems | |
| WO2023205744A1 (en) | Programmable gene insertion compositions | |
| WO2023225670A2 (en) | Ex vivo programmable gene insertion | |
| WO2023215831A1 (en) | Guide rna compositions for programmable gene insertion | |
| EP3704238B1 (en) | Engineered nucleases that target human and canine factor viii genes as a treatment for hemophilia a | |
| US20240110201A1 (en) | Compositions and Methods for Treating Hereditary Angioedema | |
| EP4615424A1 (en) | Rna compositions comprising lipid nanoparticles or lipid reconstructed natural messenger packs | |
| WO2024138194A1 (en) | Platforms, compositions, and methods for in vivo programmable gene insertion | |
| US20240279649A1 (en) | Gene editing for expression of functional factor viii for the treatment of hemophilia | |
| WO2024234006A1 (en) | Systems, compositions, and methods for targeting liver sinusodial endothelial cells (lsecs) | |
| AU2023275457A1 (en) | Gene therapy compositions and methods of use thereof | |
| WO2025050069A1 (en) | Programmable gene insertion using engineered integration enzymes | |
| CN118829727A (en) | A single construct platform for simultaneous delivery of gene editing machinery and nucleic acid cargo | |
| WO2025137461A1 (en) | Nucleic acid binding agents and uses thereof | |
| WO2023225471A2 (en) | Helitron compositions and methods | |
| WO2025080867A1 (en) | Ionizable lipids for improved mrna delivery | |
| HK40005508A (en) | Lipid nanoparticle formulations for crispr/cas components |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application | Ref document number:24735396 Country of ref document:EP Kind code of ref document:A1 |