WO2021232789A1 - Mirna-disease association prediction method, system, terminal, and storage medium - Google Patents
Mirna-disease association prediction method, system, terminal, and storage mediumDownload PDFInfo
- Publication number
- WO2021232789A1 WO2021232789A1PCT/CN2020/139689CN2020139689WWO2021232789A1WO 2021232789 A1WO2021232789 A1WO 2021232789A1CN 2020139689 WCN2020139689 WCN 2020139689WWO 2021232789 A1WO2021232789 A1WO 2021232789A1
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- mirna
- disease
- matrix
- similarity matrix
- similarity
- Prior art date
- Legal status (The legal status is an assumption and is not a legal conclusion. Google has not performed a legal analysis and makes no representation as to the accuracy of the status listed.)
- Ceased
Links
Images
Classifications
- G—PHYSICS
- G16—INFORMATION AND COMMUNICATION TECHNOLOGY [ICT] SPECIALLY ADAPTED FOR SPECIFIC APPLICATION FIELDS
- G16B—BIOINFORMATICS, i.e. INFORMATION AND COMMUNICATION TECHNOLOGY [ICT] SPECIALLY ADAPTED FOR GENETIC OR PROTEIN-RELATED DATA PROCESSING IN COMPUTATIONAL MOLECULAR BIOLOGY
- G16B20/00—ICT specially adapted for functional genomics or proteomics, e.g. genotype-phenotype associations
- G—PHYSICS
- G06—COMPUTING OR CALCULATING; COUNTING
- G06F—ELECTRIC DIGITAL DATA PROCESSING
- G06F18/00—Pattern recognition
- G06F18/20—Analysing
- G06F18/22—Matching criteria, e.g. proximity measures
- G—PHYSICS
- G16—INFORMATION AND COMMUNICATION TECHNOLOGY [ICT] SPECIALLY ADAPTED FOR SPECIFIC APPLICATION FIELDS
- G16B—BIOINFORMATICS, i.e. INFORMATION AND COMMUNICATION TECHNOLOGY [ICT] SPECIALLY ADAPTED FOR GENETIC OR PROTEIN-RELATED DATA PROCESSING IN COMPUTATIONAL MOLECULAR BIOLOGY
- G16B40/00—ICT specially adapted for biostatistics; ICT specially adapted for bioinformatics-related machine learning or data mining, e.g. knowledge discovery or pattern finding
- G—PHYSICS
- G16—INFORMATION AND COMMUNICATION TECHNOLOGY [ICT] SPECIALLY ADAPTED FOR SPECIFIC APPLICATION FIELDS
- G16H—HEALTHCARE INFORMATICS, i.e. INFORMATION AND COMMUNICATION TECHNOLOGY [ICT] SPECIALLY ADAPTED FOR THE HANDLING OR PROCESSING OF MEDICAL OR HEALTHCARE DATA
- G16H50/00—ICT specially adapted for medical diagnosis, medical simulation or medical data mining; ICT specially adapted for detecting, monitoring or modelling epidemics or pandemics
- G16H50/70—ICT specially adapted for medical diagnosis, medical simulation or medical data mining; ICT specially adapted for detecting, monitoring or modelling epidemics or pandemics for mining of medical data, e.g. analysing previous cases of other patients
Definitions
- the embodiments of the present applicationbelong to the technical field of bioinformatics, and particularly relate to a miRNA-disease association prediction method, system, terminal, and storage medium.
- miRNAsare small endogenous non-coding RNAs with a length of about 22 nucleotides, which inhibit the expression of target genes by inducing messenger RNA degradation, translation inhibition or other morphological regulation mechanisms.
- a large amount of research evidenceshows that miRNAs play an important role in many biological processes. MiRNA dysfunction and miRNA mutations can lead to various diseases. Therefore, identifying the interaction between miRNAs and diseases is helpful for humans to understand the mechanism of the disease, thereby providing help for the prevention and treatment of the disease.
- the patentdiscloses a disease-associated miRNA prediction method and system based on self-avoidance random walks.
- the patentuses self-avoidance random walks to traverse the disease-miRNA bipartite graph and uses the two attributes of self-avoidance random walks (two The ratio of the transition probability between each node to the average step size) is used to measure the degree of association between nodes, so as to realize the association prediction between disease and miRNA.
- This methodcannot be used to predict the association between diseases and miRNAs for which there is no known association.
- the embodiments of the applicationprovide a miRNA-disease association prediction method, system, terminal, and storage medium, which are intended to at least to a certain extent solve the problem of predicting miRNA-disease association calculation methods in the prior art, which only considers the relationship between miRNA and disease.
- One-sided informationinability to fully characterize the complex relationship between miRNAs and diseases, low prediction accuracy, and technical problems that cannot be used to predict the association between diseases and miRNAs without known associations.
- a miRNA-disease association prediction methodincludes the following steps:
- Step aConstruct miRNA-disease association matrix, miRNA similarity matrix and disease similarity matrix based on miRNA-disease related data
- Step bconstruct a heterogeneous network according to the miRNA-disease association matrix, miRNA similarity matrix and disease similarity matrix;
- Step cUse a neural network to learn the topology information of the heterogeneous network, calculate the optimal parameters of the heterogeneous network through topology maintenance, and reconstruct the heterogeneous network according to the optimal parameters; the reconstructed heterogeneous network
- the structural networkis the correlation score matrix between miRNA and disease.
- the technical solution adopted in the embodiment of the applicationfurther includes: in the step a, the miRNA-disease related data includes miRNA-disease related data, gene function information, miRNA-target related information, miRNA family and cluster information, and miRNA function Similarity data.
- the technical solution adopted in the embodiment of the present applicationfurther includes: in the step a, the construction of the miRNA-disease association matrix is specifically:
- a miRNA-disease association matrixis constructed according to the miRNA-disease association data.
- the technical solution adopted in the embodiment of the present applicationfurther includes: in the step a, the construction of the miRNA similarity matrix is specifically:
- miRNA similarity matrixis constructed.
- the technical solution adopted in the embodiment of the applicationfurther includes: the construction of the miRNA similarity matrix further includes:
- the above four similarity matricesare weighted and summed to obtain the miRNA similarity matrix.
- the technical solution adopted in the embodiment of the present applicationfurther includes: in the step a, the constructing a disease similarity matrix is specifically:
- a disease similarity matrixis constructed based on the gene function information.
- the technical solution adopted in the embodiment of the present applicationfurther includes: in the step b, the constructing a heterogeneous network specifically includes:
- the standardized miRNA-disease association matrix, the miRNA similarity matrix and the disease similarity matrixare combined into a heterogeneous network.
- the technical solution adopted in the embodiment of the present applicationfurther includes: in the step c, the calculation of the optimal parameters of the heterogeneous network through topology preservation includes:
- MSrepresents the miRNA similarity matrix
- MDrepresents the mirna-disease association matrix
- DSrepresents the disease similarity matrix
- Frepresents the feature matrix of the node
- the number of rowsis the number of nodes
- the number of columnsis the dimension of the feature vector
- F TRepresents the transposed matrix of F.
- a miRNA-disease association prediction systemincluding:
- Heterogeneous network construction moduleused to construct a heterogeneous network according to the miRNA-disease association matrix, miRNA similarity matrix and disease similarity matrix;
- Feature extraction moduleused to learn the topology information of the heterogeneous network by using a neural network
- Network parameter calculation moduleused to calculate the optimal parameters of the heterogeneous network through topology maintenance
- Network reconstruction moduleused to reconstruct the heterogeneous network according to the optimal parameters; the reconstructed heterogeneous network is the correlation score matrix between miRNA and disease.
- a terminalincludes a processor and a memory coupled to the processor, wherein:
- the memorystores program instructions for realizing the miRNA-disease association prediction method
- the processoris configured to execute the program instructions stored in the memory to control miRNA-disease association prediction.
- Another technical solution adopted by the embodiment of the present applicationis: a storage medium storing program instructions executable by a processor, and the program instructions are used to execute the miRNA-disease association prediction method.
- the beneficial effects produced by the embodiments of the present applicationare: the miRNA-disease association prediction method, system, terminal and storage medium of the embodiments of the present application are based on miRNA similarity network, disease similarity network and experimentally verified miRNA -Disease association network constructs a heterogeneous network, and then uses neural network to learn the topology information of the heterogeneous network, extracts the feature representation of miRNA and disease, calculates the optimal parameters of the heterogeneous network, and finally reconstructs the heterogeneous network, which improves the prediction of miRNA and disease The accuracy of the relevance.
- This applicationsolves the problems of high cost and time-consuming biological experiment methods, improves the effect of miRNA-disease association prediction, is practical, and can be used for association prediction between diseases and miRNAs that have no known association.
- Fig. 1is a flowchart of a miRNA-disease association prediction method according to the first embodiment of the present application
- FIG. 2is a schematic diagram of the prediction results of the miRNA-disease association prediction method according to the second embodiment of the application;
- Figure 3is a schematic structural diagram of a miRNA-disease association prediction system according to an embodiment of the present application.
- FIG. 4is a schematic diagram of a terminal structure according to an embodiment of the application.
- FIG. 5is a schematic structural diagram of a storage medium according to an embodiment of the application.
- FIG. 1is a flowchart of a miRNA-disease association prediction method according to an embodiment of the present application.
- the miRNA-disease association prediction method of the embodiment of the applicationincludes the following steps:
- Step 100Obtain multi-source information related to miRNA-disease from the database
- the obtained multi-source informationspecifically includes: downloading known miRNA-disease related data from the human miRNA-disease database (HMDDv2.0); downloading gene function information from the HumanNet database; downloading miRNA-target related information from the miRTarBase database (Target gene); Download miRNA family and cluster information from miRBase database; Download miRNA functional similarity data (http://www.cuilab.cn/files/images/cuilab/misim.zip).
- Step 200construct a miRNA-disease association matrix MD according to the miRNA-disease association data
- step 200the construction of the miRNA-disease association matrix is specifically as follows: if a disease is associated with a miRNA, an edge is added between the two nodes.
- Step 300construct a miRNA similarity matrix MS according to target gene information, miRNA family information, miRNA cluster information, and miRNA functional similarity data;
- step 300the construction method of the miRNA similarity matrix specifically includes:
- Step 301Calculate the target gene-based similarity matrix RST based on the target gene information, and the matrix represents the number of target genes commonly associated with miRNAs;
- Step 302Calculate according to the miRNA family information to obtain a similarity matrix RSF based on the miRNA family information
- Step 303Calculate according to the miRNA cluster information to obtain a similarity matrix RSC based on the miRNA cluster information
- Step 304Calculate the similarity matrix RSD based on the miRNA functional similarity data based on the miRNA functional similarity data;
- Step 305Then, the above four similarity matrices are weighted and summed to construct the final miRNA similarity matrix:
- Step 400construct a disease similarity matrix DS based on gene function information
- step 400genes with similar functions can regulate similar diseases. Therefore, this application uses gene function information to construct a disease similarity matrix. Information contains two genes or gene set from the database downloads HumanNet gene functions of the log-likelihood score LLS, d i and d j disease similarity DS (d i, d i) is calculated as follows:
- Formula (2), S (d i) and S (d j)respectively set with d i and d j of disease-related genes.
- LLS(x,S(d i ))is the log-likelihood score of gene x and gene set S(d i ).
- Step 500Combine the miRNA-disease association matrix, miRNA similarity matrix and disease similarity matrix to construct a heterogeneous network
- the heterogeneous network construction methodspecifically includes:
- Col(A)represents the number of columns of matrix A
- (i,j)represents the i-th row and jth column of the matrix
- (i,k)represents the i-th row and kth column of the matrix.
- Step 600Use the neural network to learn the topology information of the heterogeneous network, and extract the deep feature representation of the node;
- step 600the information of neighboring nodes is gathered, the network topology structure information can be learned, and the deep feature representation of the node can be obtained.
- the deep feature representation of the nodeFor any node u of type t(t ⁇ NT), its deep feature representation is obtained by fusing the information of neighbor nodes and itself, and then standardizing:
- EM' tis the deep feature representation of the node belonging to type t
- EM tis the feature representation of the node randomly initialized
- E'(u)is the final node feature representation after standardization
- vis the same as u
- W s ⁇ R d ⁇ d , b s ⁇ R d , W 0 ⁇ R 2d ⁇ d and b 0 ⁇ R dare the training parameters of the heterogeneous network
- ⁇ ( ⁇ )is For the activation function in the heterogeneous network, the RELU function is used in the embodiment of this application.
- Step 700Maintain and calculate network parameters through the topology
- step 700the embodiment of the present application obtains the optimal parameter by minimizing the mean square error between the reconstructed matrix and the original matrix, and specifically obtains the optimal parameter by means of gradient descent:
- MSrepresents the miRNA similarity matrix
- MDrepresents the mirna-disease association matrix
- DSrepresents the disease similarity matrix
- Frepresents the feature matrix of the node
- the number of rowsis the number of nodes
- the number of columnsis the dimension of the feature vector
- F Trepresents the transposed matrix of F.
- Step 800Rebuild the heterogeneous network according to the optimal parameters
- step 800after the parameter training is completed, the correlation score between each miRNA and the disease can be obtained by reconstructing the heterogeneous network:
- the reconstructed heterogeneous network MD'is the correlation score matrix between miRNA and disease. The higher the score, the stronger the correlation between miRNA and disease.
- the second embodiment of the present applicationselects the disclosed data as test data, and uses a 5-fold cross-validation method to evaluate the technical solution of the present application. Specifically:
- S3Construct a miRNA similarity matrix MS based on target gene information, miRNA family information, miRNA cluster information, and miRNA functional similarity data;
- the miRNA similarity matrix RSFis calculated based on the miRNA family information.
- the miRNAs in the databelong to 299 different families. If two miRNAs belong to the same family, their value in the similarity matrix is set to 1, and vice versa. Is 0; the miRNA similarity matrix RSC calculated based on the miRNA cluster information, the miRNAs in the data belong to 153 different clusters, the same as the matrix RSF, if two miRNAs belong to the same cluster, their values in the similarity matrix are set Set to 1, otherwise set to 0.
- FIG. 3is a schematic structural diagram of a miRNA-disease association prediction system according to an embodiment of the present application.
- the miRNA-disease association prediction system of the embodiment of the applicationincludes:
- Data acquisition moduleused to acquire multi-source information related to miRNA-disease from the database; the acquired multi-source information specifically includes: downloading known miRNA-disease-related data from the human miRNA-disease database (HMDDv2.0) Download gene function information from HumanNet database; download miRNA-target association information (target gene) from miRTarBase database; download miRNA family and cluster information from miRBase database; download miRNA functional similarity data (http://www.cuilab.cn /files/images/cuilab/misim.zip).
- HMDDv2.0Human miRNA-disease database
- the first matrix building moduleused to construct a miRNA-disease association matrix MD based on the miRNA-disease association data; among them, the miRNA-disease association matrix construction method is specifically: if a disease is associated with a miRNA, then between two nodes Add an edge in between.
- the second matrix construction moduleused to construct a miRNA similarity matrix MS based on target gene information, miRNA family information, miRNA cluster information, and miRNA functional similarity data; among them, the construction of miRNA similarity matrix specifically includes:
- Target gene-based similarity matrix RSTCalculate the target gene-based similarity matrix RST based on the target gene information, which represents the number of target genes that are commonly associated between miRNAs;
- the third matrix construction moduleis used to construct a disease similarity matrix DS based on gene function information; among them, genes with similar functions can regulate similar diseases. Therefore, this application uses gene function information to construct a disease similarity matrix.
- Informationcontains two genes or gene set from the database downloads HumanNet gene functions of the log-likelihood score LLS, d i and d j disease similarity DS (d i, d i) is calculated as follows:
- Formula (2), S (d i) and S (d j)respectively set with d i and d j of disease-related genes.
- LLS(x,S(d i ))is the log-likelihood score of gene x and gene set S(d i ).
- Heterogeneous network building moduleused to merge miRNA-disease association matrix, miRNA similarity matrix and disease similarity matrix to construct a heterogeneous network; among them, the heterogeneous network construction methods include:
- Col(A)represents the number of columns of matrix A
- (i,j)represents the i-th row and jth column of the matrix
- (i,k)represents the i-th row and kth column of the matrix.
- Feature extraction moduleused to use neural network to learn the topology information of heterogeneous networks and extract the deep feature representations of nodes; among them, gather the information of neighbor nodes to learn network topology structure information and obtain deep feature representations of nodes. For any node u of type t(t ⁇ NT), its deep feature representation is obtained by fusing the information of neighbor nodes and itself, and then standardizing:
- EM' tis the deep feature representation of the node belonging to type t
- EM tis the feature representation of the node randomly initialized
- E'(u)is the final node feature representation after standardization
- vis the same as u
- W s ⁇ R d ⁇ d , b s ⁇ R d , W 0 ⁇ R 2d ⁇ d and b 0 ⁇ R dare the training parameters of the heterogeneous network
- ⁇ ( ⁇ )is For the activation function in the heterogeneous network, the RELU function is used in the embodiment of this application.
- Network parameter calculation moduleused to maintain and calculate network parameters through topology; among them, this application obtains the optimal parameters by minimizing the mean square error between the reconstructed matrix and the original matrix, specifically the optimal parameters are obtained by gradient descent:
- MSrepresents the miRNA similarity matrix
- MDrepresents the mirna-disease association matrix
- DSrepresents the disease similarity matrix
- Frepresents the feature matrix of the node
- the number of rowsis the number of nodes
- the number of columnsis the dimension of the feature vector
- F Trepresents the transposed matrix of F.
- Network reconstruction moduleused to reconstruct the heterogeneous network according to the optimal parameters; after the parameter training is completed, the correlation score between each miRNA and disease can be obtained by reconstructing the heterogeneous network:
- the reconstructed heterogeneous network MD'is the correlation score matrix between miRNA and disease. The higher the score, the stronger the correlation between miRNA and disease.
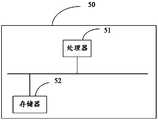
- FIG. 4is a schematic diagram of a terminal structure according to an embodiment of the application.
- the terminal 50includes a processor 51 and a memory 52 coupled to the processor 51.
- the memory 52stores program instructions for realizing the miRNA-disease association prediction method described above.
- the processor 51is configured to execute program instructions stored in the memory 52 to control miRNA-disease association prediction.
- the processor 51may also be referred to as a CPU (Central Processing Unit, central processing unit).
- the processor 51may be an integrated circuit chip with signal processing capability.
- the processor 51may also be a general-purpose processor, a digital signal processor (DSP), an application specific integrated circuit (ASIC), an off-the-shelf programmable gate array (FPGA) or other programmable logic device, a discrete gate or transistor logic device, or a discrete hardware component.
- DSPdigital signal processor
- ASICapplication specific integrated circuit
- FPGAoff-the-shelf programmable gate array
- the general-purpose processormay be a microprocessor or the processor may also be any conventional processor or the like.
- FIG. 5is a schematic structural diagram of a storage medium according to an embodiment of the application.
- the storage medium of this embodiment of the present applicationstores a program file 61 that can implement all the above methods.
- the program file 61can be stored in the above storage medium in the form of a software product, and includes several instructions to enable a computer device (which can It is a personal computer, a server, or a network device, etc.) or a processor (processor) that executes all or part of the steps of the various embodiments of the present invention.
- the aforementioned storage mediainclude: U disk, mobile hard disk, read-only memory (ROM, Read-Only Memory), random access memory (RAM, Random Access Memory), magnetic disks or optical disks and other media that can store program codes. , Or terminal devices such as computers, servers, mobile phones, and tablets.
- the miRNA-disease association prediction method, system, terminal and storage medium of the embodiments of the applicationconstruct a heterogeneous network based on the miRNA similarity network, the disease similarity network and the experimentally verified miRNA-disease association network, and then use the neural network to learn the heterogeneity
- the topology information of the network, the feature representation of miRNA and diseaseare extracted, the optimal parameters of the heterogeneous network are calculated, and the heterogeneous network is finally reconstructed, which improves the accuracy of predicting the correlation between miRNA and disease.
- This applicationsolves the problems of high cost and time-consuming biological experiment methods, improves the effect of miRNA-disease association prediction, is practical, and can be used for association prediction between diseases and miRNAs that have no known association.
Landscapes
- Engineering & Computer Science (AREA)
- Health & Medical Sciences (AREA)
- Life Sciences & Earth Sciences (AREA)
- Physics & Mathematics (AREA)
- Data Mining & Analysis (AREA)
- Medical Informatics (AREA)
- Bioinformatics & Cheminformatics (AREA)
- Theoretical Computer Science (AREA)
- Evolutionary Biology (AREA)
- Public Health (AREA)
- General Health & Medical Sciences (AREA)
- Bioinformatics & Computational Biology (AREA)
- Evolutionary Computation (AREA)
- Epidemiology (AREA)
- Biotechnology (AREA)
- Databases & Information Systems (AREA)
- Spectroscopy & Molecular Physics (AREA)
- Biophysics (AREA)
- Artificial Intelligence (AREA)
- Computer Vision & Pattern Recognition (AREA)
- Software Systems (AREA)
- Genetics & Genomics (AREA)
- Molecular Biology (AREA)
- Proteomics, Peptides & Aminoacids (AREA)
- Analytical Chemistry (AREA)
- Chemical & Material Sciences (AREA)
- Bioethics (AREA)
- Biomedical Technology (AREA)
- Pathology (AREA)
- Primary Health Care (AREA)
- General Engineering & Computer Science (AREA)
- General Physics & Mathematics (AREA)
- Measuring Or Testing Involving Enzymes Or Micro-Organisms (AREA)
Abstract
Description
Translated fromChinese本申请实施例属于生物信息学技术领域,特别涉及一种miRNA-疾病关联预测方法、系统、终端以及存储介质。The embodiments of the present application belong to the technical field of bioinformatics, and particularly relate to a miRNA-disease association prediction method, system, terminal, and storage medium.
miRNAs是一类长度约为22个核苷酸的微小内源性非编码RNA,通过诱导信使RNA降解、翻译抑制或其它形态调节机制来抑制靶基因的表达。大量研究证据显示,miRNA在许多生物进程中发挥重要作用,miRNA功能失调和miRNA突变会导致各种疾病的发生。因此,识别miRNA与疾病之间的相互作用关系有利于人类理解疾病机制,从而为疾病的预防和治疗提供帮助。miRNAs are small endogenous non-coding RNAs with a length of about 22 nucleotides, which inhibit the expression of target genes by inducing messenger RNA degradation, translation inhibition or other morphological regulation mechanisms. A large amount of research evidence shows that miRNAs play an important role in many biological processes. MiRNA dysfunction and miRNA mutations can lead to various diseases. Therefore, identifying the interaction between miRNAs and diseases is helpful for humans to understand the mechanism of the disease, thereby providing help for the prevention and treatment of the disease.
生物实验的方法需要大量的资源和时间成本,因此,许多预测miRNA-疾病关联的计算方法被提出。现有技术中,预测miRNA-疾病关联的计算方法主要包括基于机器学习的方法和基于网络的方法。然而上述两种方法只考虑了miRNA与疾病之间片面的信息,不能充分表征miRNA与疾病之间的复杂关系,且预测准确度有待提高。因此,有必要设计一种能充分学习到miRNA和疾病关系的方法。Biological experimental methods require a lot of resources and time costs. Therefore, many computational methods for predicting miRNA-disease associations have been proposed. In the prior art, calculation methods for predicting miRNA-disease associations mainly include machine learning-based methods and network-based methods. However, the above two methods only consider the one-sided information between miRNA and disease, and cannot fully characterize the complex relationship between miRNA and disease, and the prediction accuracy needs to be improved. Therefore, it is necessary to design a method that can fully learn the relationship between miRNA and disease.
专利(CN109256215A)公开了一种基于自回避随机游走的疾病关联miRNA预测方法及系统,该专利利用自回避随机游走遍历疾病-miRNA二分 图,使用自回避随机游走的两个属性(两个节点之间的转移概率与平均步长)之比来度量节点间的关联度,实现疾病与miRNA之间的关联预测。该方法并不能用于没有已知关联的疾病和miRNA之间关联预测。The patent (CN109256215A) discloses a disease-associated miRNA prediction method and system based on self-avoidance random walks. The patent uses self-avoidance random walks to traverse the disease-miRNA bipartite graph and uses the two attributes of self-avoidance random walks (two The ratio of the transition probability between each node to the average step size) is used to measure the degree of association between nodes, so as to realize the association prediction between disease and miRNA. This method cannot be used to predict the association between diseases and miRNAs for which there is no known association.
发明内容Summary of the invention
本申请实施例提供了一种miRNA-疾病关联预测方法、系统、终端以及存储介质,旨在至少在一定程度上解决现有技术中的预测miRNA-疾病关联计算方法只考虑了miRNA与疾病之间片面的信息、不能充分表征miRNA与疾病之间的复杂关系、预测准确度较低,以及不能用于没有已知关联的疾病和miRNA之间的关联预测的技术问题。The embodiments of the application provide a miRNA-disease association prediction method, system, terminal, and storage medium, which are intended to at least to a certain extent solve the problem of predicting miRNA-disease association calculation methods in the prior art, which only considers the relationship between miRNA and disease. One-sided information, inability to fully characterize the complex relationship between miRNAs and diseases, low prediction accuracy, and technical problems that cannot be used to predict the association between diseases and miRNAs without known associations.
为了解决上述问题,本申请实施例提供了如下技术方案:In order to solve the foregoing problems, the embodiments of the present application provide the following technical solutions:
一种miRNA-疾病关联预测方法,包括以下步骤:A miRNA-disease association prediction method includes the following steps:
步骤a:根据miRNA-疾病相关数据构建miRNA-疾病关联矩阵、miRNA相似度矩阵以及疾病相似度矩阵;Step a: Construct miRNA-disease association matrix, miRNA similarity matrix and disease similarity matrix based on miRNA-disease related data;
步骤b:根据所述miRNA-疾病关联矩阵、miRNA相似度矩阵和疾病相似度矩阵构建异构网络;Step b: construct a heterogeneous network according to the miRNA-disease association matrix, miRNA similarity matrix and disease similarity matrix;
步骤c:采用神经网络学习所述异构网络的拓扑信息,通过拓扑保持计算所述异构网络的最优参数,并根据所述最优参数重建所述异构网络;所述重建后的异构网络即为miRNA与疾病的关联得分矩阵。Step c: Use a neural network to learn the topology information of the heterogeneous network, calculate the optimal parameters of the heterogeneous network through topology maintenance, and reconstruct the heterogeneous network according to the optimal parameters; the reconstructed heterogeneous network The structural network is the correlation score matrix between miRNA and disease.
本申请实施例采取的技术方案还包括:所述步骤a中,所述miRNA-疾病相关数据包括miRNA-疾病关联数据、基因功能信息、miRNA-target关联信息、miRNA家族和簇的信息以及miRNA功能相似度数据。The technical solution adopted in the embodiment of the application further includes: in the step a, the miRNA-disease related data includes miRNA-disease related data, gene function information, miRNA-target related information, miRNA family and cluster information, and miRNA function Similarity data.
本申请实施例采取的技术方案还包括:在所述步骤a中,所述构建miRNA-疾病关联矩阵具体为:The technical solution adopted in the embodiment of the present application further includes: in the step a, the construction of the miRNA-disease association matrix is specifically:
根据所述miRNA-疾病关联数据构建miRNA-疾病关联矩阵。A miRNA-disease association matrix is constructed according to the miRNA-disease association data.
本申请实施例采取的技术方案还包括:在所述步骤a中,所述构建miRNA相似度矩阵具体为:The technical solution adopted in the embodiment of the present application further includes: in the step a, the construction of the miRNA similarity matrix is specifically:
根据所述miRNA-target关联信息、miRNA家族信息、miRNA簇信息以及miRNA功能相似度数据构建miRNA相似度矩阵。According to the miRNA-target association information, miRNA family information, miRNA cluster information, and miRNA functional similarity data, a miRNA similarity matrix is constructed.
本申请实施例采取的技术方案还包括:所述构建miRNA相似度矩阵还包括:The technical solution adopted in the embodiment of the application further includes: the construction of the miRNA similarity matrix further includes:
根据所述miRNA-target关联信息计算得到基于靶基因的相似度矩阵;Calculating a similarity matrix based on the target gene according to the miRNA-target association information;
根据所述miRNA家族信息计算得到基于miRNA家族信息的相似度矩阵;Calculating a similarity matrix based on the miRNA family information according to the miRNA family information;
根据miRNA簇信息计算得到基于miRNA簇信息的相似度矩阵;Calculate the similarity matrix based on the miRNA cluster information according to the miRNA cluster information;
根据miRNA功能相似度数据计算得到基于miRNA功能相似度数据的相似度矩阵;Calculate the similarity matrix based on the miRNA functional similarity data based on the miRNA functional similarity data;
将上述四种相似度矩阵加权求和,得到miRNA相似度矩阵。The above four similarity matrices are weighted and summed to obtain the miRNA similarity matrix.
本申请实施例采取的技术方案还包括:在所述步骤a中,所述构建疾病相似度矩阵具体为:The technical solution adopted in the embodiment of the present application further includes: in the step a, the constructing a disease similarity matrix is specifically:
基于所述基因功能信息构建疾病相似度矩阵。A disease similarity matrix is constructed based on the gene function information.
本申请实施例采取的技术方案还包括:在所述步骤b中,所述构建异构网络具体包括:The technical solution adopted in the embodiment of the present application further includes: in the step b, the constructing a heterogeneous network specifically includes:
将所述miRNA-疾病关联矩阵、所述miRNA相似度矩阵和所述疾病相似度矩阵标准化;Standardizing the miRNA-disease association matrix, the miRNA similarity matrix and the disease similarity matrix;
将标准化后的miRNA-疾病关联矩阵、所述miRNA相似度矩阵和所述疾病相似度矩阵合并成一个异构网络。The standardized miRNA-disease association matrix, the miRNA similarity matrix and the disease similarity matrix are combined into a heterogeneous network.
本申请实施例采取的技术方案还包括:在所述步骤c中,所述通过拓扑保持计算所述异构网络的最优参数包括:The technical solution adopted in the embodiment of the present application further includes: in the step c, the calculation of the optimal parameters of the heterogeneous network through topology preservation includes:
通过梯度下降方式得到网络最优参数:Obtain the optimal parameters of the network through gradient descent:
上式中,MS表示miRNA相似度矩阵,MD表示mirna-疾病关联矩阵,DS表示疾病相似度矩阵,F表示节点的特征矩阵,行数为节点个数,列数为特征向量的维度,FT表示F的转置矩阵。In the above formula, MS represents the miRNA similarity matrix, MD represents the mirna-disease association matrix, DS represents the disease similarity matrix, F represents the feature matrix of the node, the number of rows is the number of nodes, the number of columns is the dimension of the feature vector, FT Represents the transposed matrix of F.
本申请实施例采取的另一技术方案为:一种miRNA-疾病关联预测系统,包括:Another technical solution adopted in the embodiment of the present application is: a miRNA-disease association prediction system, including:
根据miRNA-疾病相关数据构建miRNA-疾病关联矩阵、miRNA相似度矩阵以及疾病相似度矩阵的第一矩阵构建模块、第二矩阵构建模块以及第三矩阵构建模块;以及Constructing the miRNA-disease association matrix, the miRNA similarity matrix, and the first matrix building module, the second matrix building module, and the third matrix building module of the disease similarity matrix according to the miRNA-disease related data; and
异构网络构建模块:用于根据所述miRNA-疾病关联矩阵、miRNA相似度矩阵和疾病相似度矩阵构建异构网络;Heterogeneous network construction module: used to construct a heterogeneous network according to the miRNA-disease association matrix, miRNA similarity matrix and disease similarity matrix;
特征提取模块:用于采用神经网络学习所述异构网络的拓扑信息;Feature extraction module: used to learn the topology information of the heterogeneous network by using a neural network;
网络参数计算模块:用于通过拓扑保持计算所述异构网络最优参数;Network parameter calculation module: used to calculate the optimal parameters of the heterogeneous network through topology maintenance;
网络重建模块:用于根据所述最优参数重建所述异构网络;所述重建后的异构网络即为miRNA与疾病的关联得分矩阵。Network reconstruction module: used to reconstruct the heterogeneous network according to the optimal parameters; the reconstructed heterogeneous network is the correlation score matrix between miRNA and disease.
本申请实施例采取的又一技术方案为:一种终端,所述终端包括处理器、与所述处理器耦接的存储器,其中,Another technical solution adopted by the embodiment of the present application is: a terminal, the terminal includes a processor and a memory coupled to the processor, wherein:
所述存储器存储有用于实现所述miRNA-疾病关联预测方法的程序指令;The memory stores program instructions for realizing the miRNA-disease association prediction method;
所述处理器用于执行所述存储器存储的所述程序指令以控制miRNA-疾病关联预测。The processor is configured to execute the program instructions stored in the memory to control miRNA-disease association prediction.
本申请实施例采取的又一技术方案为:一种存储介质,存储有处理器可运行的程序指令,所述程序指令用于执行所述miRNA-疾病关联预测方法。Another technical solution adopted by the embodiment of the present application is: a storage medium storing program instructions executable by a processor, and the program instructions are used to execute the miRNA-disease association prediction method.
相对于现有技术,本申请实施例产生的有益效果在于:本申请实施例的miRNA-疾病关联预测方法、系统、终端以及存储介质基于miRNA相似度网络、疾病的相似度网络和实验验证的miRNA-疾病关联网络构建异构网络,然后采用神经网络学习异构网络的拓扑信息,提取miRNA和疾病的特征表示,计算异构网络的最优参数,最后重建异构网络,提高了预测miRNA和疾病关联性的准确率。本申请解决了生物实验方法成本昂贵和耗时的问题,提高了miRNA-疾病关联预测的效果,具有实用性,并可用于没有已知关联的疾病和miRNA之间的关联预测。Compared with the prior art, the beneficial effects produced by the embodiments of the present application are: the miRNA-disease association prediction method, system, terminal and storage medium of the embodiments of the present application are based on miRNA similarity network, disease similarity network and experimentally verified miRNA -Disease association network constructs a heterogeneous network, and then uses neural network to learn the topology information of the heterogeneous network, extracts the feature representation of miRNA and disease, calculates the optimal parameters of the heterogeneous network, and finally reconstructs the heterogeneous network, which improves the prediction of miRNA and disease The accuracy of the relevance. This application solves the problems of high cost and time-consuming biological experiment methods, improves the effect of miRNA-disease association prediction, is practical, and can be used for association prediction between diseases and miRNAs that have no known association.
图1是本申请第一实施例的miRNA-疾病关联预测方法的流程图;Fig. 1 is a flowchart of a miRNA-disease association prediction method according to the first embodiment of the present application;
图2为本申请第二实施例的miRNA-疾病关联预测方法的预测结果示意图;2 is a schematic diagram of the prediction results of the miRNA-disease association prediction method according to the second embodiment of the application;
图3是本申请实施例的miRNA-疾病关联预测系统的结构示意图;Figure 3 is a schematic structural diagram of a miRNA-disease association prediction system according to an embodiment of the present application;
图4为本申请实施例的终端结构示意图;FIG. 4 is a schematic diagram of a terminal structure according to an embodiment of the application;
图5为本申请实施例的存储介质的结构示意图。FIG. 5 is a schematic structural diagram of a storage medium according to an embodiment of the application.
为了使本申请的目的、技术方案及优点更加清楚明白,以下结合附图及实施例,对本申请进行进一步详细说明。应当理解,此处所描述的具体实施例仅用以解释本申请,并不用于限定本申请。In order to make the purpose, technical solutions, and advantages of this application clearer and clearer, the following further describes the application in detail with reference to the accompanying drawings and embodiments. It should be understood that the specific embodiments described here are only used to explain the application, and are not used to limit the application.
请参阅图1,是本申请实施例的miRNA-疾病关联预测方法的流程图。本申请实施例的miRNA-疾病关联预测方法包括以下步骤:Please refer to FIG. 1, which is a flowchart of a miRNA-disease association prediction method according to an embodiment of the present application. The miRNA-disease association prediction method of the embodiment of the application includes the following steps:
步骤100:从数据库中获取与miRNA-疾病相关的多源信息;Step 100: Obtain multi-source information related to miRNA-disease from the database;
步骤100中,获取的多源信息具体包括:从人类miRNA-疾病数据库(HMDDv2.0)下载已知的miRNA-疾病关联数据;从HumanNet数据库下载基因功能信息;从miRTarBase数据库下载miRNA-target关联信息(靶基因);从miRBase数据库下载miRNA家族和簇的信息;下载miRNA功能相似度数据(http://www.cuilab.cn/files/images/cuilab/misim.zip)。In
步骤200:根据miRNA-疾病关联数据构建miRNA-疾病关联矩阵MD;Step 200: construct a miRNA-disease association matrix MD according to the miRNA-disease association data;
步骤200中,miRNA-疾病关联矩阵构建方式具体为:如果一种疾病和一种miRNA关联,则在两个节点之间加一条边。In
步骤300:根据靶基因信息、miRNA家族信息、miRNA簇信息以及miRNA功能相似度数据构建miRNA相似度矩阵MS;Step 300: construct a miRNA similarity matrix MS according to target gene information, miRNA family information, miRNA cluster information, and miRNA functional similarity data;
步骤300中,miRNA相似度矩阵的构建方式具体包括:In
步骤301:根据靶基因信息计算得到基于靶基因的相似度矩阵RST,该矩阵代表miRNA之间共同关联的靶基因的数量;Step 301: Calculate the target gene-based similarity matrix RST based on the target gene information, and the matrix represents the number of target genes commonly associated with miRNAs;
步骤302:根据miRNA家族信息计算得到基于miRNA家族信息的相似度矩阵RSF;Step 302: Calculate according to the miRNA family information to obtain a similarity matrix RSF based on the miRNA family information;
步骤303:根据miRNA簇信息计算得到基于miRNA簇信息的相似度矩阵RSC;Step 303: Calculate according to the miRNA cluster information to obtain a similarity matrix RSC based on the miRNA cluster information;
步骤304:根据miRNA功能相似度数据计算得到基于miRNA功能相似度数据的相似度矩阵RSD;Step 304: Calculate the similarity matrix RSD based on the miRNA functional similarity data based on the miRNA functional similarity data;
步骤305:然将上述四种相似度矩阵加权求和,构建最终的miRNA相似度矩阵:Step 305: Then, the above four similarity matrices are weighted and summed to construct the final miRNA similarity matrix:
MS(ri,rj)=α·RST(ri,rj)+β·RSF(rir,j)+γ·RSC(ri,rj)+θ·RSD(ri,rj) (1)MS(ri ,rj )=α·RST(ri ,rj )+β·RSF(ri r,j )+γ·RSC(ri ,rj )+θ·RSD(ri ,rj ) (1)
式(1)中,MS(ri,rj)表示miRNAri和miRNArj的相似度;α=0.2,β=0.1,γ=0.2,θ=0.55为矩阵权重。In formula (1), MS(ri , rj ) representsthe similarity between miRNAr i and miRNArj ; α=0.2, β=0.1, γ=0.2, θ=0.55 are matrix weights.
步骤400:基于基因功能信息构建疾病相似度矩阵DS;Step 400: construct a disease similarity matrix DS based on gene function information;
步骤400中,功能相似的基因可以调节相似的疾病,因此,本申请采用基因功能信息构建疾病的相似度矩阵。从HumanNet数据库下载的基因功能信息包含两个基因或基因集的对数似然得分LLS,疾病di和dj的相似度DS(di,di)计算公式如下:In step 400, genes with similar functions can regulate similar diseases. Therefore, this application uses gene function information to construct a disease similarity matrix. Information contains two genes or gene set from the database downloads HumanNet gene functions of the log-likelihood score LLS, di and dj disease similarity DS (di, di) is calculated as follows:
式(2)中,S(di)和S(dj)分别表示跟疾病di和dj相关的基因的集合。LLS(x,S(di))为基因x与基因集S(di)的对数似然得分。Formula (2), S (di) and S (dj) respectively set with di and dj of disease-related genes. LLS(x,S(di )) is the log-likelihood score of gene x and gene set S(di ).
步骤500:合并miRNA-疾病关联矩阵、miRNA相似度矩阵和疾病相似度矩阵,构建异构网络;Step 500: Combine the miRNA-disease association matrix, miRNA similarity matrix and disease similarity matrix to construct a heterogeneous network;
步骤500中,异构网络构建方式具体包括:In step 500, the heterogeneous network construction method specifically includes:
首先,将miRNA-疾病关联矩阵、miRNA相似度矩阵和疾病相似度矩阵标准化;假设A表示任意一个矩阵,A’表示标准化之后的矩阵,矩阵标准化公式可以表示成:First, standardize the miRNA-disease association matrix, miRNA similarity matrix, and disease similarity matrix; suppose A represents any matrix, and A'represents the standardized matrix. The matrix standardization formula can be expressed as:
式(3)中,Col(A)表示矩阵A的列数,(i,j)表示矩阵的第i行第j列,(i,k)表示矩阵的第i行第k列。In formula (3), Col(A) represents the number of columns of matrix A, (i,j) represents the i-th row and jth column of the matrix, and (i,k) represents the i-th row and kth column of the matrix.
然后,将三个矩阵合并成一个异构网络G=(V,E),V是节点的集合,包含两种类型的节点,NT={miRNA,疾病};E是边的集合,包含三种类型的边,ET={miRNA-miRNA,miRNA-疾病,疾病-疾病}。Then, merge the three matrices into a heterogeneous network G=(V,E), V is a set of nodes, including two types of nodes, NT={miRNA, disease}; E is a set of edges, including three types Type side, ET={miRNA-miRNA, miRNA-disease, disease-disease}.
步骤600:采用神经网络学习异构网络的拓扑信息,提取节点的深层特征表示;Step 600: Use the neural network to learn the topology information of the heterogeneous network, and extract the deep feature representation of the node;
步骤600中,汇聚邻居节点的信息,可以学习到网络拓扑结构信息,获得节点深层特征表示。对于任意属于类型t(t∈NT)的节点u,其深层特征表示通过融合邻居节点和其本身的信息,然后再标准化获得:In step 600, the information of neighboring nodes is gathered, the network topology structure information can be learned, and the deep feature representation of the node can be obtained. For any node u of type t(t∈NT), its deep feature representation is obtained by fusing the information of neighbor nodes and itself, and then standardizing:
式(4)中,EM′t为属于类型t的节点的深层特征表示,EMt为节点随机初始化的特征表示,E′(u)为标准化之后的最终的节点特征表示,v为与u由属 于类型t的边相连的邻居节点,Ws∈Rd×d、bs∈Rd、W0∈R2d×d和b0∈Rd为异构网络的训练参数,σ(·)为异构网络中的激活函数,本申请实施例中采用RELU函数。In formula (4), EM't is the deep feature representation of the node belonging to type t, EMt is the feature representation of the node randomly initialized, E'(u) is the final node feature representation after standardization, v is the same as u Neighbor nodes connected by edges of type t, Ws ∈Rd×d , bs ∈Rd , W0 ∈R2d×d and b0 ∈Rd are the training parameters of the heterogeneous network, and σ(·) is For the activation function in the heterogeneous network, the RELU function is used in the embodiment of this application.
步骤700:通过拓扑保持计算网络参数;Step 700: Maintain and calculate network parameters through the topology;
步骤700中,本申请实施例通过最小化重建矩阵与原矩阵之间的均方误差求得最优参数,具体为通过梯度下降的方式得到最优参数:In step 700, the embodiment of the present application obtains the optimal parameter by minimizing the mean square error between the reconstructed matrix and the original matrix, and specifically obtains the optimal parameter by means of gradient descent:
式(5)中,MS表示miRNA相似度矩阵,MD表示mirna-疾病关联矩阵,DS表示疾病相似度矩阵,F表示节点的特征矩阵,行数为节点个数,列数为特征向量的维度,FT表示F的转置矩阵。In formula (5), MS represents the miRNA similarity matrix, MD represents the mirna-disease association matrix, DS represents the disease similarity matrix, F represents the feature matrix of the node, the number of rows is the number of nodes, and the number of columns is the dimension of the feature vector. FT represents the transposed matrix of F.
步骤800:根据最优参数重建异构网络;Step 800: Rebuild the heterogeneous network according to the optimal parameters;
步骤800中,参数训练完成后,每种miRNA和疾病之间的关联得分可以通过重建异构网络得到:In step 800, after the parameter training is completed, the correlation score between each miRNA and the disease can be obtained by reconstructing the heterogeneous network:
MD‘=(MDreeconstruct+DMTreeconstruct)/2 (7)MD'=(MDreeconstruct +DMTreeconstruct )/2 (7)
重建后的异构网络MD‘即为miRNA与疾病的关联得分矩阵,得分越高,则miRNA与疾病之间的关联性越强。The reconstructed heterogeneous network MD' is the correlation score matrix between miRNA and disease. The higher the score, the stronger the correlation between miRNA and disease.
实施例一:Example one:
以下为本申请第二实施例的miRNA-疾病关联预测方法的实现步骤。本申请第二实施例选取公开的数据为测试数据,并采用5折交叉验证的方法对本申请技术方案进行评估。具体包括:The following are the implementation steps of the miRNA-disease association prediction method according to the second embodiment of the application. The second embodiment of the present application selects the disclosed data as test data, and uses a 5-fold cross-validation method to evaluate the technical solution of the present application. Specifically:
S1:从数据库中获取与miRNA-疾病相关的多源信息;S1: Obtain multi-source information related to miRNA-disease from the database;
S2:根据miRNA-疾病关联数据构建miRNA-疾病关联矩阵MD;S2: Construct miRNA-disease association matrix MD based on miRNA-disease association data;
该步骤中,从测试数据中去掉重复数据后,共有577种miRNA和336种疾病,6441个关联。在构建miRNA-疾病关联矩阵时,将所有关联随机分成5份,其中4份作为训练集,1份作为测试集,测试集中的关联在miRNA-疾病关联矩阵中设为0,循环进行5次,直到所有关联都被测试。In this step, after removing duplicate data from the test data, there are a total of 577 miRNAs, 336 diseases, and 6,441 associations. When constructing the miRNA-disease association matrix, all associations are randomly divided into 5 parts, 4 parts are used as the training set and 1 part is used as the test set. The association in the test set is set to 0 in the miRNA-disease association matrix, and the cycle is repeated 5 times. Until all associations are tested.
S3:根据靶基因信息、miRNA家族信息、miRNA簇信息以及miRNA功能相似度数据构建miRNA相似度矩阵MS;S3: Construct a miRNA similarity matrix MS based on target gene information, miRNA family information, miRNA cluster information, and miRNA functional similarity data;
本步骤中,基于miRNA家族信息计算的miRNA相似度矩阵RSF,数据中的miRNA属于299个不同的家族,如果两个miRNA属于同一个家族,它们在相似度矩阵中的值设为1,反之设为0;基于miRNA簇的信息计算的miRNA相似度矩阵RSC,数据中的miRNA属于153个不同的簇,同矩阵RSF一样,如果两个miRNA属于同一个簇,它们在相似度矩阵中的值设为1,反之设为0。In this step, the miRNA similarity matrix RSF is calculated based on the miRNA family information. The miRNAs in the data belong to 299 different families. If two miRNAs belong to the same family, their value in the similarity matrix is set to 1, and vice versa. Is 0; the miRNA similarity matrix RSC calculated based on the miRNA cluster information, the miRNAs in the data belong to 153 different clusters, the same as the matrix RSF, if two miRNAs belong to the same cluster, their values in the similarity matrix are set Set to 1, otherwise set to 0.
S4:构建疾病相似度矩阵DS,得到336种疾病之间的112,896个关联相似度;S4: Construct the disease similarity matrix DS to obtain 112,896 association similarities among 336 diseases;
S5:构建异构网络;S5: Build a heterogeneous network;
S6:训练神经网络;S6: Training neural network;
S7:重建异构网络;S7: Rebuild a heterogeneous network;
S8:结果评估;S8: Result evaluation;
将MD‘中训练集关联的得分与其真实标签对比,通过计算不同阈值下的假阳性率(FPR)和真阳性率(TPR),绘制了接收者操作特征曲线(ROC) 并采用AUC(ROC下曲线面积)值来评价预测的结果,如图2所示。预测结果表明本申请可以提高miRNA-疾病关联预测的效果,具有较高的实用性。Comparing the scores associated with the training set in MD' with the real labels, by calculating the false positive rate (FPR) and true positive rate (TPR) under different thresholds, the receiver operating characteristic curve (ROC) is drawn and AUC (under ROC) is used. (Curve area) value to evaluate the predicted results, as shown in Figure 2. The prediction result shows that the application can improve the effect of miRNA-disease association prediction, and has high practicability.
实施例二:Embodiment two:
为了进一步评估本申请预测miRNA-疾病关联的真实性能,以下实施例针对胰腺癌进行了案例研究。在案例中,将所有跟胰腺癌相关的已知关联都标记为未知,然后研究模型找出这些关联的miRNA的能力。选择预测得分最高的前50个miRNA在dbDEMC2和PhenomiR2数据库中进行验证,数据库存储了实验证实的miRNA和疾病的关联。从验证结果可以看出,通过本申请预测得到的与胰腺癌相关的50个miRNA(如表1所示)均在数据库中找到,进一步证明了模型的实用性,模型可以用于没有任何已知关联的疾病的相关miRNA预测。In order to further evaluate the true performance of this application for predicting miRNA-disease associations, the following examples have conducted case studies on pancreatic cancer. In the case, all known associations related to pancreatic cancer are marked as unknown, and the model's ability to find these associated miRNAs is then studied. The top 50 miRNAs with the highest prediction scores are selected for verification in the dbDEMC2 and PhenomiR2 databases, which store experimentally confirmed associations between miRNAs and diseases. It can be seen from the verification results that the 50 miRNAs (shown in Table 1) related to pancreatic cancer predicted by this application are all found in the database, which further proves the practicality of the model and that the model can be used without any known Related miRNA predictions for related diseases.
请参阅图3,是本申请实施例的miRNA-疾病关联预测系统的结构示意图。本申请实施例的miRNA-疾病关联预测系统包括:Please refer to FIG. 3, which is a schematic structural diagram of a miRNA-disease association prediction system according to an embodiment of the present application. The miRNA-disease association prediction system of the embodiment of the application includes:
数据获取模块:用于从数据库中获取与miRNA-疾病相关的多源信息;其中,获取的多源信息具体包括:从人类miRNA-疾病数据库(HMDDv2.0)下载已知的miRNA-疾病关联数据;从HumanNet数据库下载基因功能信息;从miRTarBase数据库下载miRNA-target关联信息(靶基因);从miRBase数据库下载miRNA家族和簇的信息;下载miRNA功能相似度数据(http://www.cuilab.cn/files/images/cuilab/misim.zip)。Data acquisition module: used to acquire multi-source information related to miRNA-disease from the database; the acquired multi-source information specifically includes: downloading known miRNA-disease-related data from the human miRNA-disease database (HMDDv2.0) Download gene function information from HumanNet database; download miRNA-target association information (target gene) from miRTarBase database; download miRNA family and cluster information from miRBase database; download miRNA functional similarity data (http://www.cuilab.cn /files/images/cuilab/misim.zip).
第一矩阵构建模块:用于根据miRNA-疾病关联数据构建miRNA-疾病关联矩阵MD;其中,miRNA-疾病关联矩阵构建方式具体为:如果一种疾病和一种miRNA关联,则在两个节点之间加一条边。The first matrix building module: used to construct a miRNA-disease association matrix MD based on the miRNA-disease association data; among them, the miRNA-disease association matrix construction method is specifically: if a disease is associated with a miRNA, then between two nodes Add an edge in between.
第二矩阵构建模块:用于根据靶基因信息、miRNA家族信息、miRNA簇信息以及miRNA功能相似度数据构建miRNA相似度矩阵MS;其中,miRNA相似度矩阵的构建方式具体包括:The second matrix construction module: used to construct a miRNA similarity matrix MS based on target gene information, miRNA family information, miRNA cluster information, and miRNA functional similarity data; among them, the construction of miRNA similarity matrix specifically includes:
根据靶基因信息计算得到基于靶基因的相似度矩阵RST,该矩阵代表miRNA之间共同关联的靶基因的数量;Calculate the target gene-based similarity matrix RST based on the target gene information, which represents the number of target genes that are commonly associated between miRNAs;
根据miRNA家族信息计算得到基于miRNA家族信息的相似度矩阵RSF;Calculate the similarity matrix RSF based on the miRNA family information according to the miRNA family information;
根据miRNA簇信息计算得到基于miRNA簇信息的相似度矩阵RSC;Calculate the similarity matrix RSC based on the miRNA cluster information according to the miRNA cluster information;
根据miRNA功能相似度数据计算得到基于miRNA功能相似度数据的相似度矩阵RSD;Calculate the similarity matrix RSD based on the miRNA functional similarity data based on the miRNA functional similarity data;
然将上述四种相似度矩阵加权求和,构建最终的miRNA相似度矩阵:Then, the above four similarity matrices are weighted and summed to construct the final miRNA similarity matrix:
MS(ri,rj)=α·RST(ri,rj)+β·RSF(ri,rj)+γ·RSC(ri,rj)+θ·RSD(ri,rj) (1)MS(ri ,rj )=α·RST(ri ,rj )+β·RSF(ri ,rj )+γ·RSC(ri ,rj )+θ·RSD(ri ,rj ) (1)
式(1)中,MS(ri,rj)表示miRNAri和miRNArj的相似度;α=0.2,β=0.1,γ=0.2,θ=0.55为矩阵权重。In formula (1), MS(ri , rj ) representsthe similarity between miRNAr i and miRNArj ; α=0.2, β=0.1, γ=0.2, θ=0.55 are matrix weights.
第三矩阵构建模块:用于基于基因功能信息构建疾病相似度矩阵DS;其中,功能相似的基因可以调节相似的疾病,因此,本申请采用基因功能信息构建疾病的相似度矩阵。从HumanNet数据库下载的基因功能信息包含两个基因或基因集的对数似然得分LLS,疾病di和dj的相似度DS(di,di)计算公式如下:The third matrix construction module is used to construct a disease similarity matrix DS based on gene function information; among them, genes with similar functions can regulate similar diseases. Therefore, this application uses gene function information to construct a disease similarity matrix. Information contains two genes or gene set from the database downloads HumanNet gene functions of the log-likelihood score LLS, di and dj disease similarity DS (di, di) is calculated as follows:
式(2)中,S(di)和S(dj)分别表示跟疾病di和dj相关的基因的集合。LLS(x,S(di))为基因x与基因集S(di)的对数似然得分。Formula (2), S (di) and S (dj) respectively set with di and dj of disease-related genes. LLS(x,S(di )) is the log-likelihood score of gene x and gene set S(di ).
异构网络构建模块:用于合并miRNA-疾病关联矩阵、miRNA相似度矩阵和疾病相似度矩阵,构建异构网络;其中,异构网络构建方式具体包括:Heterogeneous network building module: used to merge miRNA-disease association matrix, miRNA similarity matrix and disease similarity matrix to construct a heterogeneous network; among them, the heterogeneous network construction methods include:
首先,将miRNA-疾病关联矩阵、miRNA相似度矩阵和疾病相似度矩阵标准化;假设A表示任意一个矩阵,A’表示标准化之后的矩阵,矩阵标准化公式可以表示成:First, standardize the miRNA-disease association matrix, miRNA similarity matrix, and disease similarity matrix; suppose A represents any matrix, and A'represents the standardized matrix. The matrix standardization formula can be expressed as:
式(3)中,Col(A)表示矩阵A的列数,(i,j)表示矩阵的第i行第j列,(i,k)表示矩阵的第i行第k列。In formula (3), Col(A) represents the number of columns of matrix A, (i,j) represents the i-th row and jth column of the matrix, and (i,k) represents the i-th row and kth column of the matrix.
然后,将三个矩阵合并成一个异构网络G=(V,E),V是节点的集合,包含两种类型的节点,NT={miRNA,疾病};E是边的集合,包含三种类型的边,ET={miRNA-miRNA,miRNA-疾病,疾病-疾病}。Then, merge the three matrices into a heterogeneous network G=(V,E), V is a set of nodes, including two types of nodes, NT={miRNA, disease}; E is a set of edges, including three types Type side, ET={miRNA-miRNA, miRNA-disease, disease-disease}.
特征提取模块:用于采用神经网络学习异构网络的拓扑信息,提取节点的深层特征表示;其中,汇聚邻居节点的信息,可以学习到网络拓扑结构信息,获得节点深层特征表示。对于任意属于类型t(t∈NT)的节点u,其深层特征表示通过融合邻居节点和其本身的信息,然后再标准化获得:Feature extraction module: used to use neural network to learn the topology information of heterogeneous networks and extract the deep feature representations of nodes; among them, gather the information of neighbor nodes to learn network topology structure information and obtain deep feature representations of nodes. For any node u of type t(t∈NT), its deep feature representation is obtained by fusing the information of neighbor nodes and itself, and then standardizing:
式(4)中,EM′t为属于类型t的节点的深层特征表示,EMt为节点随机初始化的特征表示,E′(u)为标准化之后的最终的节点特征表示,v为与u由属于类型t的边相连的邻居节点,Ws∈Rd×d、bs∈Rd、W0∈R2d×d和b0∈Rd为异构网络的训练参数,σ(·)为异构网络中的激活函数,本申请实施例中采用RELU函数。In formula (4), EM't is the deep feature representation of the node belonging to type t, EMt is the feature representation of the node randomly initialized, E'(u) is the final node feature representation after standardization, v is the same as u Neighbor nodes connected by edges of type t, Ws ∈Rd×d , bs ∈Rd , W0 ∈R2d×d and b0 ∈Rd are the training parameters of the heterogeneous network, and σ(·) is For the activation function in the heterogeneous network, the RELU function is used in the embodiment of this application.
网络参数计算模块:用于通过拓扑保持计算网络参数;其中,本申请通过最小化重建矩阵与原矩阵之间的均方误差求得最优参数,具体为通过梯度下降的方式得到最优参数:Network parameter calculation module: used to maintain and calculate network parameters through topology; among them, this application obtains the optimal parameters by minimizing the mean square error between the reconstructed matrix and the original matrix, specifically the optimal parameters are obtained by gradient descent:
式(5)中,MS表示miRNA相似度矩阵,MD表示mirna-疾病关联矩阵,DS表示疾病相似度矩阵,F表示节点的特征矩阵,行数为节点个数,列数为特征向量的维度,FT表示F的转置矩阵。In formula (5), MS represents the miRNA similarity matrix, MD represents the mirna-disease association matrix, DS represents the disease similarity matrix, F represents the feature matrix of the node, the number of rows is the number of nodes, and the number of columns is the dimension of the feature vector. FT represents the transposed matrix of F.
网络重建模块:用于根据最优参数重建异构网络;参数训练完成后,每种miRNA和疾病之间的关联得分可以通过重建异构网络得到:Network reconstruction module: used to reconstruct the heterogeneous network according to the optimal parameters; after the parameter training is completed, the correlation score between each miRNA and disease can be obtained by reconstructing the heterogeneous network:
MD‘=(MDreeconstruct+DMTreeconstruct)/2 (7)MD'=(MDreeconstruct +DMTreeconstruct )/2 (7)
重建后的异构网络MD‘即为miRNA与疾病的关联得分矩阵,得分越高,则miRNA与疾病之间的关联性越强。The reconstructed heterogeneous network MD' is the correlation score matrix between miRNA and disease. The higher the score, the stronger the correlation between miRNA and disease.
请参阅图4,为本申请实施例的终端结构示意图。该终端50包括处理器51、与处理器51耦接的存储器52。Please refer to FIG. 4, which is a schematic diagram of a terminal structure according to an embodiment of the application. The terminal 50 includes a
存储器52存储有用于实现上述miRNA-疾病关联预测方法的程序指令。The
处理器51用于执行存储器52存储的程序指令以控制miRNA-疾病关联预测。The
其中,处理器51还可以称为CPU(Central Processing Unit,中央处理单元)。处理器51可能是一种集成电路芯片,具有信号的处理能力。处理器51还可以是通用处理器、数字信号处理器(DSP)、专用集成电路(ASIC)、现成可编程门阵列(FPGA)或者其他可编程逻辑器件、分立门或者晶体管逻辑器件、分立硬件组件。通用处理器可以是微处理器或者该处理器也可以是任何常规的处理器等。The
请参阅图5,为本申请实施例的存储介质的结构示意图。本申请实施例的存储介质存储有能够实现上述所有方法的程序文件61,其中,该程序文件61可以以软件产品的形式存储在上述存储介质中,包括若干指令用以使得一台计算机设备(可以是个人计算机,服务器,或者网络设备等)或处理器(processor)执行本发明各个实施方式方法的全部或部分步骤。而前述的存储介质包括:U盘、移动硬盘、只读存储器(ROM,Read-Only Memory)、随机存取存储器(RAM,Random Access Memory)、磁碟或者光盘等各种可以存储程序代码的介质,或者是计算机、服务器、手机、平板等终端设备。Please refer to FIG. 5, which is a schematic structural diagram of a storage medium according to an embodiment of the application. The storage medium of this embodiment of the present application stores a
本申请实施例的miRNA-疾病关联预测方法、系统、终端以及存储介质基于miRNA相似度网络、疾病的相似度网络和实验验证的miRNA-疾病关联网络构建异构网络,然后采用神经网络学习异构网络的拓扑信息,提取miRNA和疾病的特征表示,计算异构网络的最优参数,最后重建异构网络,提高了预测miRNA和疾病关联性的准确率。本申请解决了生物实验方法成本昂贵和耗时的问题,提高了miRNA-疾病关联预测的效果,具有实用性,并可用于没有已知关联的疾病和miRNA之间的关联预测。The miRNA-disease association prediction method, system, terminal and storage medium of the embodiments of the application construct a heterogeneous network based on the miRNA similarity network, the disease similarity network and the experimentally verified miRNA-disease association network, and then use the neural network to learn the heterogeneity The topology information of the network, the feature representation of miRNA and disease are extracted, the optimal parameters of the heterogeneous network are calculated, and the heterogeneous network is finally reconstructed, which improves the accuracy of predicting the correlation between miRNA and disease. This application solves the problems of high cost and time-consuming biological experiment methods, improves the effect of miRNA-disease association prediction, is practical, and can be used for association prediction between diseases and miRNAs that have no known association.
对所公开的实施例的上述说明,使本领域专业技术人员能够实现或使用本申请。对这些实施例的多种修改对本领域的专业技术人员来说将是显而易见的,本申请中所定义的一般原理可以在不脱离本申请的精神或范围的情况下,在其它实施例中实现。因此,本申请将不会被限制于本申请所示的这些实施例,而是要符合与本申请所公开的原理和新颖特点相一致的最宽的范围。The foregoing description of the disclosed embodiments enables those skilled in the art to implement or use this application. Various modifications to these embodiments will be obvious to those skilled in the art, and the general principles defined in this application can be implemented in other embodiments without departing from the spirit or scope of this application. Therefore, this application will not be limited to the embodiments shown in this application, but should conform to the widest scope consistent with the principles and novel features disclosed in this application.
Claims (11)
Translated fromChineseApplications Claiming Priority (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| CN202010438557.9 | 2020-05-21 | ||
| CN202010438557.9ACN111681705B (en) | 2020-05-21 | 2020-05-21 | MiRNA-disease association prediction method, system, terminal and storage medium |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| WO2021232789A1true WO2021232789A1 (en) | 2021-11-25 |
Family
ID=72434200
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/CN2020/139689CeasedWO2021232789A1 (en) | 2020-05-21 | 2020-12-25 | Mirna-disease association prediction method, system, terminal, and storage medium |
Country Status (2)
| Country | Link |
|---|---|
| CN (1) | CN111681705B (en) |
| WO (1) | WO2021232789A1 (en) |
Cited By (11)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN114093527A (en)* | 2021-12-01 | 2022-02-25 | 中国科学院新疆理化技术研究所 | A method and system for drug relocation based on spatial similarity constraints and non-negative matrix factorization |
| CN114613452A (en)* | 2022-03-08 | 2022-06-10 | 电子科技大学 | Drug relocation method and system based on drug classification map neural network |
| CN114613437A (en)* | 2022-03-08 | 2022-06-10 | 电子科技大学 | A method and system for predicting the association between miRNA and disease based on heterogeneous graph |
| CN114613438A (en)* | 2022-03-08 | 2022-06-10 | 电子科技大学 | A miRNA-disease association prediction method and system |
| CN115359903A (en)* | 2022-08-17 | 2022-11-18 | 常州大学 | miRNA-disease prediction method based on sparse learning and random walk |
| CN116189780A (en)* | 2023-02-14 | 2023-05-30 | 广东工业大学 | MiRNA-disease association prediction method based on Logistic function filling similarity matrix |
| CN116467465A (en)* | 2023-04-18 | 2023-07-21 | 平安科技(深圳)有限公司 | Text labeling method, device, and computer equipment based on knowledge graph |
| CN117238377A (en)* | 2023-05-09 | 2023-12-15 | 常州大学 | miRNA and disease association prediction method based on graph rolling network |
| CN117316268A (en)* | 2023-08-25 | 2023-12-29 | 常州大学 | Cross-modal and graph convolution-based miRNA and disease prediction method |
| CN117393143A (en)* | 2023-10-11 | 2024-01-12 | 哈尔滨工业大学(深圳)(哈尔滨工业大学深圳科技创新研究院) | Circular RNA-disease association prediction method based on graph representation learning |
| CN119541653A (en)* | 2024-11-22 | 2025-02-28 | 华中科技大学 | A method for constructing a miRNA-disease association prediction model based on metapathway embedding |
Families Citing this family (11)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN111681705B (en)* | 2020-05-21 | 2024-05-24 | 中国科学院深圳先进技术研究院 | MiRNA-disease association prediction method, system, terminal and storage medium |
| CN112183837A (en)* | 2020-09-22 | 2021-01-05 | 曲阜师范大学 | A prediction method of miRNA-disease association based on autoencoding model |
| CN112364880B (en)* | 2020-11-30 | 2022-06-14 | 腾讯科技(深圳)有限公司 | Omics data processing method, device, equipment and medium based on graph neural network |
| IL289151B2 (en)* | 2020-12-23 | 2025-08-01 | Bgi Genomics Co Ltd | Method and device for determining a degree of gene association |
| CN112667772B (en)* | 2020-12-23 | 2023-04-07 | 深圳华大基因科技服务有限公司 | Method and device for determining gene association degree |
| CN112784913B (en)* | 2021-01-29 | 2023-07-25 | 湖南大学 | A miRNA-disease association prediction method and device based on graph neural network fusion of multi-view information |
| CN112951320B (en)* | 2021-03-03 | 2023-05-16 | 深圳大学 | Biomedical network association prediction method based on ensemble learning |
| CN112951328B (en)* | 2021-03-03 | 2022-04-22 | 湖南大学 | miRNA-gene relationship prediction method and system based on deep learning heterogeneous information network |
| CN113838527B (en)* | 2021-09-26 | 2023-09-01 | 平安科技(深圳)有限公司 | Method and device for generating target gene prediction model and storage medium |
| CN114171124A (en)* | 2021-11-22 | 2022-03-11 | 湖南大学 | Regression-based disease and miRNA (micro ribonucleic acid) associated prediction method with excellent performance |
| CN119649913A (en)* | 2024-11-28 | 2025-03-18 | 长沙理工大学 | A disease gene identification method and system based on heterogeneous network impulse dynamics |
Citations (5)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US20140180599A1 (en)* | 2012-12-20 | 2014-06-26 | Samsung Electronics Co., Ltd. | Methods and apparatus for analyzing genetic information |
| CN109243538A (en)* | 2018-07-19 | 2019-01-18 | 长沙学院 | A kind of method and system of predictive disease and LncRNA incidence relation |
| CN110400600A (en)* | 2019-08-01 | 2019-11-01 | 枣庄学院 | A miRNA-disease correlation prediction method based on rotation forest algorithm |
| CN110853763A (en)* | 2019-11-11 | 2020-02-28 | 湖南城市学院 | Fusion attribute-based miRNA-disease association identification method and system |
| CN111681705A (en)* | 2020-05-21 | 2020-09-18 | 中国科学院深圳先进技术研究院 | A miRNA-disease association prediction method, system, terminal and storage medium |
Family Cites Families (9)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US11037684B2 (en)* | 2014-11-14 | 2021-06-15 | International Business Machines Corporation | Generating drug repositioning hypotheses based on integrating multiple aspects of drug similarity and disease similarity |
| WO2018049506A1 (en)* | 2016-09-14 | 2018-03-22 | Ontario Institute For Cancer Research (Oicr) | Mirna prostate cancer marker |
| CN107506608B (en)* | 2017-09-29 | 2020-09-29 | 杭州电子科技大学 | An improved collaborative filtering-based miRNA-disease association prediction method |
| CN107506617B (en)* | 2017-09-29 | 2020-07-21 | 杭州电子科技大学 | Semi-local social information miRNA-disease association prediction method |
| CN107862179A (en)* | 2017-11-06 | 2018-03-30 | 中南大学 | A kind of miRNA disease association Relationship Prediction methods decomposed based on similitude and logic matrix |
| EP3550568B8 (en)* | 2018-04-07 | 2024-08-14 | Tata Consultancy Services Limited | Graph convolution based gene prioritization on heterogeneous networks |
| CN109935332A (en)* | 2019-03-01 | 2019-06-25 | 桂林电子科技大学 | A miRNA-disease association prediction method based on double random walk model |
| CN110782948A (en)* | 2019-10-18 | 2020-02-11 | 湖南大学 | Predicting potential associations of miRNAs with diseases based on constrained probability matrix factorization |
| CN111161796B (en)* | 2019-12-30 | 2024-04-16 | 中南大学 | Method and system for predicting PD potential gene and miRNA |
- 2020
- 2020-05-21CNCN202010438557.9Apatent/CN111681705B/enactiveActive
- 2020-12-25WOPCT/CN2020/139689patent/WO2021232789A1/ennot_activeCeased
Patent Citations (5)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US20140180599A1 (en)* | 2012-12-20 | 2014-06-26 | Samsung Electronics Co., Ltd. | Methods and apparatus for analyzing genetic information |
| CN109243538A (en)* | 2018-07-19 | 2019-01-18 | 长沙学院 | A kind of method and system of predictive disease and LncRNA incidence relation |
| CN110400600A (en)* | 2019-08-01 | 2019-11-01 | 枣庄学院 | A miRNA-disease correlation prediction method based on rotation forest algorithm |
| CN110853763A (en)* | 2019-11-11 | 2020-02-28 | 湖南城市学院 | Fusion attribute-based miRNA-disease association identification method and system |
| CN111681705A (en)* | 2020-05-21 | 2020-09-18 | 中国科学院深圳先进技术研究院 | A miRNA-disease association prediction method, system, terminal and storage medium |
Cited By (13)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN114093527A (en)* | 2021-12-01 | 2022-02-25 | 中国科学院新疆理化技术研究所 | A method and system for drug relocation based on spatial similarity constraints and non-negative matrix factorization |
| CN114613437B (en)* | 2022-03-08 | 2023-05-26 | 电子科技大学 | Method and system for predicting association of miRNA and diseases based on different patterns |
| CN114613437A (en)* | 2022-03-08 | 2022-06-10 | 电子科技大学 | A method and system for predicting the association between miRNA and disease based on heterogeneous graph |
| CN114613438A (en)* | 2022-03-08 | 2022-06-10 | 电子科技大学 | A miRNA-disease association prediction method and system |
| CN114613452B (en)* | 2022-03-08 | 2023-04-28 | 电子科技大学 | Drug repositioning method and system based on drug classification graph neural network |
| CN114613452A (en)* | 2022-03-08 | 2022-06-10 | 电子科技大学 | Drug relocation method and system based on drug classification map neural network |
| CN115359903A (en)* | 2022-08-17 | 2022-11-18 | 常州大学 | miRNA-disease prediction method based on sparse learning and random walk |
| CN116189780A (en)* | 2023-02-14 | 2023-05-30 | 广东工业大学 | MiRNA-disease association prediction method based on Logistic function filling similarity matrix |
| CN116467465A (en)* | 2023-04-18 | 2023-07-21 | 平安科技(深圳)有限公司 | Text labeling method, device, and computer equipment based on knowledge graph |
| CN117238377A (en)* | 2023-05-09 | 2023-12-15 | 常州大学 | miRNA and disease association prediction method based on graph rolling network |
| CN117316268A (en)* | 2023-08-25 | 2023-12-29 | 常州大学 | Cross-modal and graph convolution-based miRNA and disease prediction method |
| CN117393143A (en)* | 2023-10-11 | 2024-01-12 | 哈尔滨工业大学(深圳)(哈尔滨工业大学深圳科技创新研究院) | Circular RNA-disease association prediction method based on graph representation learning |
| CN119541653A (en)* | 2024-11-22 | 2025-02-28 | 华中科技大学 | A method for constructing a miRNA-disease association prediction model based on metapathway embedding |
Also Published As
| Publication number | Publication date |
|---|---|
| CN111681705B (en) | 2024-05-24 |
| CN111681705A (en) | 2020-09-18 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| WO2021232789A1 (en) | Mirna-disease association prediction method, system, terminal, and storage medium | |
| CN109935332A (en) | A miRNA-disease association prediction method based on double random walk model | |
| Knag et al. | A sparse coding neural network ASIC with on-chip learning for feature extraction and encoding | |
| CN113299338B (en) | Synthetic lethal gene pair prediction method, system, terminal and medium based on knowledge graph | |
| Zeng et al. | Deep collaborative filtering for prediction of disease genes | |
| CN112420126A (en) | A drug target prediction method based on multi-source data fusion and network structure perturbation | |
| CN115602243A (en) | A prediction method of disease association information based on multi-similarity fusion | |
| CN110782948A (en) | Predicting potential associations of miRNAs with diseases based on constrained probability matrix factorization | |
| WO2018036547A1 (en) | Data processing method and device thereof | |
| CN114913916A (en) | Drug repositioning method for predicting new coronavirus-adapted drugs | |
| CN115019878B (en) | A drug discovery method based on graph representation and deep learning | |
| CN108920895A (en) | A kind of incidence relation prediction technique of circular rna and disease | |
| CN115985520B (en) | Prediction method of drug-disease association based on graph regularized matrix factorization | |
| CN116646002A (en) | Multi-non-coding RNA and disease association prediction method, device, equipment and medium | |
| Lee et al. | SOMEA: self-organizing map based extraction algorithm for DNA motif identification with heterogeneous model | |
| Wu et al. | Leveraging multiple gene networks to prioritize GWAS candidate genes via network representation learning | |
| CN117423389A (en) | Disease-associated circRNA prediction method based on graph attention random walk | |
| Zhao et al. | Protein function prediction with functional and topological knowledge of gene ontology | |
| Dong et al. | Neighborhood contrastive learning-based graph neural network for bug triaging | |
| Lin et al. | Computing the diffusion state distance on graphs via algebraic multigrid and random projections | |
| CN114678064A (en) | Drug target interaction prediction method based on network characterization learning | |
| US20240281641A1 (en) | Model Weight Obtaining Method and Related System | |
| CN112951320B (en) | Biomedical network association prediction method based on ensemble learning | |
| CN116646012A (en) | LncRNA-disease association prediction method based on unbalanced neighbor constrained random walk | |
| JP6172315B2 (en) | Method and apparatus for mixed model selection |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application | Ref document number:20936560 Country of ref document:EP Kind code of ref document:A1 | |
| NENP | Non-entry into the national phase | Ref country code:DE | |
| 122 | Ep: pct application non-entry in european phase | Ref document number:20936560 Country of ref document:EP Kind code of ref document:A1 | |
| 122 | Ep: pct application non-entry in european phase | Ref document number:20936560 Country of ref document:EP Kind code of ref document:A1 | |
| 32PN | Ep: public notification in the ep bulletin as address of the adressee cannot be established | Free format text:NOTING OF LOSS OF RIGHTS PURSUANT TO RULE 112(1) EPC (EPO FORM 1205A DATED 19/04/2023) |