CN114464250A - Gene stability screening method and system based on Ito quantum annealing - Google Patents
Gene stability screening method and system based on Ito quantum annealingDownload PDFInfo
- Publication number
- CN114464250A CN114464250ACN202210179637.6ACN202210179637ACN114464250ACN 114464250 ACN114464250 ACN 114464250ACN 202210179637 ACN202210179637 ACN 202210179637ACN 114464250 ACN114464250 ACN 114464250A
- Authority
- CN
- China
- Prior art keywords
- nodes
- ising
- node
- genetic
- spin
- Prior art date
- Legal status (The legal status is an assumption and is not a legal conclusion. Google has not performed a legal analysis and makes no representation as to the accuracy of the status listed.)
- Pending
Links
Images
Classifications
- G—PHYSICS
- G16—INFORMATION AND COMMUNICATION TECHNOLOGY [ICT] SPECIALLY ADAPTED FOR SPECIFIC APPLICATION FIELDS
- G16B—BIOINFORMATICS, i.e. INFORMATION AND COMMUNICATION TECHNOLOGY [ICT] SPECIALLY ADAPTED FOR GENETIC OR PROTEIN-RELATED DATA PROCESSING IN COMPUTATIONAL MOLECULAR BIOLOGY
- G16B20/00—ICT specially adapted for functional genomics or proteomics, e.g. genotype-phenotype associations
- G—PHYSICS
- G16—INFORMATION AND COMMUNICATION TECHNOLOGY [ICT] SPECIALLY ADAPTED FOR SPECIFIC APPLICATION FIELDS
- G16B—BIOINFORMATICS, i.e. INFORMATION AND COMMUNICATION TECHNOLOGY [ICT] SPECIALLY ADAPTED FOR GENETIC OR PROTEIN-RELATED DATA PROCESSING IN COMPUTATIONAL MOLECULAR BIOLOGY
- G16B20/00—ICT specially adapted for functional genomics or proteomics, e.g. genotype-phenotype associations
- G16B20/30—Detection of binding sites or motifs
Landscapes
- Bioinformatics & Cheminformatics (AREA)
- Health & Medical Sciences (AREA)
- Life Sciences & Earth Sciences (AREA)
- Physics & Mathematics (AREA)
- Engineering & Computer Science (AREA)
- Genetics & Genomics (AREA)
- Biotechnology (AREA)
- Biophysics (AREA)
- Chemical & Material Sciences (AREA)
- Molecular Biology (AREA)
- Proteomics, Peptides & Aminoacids (AREA)
- Bioinformatics & Computational Biology (AREA)
- Analytical Chemistry (AREA)
- Evolutionary Biology (AREA)
- General Health & Medical Sciences (AREA)
- Medical Informatics (AREA)
- Spectroscopy & Molecular Physics (AREA)
- Theoretical Computer Science (AREA)
- Measuring Or Testing Involving Enzymes Or Micro-Organisms (AREA)
Abstract
Description
Translated fromChinese技术领域technical field
本发明主要涉及到基因稳定性的技术领域,更确切的说,涉及到一种基于伊辛机量子退火的基因稳定性筛选方法及相关的筛选系统。The invention mainly relates to the technical field of gene stability, more specifically, to a gene stability screening method based on Ising machine quantum annealing and a related screening system.
背景技术Background technique
从传统的桥梁设计到现代计算机系统,重复是优雅设计的标志。复用结构简化了相关物理并使复用结构更容易实现所需的功能。这一策略通常用于合成生物学,设计师选择遗传部件并将它们合理组装成遗传工程系统,例如基于细胞的疗法、微型生物化学工厂以及生物活性材料或自主生物传感器。当设计者在多个位置重复使用遗传部件,或者是选择了具有相似DNA序列的遗传部件时,可能会无意将重复的DNA引入到遗传系统中从而产生生物学或相关领域的灾难性影响。业界总是在试图摆脱这种桎梏。From traditional bridge designs to modern computer systems, repetition is the hallmark of elegant design. The multiplexed structure simplifies the associated physics and makes it easier for the multiplexed structure to implement the desired functionality. This strategy is commonly used in synthetic biology, where designers select genetic components and rationally assemble them into genetically engineered systems, such as cell-based therapies, miniature biochemical factories, and bioactive materials or autonomous biosensors. When designers reuse genetic components in multiple locations, or select genetic components with similar DNA sequences, they may inadvertently introduce duplicate DNA into the genetic system with catastrophic effects in biology or related fields. The industry is always trying to get out of this shackles.
重复DNA大概率会扰乱合成生物学的“设计、构建、测试、学习”周期。更严重的后果是在DNA片段的合成和组装过程当中产生不必要的副产物,进而引发所谓的同源重组以及顺带产生遗传的不稳定性。另外其还进一步的阻止了下一代测序(NGS)读数的正确对齐以及其他难以预料的事故,从而降低了修复错误的能力。The high probability of repeating DNA disrupts the "design, build, test, learn" cycle of synthetic biology. A more serious consequence is the generation of unwanted by-products during the synthesis and assembly of DNA fragments, leading to so-called homologous recombination and incidental genetic instability. It also further prevents the correct alignment of next-generation sequencing (NGS) reads and other unforeseen mishaps, thereby reducing the ability to repair errors.
以改造细胞来产生十种以上高价值的化学物质的实验为例,在经历数十天的压力测试中得到了某些关于基因的启发式提示:重复基因部分大大增加了接收合成DNA片段所需的时间以及导致了苯甲酰胺合成途径中四种酶的缺失。类似的实验例如群聚且有规律间隔的短回文重复序列(CRISPR)敲除以识别调节网络相互作用、集胞藻中的代谢途径工程以产生脂肪醇、在大肠杆菌中进行大规模基因电路工程工程等,这些实验几乎都表明了重复基因部分带来的各类负面弊端是难以克服的顽疾。Taking the experiment of engineering cells to produce more than ten high-value chemicals as an example, several heuristic hints about genes were obtained during the stress test over tens of days: repetitive gene parts greatly increase the amount of DNA required to receive synthetic DNA fragments. time and resulted in the deletion of four enzymes in the benzamide synthesis pathway. Similar experiments such as clustered and regularly interspaced short palindromic repeats (CRISPR) knockout to identify regulatory network interactions, engineering of metabolic pathways in Synechocystis to produce fatty alcohols, large-scale gene circuits in E. coli Engineering engineering, etc., these experiments almost all show that all kinds of negative drawbacks brought about by repetitive gene parts are stubborn diseases that are difficult to overcome.
最长重复序列是遗传系统稳定性的关键因素,称为最大共享重复长度(Lmax)。The longest repeat sequence is a key factor in the stability of a genetic system, known as the maximum shared repeat length (Lmax ).
大肠杆菌中通常一组21个碱基对(bp)的重复序列足以触发同源重组,例如可以切除两部分之间的DNA来破坏系统功能。其他生物体例如酿酒酵母、枯草芽孢杆菌和哺乳动物病毒载体,一个12-18bp重复序列长度足以进行链入侵和同源重组。DNA组装对重复序列也很敏感:20bp的重复就让初步成功率降到40%,而11bp重复片段的DNA很容易合成是其特点。总而言之,如果一个工具箱的基因部分Lmax低于10bp,那么所有的工具都可用于构建遗传系统而不引入超过10bp的DNA重复。正是鉴于基因在遗传等方面的复杂程度和后果的不可控性,所以基因稳定性筛选显得十分必要。Often a set of 21 base pair (bp) repeats in E. coli is sufficient to trigger homologous recombination, for example the DNA between the two parts can be excised to disrupt system function. In other organisms such as Saccharomyces cerevisiae, Bacillus subtilis, and mammalian viral vectors, a 12-18 bp repeat is long enough for strand invasion and homologous recombination. DNA assembly is also sensitive to repeat sequences: 20bp repeats reduce the initial success rate to 40%, while 11bp repeats are characterized by the ease of DNA synthesis. In summary, if theLmax of a gene part of a toolbox is below 10 bp, then all tools can be used to construct a genetic system without introducing DNA repeats exceeding 10 bp. It is precisely in view of the genetic complexity of genes and the uncontrollable consequences of genes, so the gene stability screening is very necessary.
发明内容SUMMARY OF THE INVENTION
本申请涉及一种基于伊辛机量子退火的基因稳定性筛选方法,包括:The present application relates to a gene stability screening method based on Ising machine quantum annealing, including:
创建重复图,将每个遗传部分用一个节点表示,当任意一对节点表征的两组遗传部分之间存在超过最大共享重复长度的共享重复序列时,定义该一对节点具有连接边;Create a repeating graph, each genetic part is represented by a node, when there is a shared repeat sequence exceeding the maximum shared repeat length between the two sets of genetic parts represented by any pair of nodes, it is defined that the pair of nodes has a connecting edge;
利用基于伊辛机的量子退火法寻找所述重复图的最小节点覆盖状态,保障每条所述连接边至少与一个指定类别的节点相连的前提下,最大限度的减少指定类别的节点。The quantum annealing method based on the Ising machine is used to find the minimum node coverage state of the repeated graph, and on the premise that each connecting edge is connected to at least one node of the specified category, the number of nodes of the specified category is minimized.
上述的方法,其中:The above method, wherein:
从所述重复图中删除指定类别的节点,保留余下节点,籍此得到与余下节点相对应的体现为稳定集合的非重复的遗传部分。Nodes of the specified class are deleted from the repetition graph, and the remaining nodes are retained, thereby obtaining a non-repetitive genetic part corresponding to the remaining nodes, which is embodied as a stable set.
上述的方法,其中:The above method, wherein:
所述的非重复的遗传部分包括启动子、核糖体结合位点、终止子。The non-repetitive genetic part includes a promoter, a ribosome binding site, and a terminator.
上述的方法,其中:The above method, wherein:
所述的非重复的遗传部分在DNA片段合成和组装过程中用于降低同源重组。The non-repetitive genetic portion serves to reduce homologous recombination during DNA fragment synthesis and assembly.
上述的方法,其中:The above method, wherein:
构建伊辛哈密顿的一个分量HA,将节点映射到分量HA的自旋,指定类别的节点的自旋取1以及非指定类别的节点的自旋取0,以限定每条所述连接边至少与一个指定类别的节点相连:Construct a componentHA of the Ising Hamiltonian, mapping the nodes to the spin of componentHA , taking the spin of 1 for nodes of the specified class and 0 for the nodes of the non-specified class, to define each said connection An edge is connected to at least one node of the specified class:
第u个自旋为xu及第v个自旋为xv,A为分量HA的预设强度系数。The u-th spin is xu and the v-th spin is xv , andA is the preset intensity coefficient of the component HA.
上述的方法,其中:The above method, wherein:
构建伊辛哈密顿的另一个分量HB,以最大限度的减少指定类别的节点:Construct another component of the Ising Hamiltonian, HB , to minimize nodes of the specified class:
其中B为分量HB的预设强度系数。whereB is the preset intensity coefficient of the component HB.
上述的方法,其中:The above method, wherein:
所述最小节点覆盖状态对应的伊辛哈密顿量HIsing为:The Ising Hamiltonian HIsing corresponding to the minimum node coverage state is:
伊辛哈密顿量HIsing等于HA+HB。The Ising Hamiltonian HIsing is equal to HA + HB .
本申请涉及一种基于伊辛机量子退火的基因稳定性筛选系统,包括:The present application relates to a gene stability screening system based on Ising machine quantum annealing, including:
分布有多个节点的重复图模块,将每个遗传部分用一个节点表示,当任意一对节点表征的两组遗传部分之间存在超过最大共享重复长度的共享重复序列时,定义该一对节点具有连接边;A repeating graph module with multiple nodes distributed, each genetic part is represented by a node, when there is a shared repeat sequence exceeding the maximum shared repeat length between the two sets of genetic parts represented by any pair of nodes, the pair of nodes is defined. has connecting edges;
伊辛机,其利用量子退火法寻找所述重复图模块的最小节点覆盖状态,保障每条所述连接边至少与一个指定类别的节点相连的前提下,最大限度的减少指定类别的节点。Ising machine, which uses quantum annealing method to find the minimum node coverage state of the repeating graph module, and minimizes the number of nodes of the specified category on the premise that each of the connecting edges is connected to at least one node of the specified category.
上述的系统,其中:The above system, wherein:
构建伊辛哈密顿的一个分量HA,将节点映射到分量HA的自旋,指定类别的节点的自旋取1以及非指定类别的节点的自旋取0,以限定每条所述连接边至少与一个指定类别的节点相连:Construct a componentHA of the Ising Hamiltonian, mapping the nodes to the spin of componentHA , taking the spin of 1 for nodes of the specified class and 0 for the nodes of the non-specified class, to define each said connection An edge is connected to at least one node of the specified class:
第u个自旋为xu及第v个自旋为xv,A为分量HA的预设强度系数。The u-th spin is xu and the v-th spin is xv , andA is the preset intensity coefficient of the component HA.
上述的系统,其中:The above system, wherein:
构建伊辛哈密顿的另一个分量HB,以最大限度的减少指定类别的节点:Construct another component of the Ising Hamiltonian, HB , to minimize nodes of the specified class:
其中B为分量HB的预设强度系数。whereB is the preset intensity coefficient of the component HB.
上述的系统,其中:The above system, wherein:
所述最小节点覆盖状态对应的伊辛哈密顿量HIsing满足HIsing=HA+HB;The Ising Hamiltonian HIsing corresponding to the minimum node coverage state satisfies HIsing =HA +HB ;
所述伊辛机对伊辛哈密顿量HIsing进行量子退火演化后,得到的自旋的基态解定义成所述最小节点覆盖状态下的节点分布。After the Ising machine performs quantum annealing evolution on the Ising Hamiltonian HIsing , the obtained ground state solution of the spin is defined as the node distribution in the minimum node coverage state.
上述的系统,其中:The above system, wherein:
从所述重复图模块中删除指定类别的节点后,仅保留余下节点,籍此得到与余下节点相对应的体现为稳定集合的非重复的遗传部分。After the nodes of the specified category are deleted from the repeating graph module, only the remaining nodes are retained, thereby obtaining a non-repetitive genetic part corresponding to the remaining nodes, which is embodied as a stable set.
当基因工程系统的遗传部分包含重复序列时,它们很容易发生故障。设计具有所需功能的许多非重复性遗传部分仍然是具有高计算复杂性的困难挑战。为克服这一挑战本申请开发了非重复部分的类计算器例如基于伊辛机量子退火的基因稳定性筛选系统,从现有工具箱中识别最大的非重复遗传部件子集,快速生成数千个高度非重复遗传部分,包括启动子和核糖体结合位点以及终止子等。非重复性的遗传部分会大大减少同源重组,从而保证更大的遗传稳定性,实现了期望的基因稳定性筛选目的。When the genetic parts of genetically engineered systems contain repetitive sequences, they are prone to malfunction. Designing many non-repetitive genetic parts with desired functions remains a difficult challenge with high computational complexity. To overcome this challenge, the present application develops non-repetitive part-like calculators, such as a gene stability screening system based on Ising machine quantum annealing, to identify the largest subset of non-repetitive genetic parts from an existing toolbox and rapidly generate thousands of A highly non-repetitive genetic part, including promoters and ribosome binding sites and terminators, etc. The non-repetitive genetic portion will greatly reduce homologous recombination, thereby ensuring greater genetic stability and achieving the desired purpose of genetic stability screening.
附图说明Description of drawings
为使上述目的和特征及优点能够更加明显易懂,下面结合附图对具体实施方式做详细的阐释,阅读以下详细说明并参照以下附图之后,本申请的特征和优势将显而易见。In order to make the above objects, features and advantages more obvious and easy to understand, the specific embodiments are explained in detail below with reference to the accompanying drawings. After reading the following detailed description and referring to the following drawings, the features and advantages of the present application will be obvious.
图1是目标即从完整的脱氧核糖核酸库中寻找到稳定的脱氧核糖核酸库。Figure 1 is the goal of finding a stable DNA library from a complete DNA library.
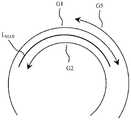
图2是两组遗传部分之间存在着超过最大共享重复长度的共享重复序列。Figure 2 shows the presence of shared repeats exceeding the maximum shared repeat length between the two sets of genetic parts.
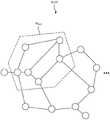
图3是创建重复图而且将每个遗传部分用重复图当中的一个节点来表示。Figure 3 creates a repetition graph and represents each genetic part as a node in the repetition graph.
图4是通过分析共享重复序列从而定义出各对节点之间是否具有连接边。Figure 4 defines whether there is a connecting edge between each pair of nodes by analyzing the shared repeat sequence.
图5是从重复图中删除指定类别的节点得到稳定集合的非重复遗传部分。Figure 5 is the non-repetitive genetic part of the stable set obtained by removing nodes of the specified class from the repetition graph.
图6是根据伊辛机的量子退火的输出从而寻找到稳定的脱氧核糖核酸库。Figure 6 is the output of quantum annealing according to the Ising machine to find a stable DNA library.
具体实施方式Detailed ways
下面将结合各实施例,对本发明的方案进行清楚完整的阐述,所描述的实施例仅是本发明用作叙述说明所用的实施例而非全部的实施例,基于该等实施例,本领域的技术人员在没有做出创造性劳动的前提下所获得的方案属于本发明的保护范围。The solution of the present invention will be clearly and completely explained below with reference to the various embodiments. The described embodiments are only the embodiments used for the description of the present invention rather than all the embodiments. The solutions obtained by the technical personnel without creative work belong to the protection scope of the present invention.
组合优化问题,在应用数学和理论计算机科学领域,指的是在一个有限的对象里集中找出最优对象的一类课题。这类问题特征是可行解的集是离散或者可以简化到离散结果并且其目标是要找到最优解。当前,常见的组合优化问题包括旅行商问题和最小生成树而行业场景领域上涉及极大规模的集成电路设计、药物设计、财务组合管理等问题。Combinatorial optimization problems, in the field of applied mathematics and theoretical computer science, refer to a class of problems that focus on finding the optimal object in a finite object. Such problems are characterized by the fact that the set of feasible solutions is discrete or can be reduced to discrete results and the goal is to find the optimal solution. At present, common combinatorial optimization problems include the traveling salesman problem and the minimum spanning tree, while the industry scenarios involve extremely large-scale integrated circuit design, drug design, financial portfolio management and other problems.
本申请将组合优化问题与基因稳定性筛选相互结合,属于全新的应用领域。The present application combines the combinatorial optimization problem with gene stability screening, which belongs to a brand new application field.
显然,无论是金融、制药或是财务领域,组合优化问题都是最贴近科技赋能日常生活与工作的实用问题之一,一旦有效解决这类问题,会迅速提高我们的日常效率,而同时经过了实验和市场论证的最优的解算工具就是“退火算法”。Obviously, whether it is in the fields of finance, pharmacy or finance, the combinatorial optimization problem is one of the practical problems that are closest to the empowerment of technology in daily life and work. Once this kind of problem is effectively solved, it will rapidly improve our daily efficiency. The optimal solution tool that has been tested and demonstrated by the market is the "annealing algorithm".
籍此,本申请面临的问题是基因稳定性筛选的组合优化,同样“退火算法”对于基因稳定性筛选解算而言依然是较好的选择。Therefore, the problem faced by this application is the combinatorial optimization of gene stability screening. Similarly, the "annealing algorithm" is still a good choice for the calculation of gene stability screening.
参见图1,广义DNA库包括了完整的DNA库,完整的DNA库作为脱氧核糖核酸库其不仅内禀的包含了所需的必要遗传部件,同时其亦包含不稳定的DNA内容。已知的遗传问题的根源之一是在多个位置重复的使用遗传部件,或选择了具有相似DNA序列的遗传部件作为工具。所以十分有必要删除不稳定DNA,保留稳定DNA库。至于如何删除不稳定的脱氧核糖核酸库的疑虑,下文会一一阐释。Referring to Figure 1, the generalized DNA library includes a complete DNA library. The complete DNA library, as a deoxyribonucleic acid library, not only inherently contains the necessary genetic components, but also contains unstable DNA content. One of the known sources of genetic problems is the repeated use of genetic components at multiple locations, or the selection of genetic components with similar DNA sequences as tools. Therefore, it is very necessary to delete the unstable DNA and keep the stable DNA pool. As for the doubts about how to delete the unstable DNA library, the following will explain them one by one.
参见图2,为了避免将重复的DNA引入到遗传系统当中从而产生生物学或遗传领域的灾难性影响,本申请引入了最长重复序列的概念。引入最长重复序列是本申请中构建重复图这个模块的前提条件,也是将基因稳定性筛选结合到量子退火的关键。而构建重复图的目的则是敲除掉那些不必要的遗传部分。Referring to FIG. 2 , in order to avoid introducing repetitive DNA into the genetic system to have catastrophic effects in the field of biology or genetics, the present application introduces the concept of the longest repeat sequence. The introduction of the longest repeat sequence is a prerequisite for the construction of the repeat map module in this application, and it is also the key to combining gene stability screening with quantum annealing. The purpose of building a repeat map is to knock out those unnecessary genetic parts.
参见图2,通常任意两组遗传部分之间存在超过最大共享重复长度(Lmax)的相关共享重复序列时。如果这个条件成立,那么认为它们之间有较强的耦合强度。重复序列是指在基因组中重复出现的DNA序列,根据其在基因组中重复频率特点又可分为串联重复序列以及分散重复序列。现举例说明,例如假设图示的两组遗传部分G1、G2之间存在超过最大共享重复长度(Lmax)的相关共享重复序列。相反的情况是,例如再假设图示的两组遗传部分G1、G5之间不存在超过最大共享重复长度(Lmax)的共享重复序列以及另两组遗传部分G2、G5之间不存在超过最大共享重复长度(Lmax)的共享重复序列。序列检查过程通常是针对碱基例如片段ATCAG等对象进行的检查。Referring to Figure 2, typically there are related shared repeats that exceed the maximum shared repeat length (Lmax ) between any two sets of genetic parts. If this condition holds, then it is considered that there is a strong coupling strength between them. Repeated sequences refer to DNA sequences that appear repeatedly in the genome, and can be divided into tandem repeats and scattered repeats according to their repeat frequency in the genome. By way of example, suppose, for example, that there are related shared repeats exceeding the maximum shared repeat length (Lmax ) between the two sets of genetic parts G1, G2 shown. On the contrary, for example, it is assumed that there is no shared repeat sequence exceeding the maximum shared repeat length (L max ) between the two groups of genetic parts G1 and G5 shown in the figure, and there is no shared repeat sequence exceeding the maximum shared repeat length (Lmax ) between the other two groups of genetic parts G2 and G5. Shared repeat sequences of shared repeat length (Lmax ). The sequence checking process is usually performed on objects such as bases such as fragment ATCAG.
参见图2,现认为C(G1,G2)=1以及C(G1,G5)=C(G2,G5)=0。例如前述的某两组遗传部分G1、G2因为满足关于Lmax的共享重复序列,则它们连接(C=1)。再如两组遗传部分G2、G5间不满足关于Lmax的共享重复序列,它们不连接(C=0)。再如两组遗传部分G1、G5间不满足关于Lmax的共享重复序列,它们不连接(C=0)。如果说最长重复序列是遗传系统稳定性的关键因素,那么这种因素体现在C值上,其也是下文描述的星状链接式重复图的设计基础。Referring to Figure 2, it is now considered that C(G1, G2)=1 and C(G1, G5)=C(G2, G5)=0. For example, the aforementioned two groups of genetic parts G1 and G2 are connected (C=1) because they satisfy the shared repeat sequence with respect toLmax . Another example is that the shared repeat sequence aboutLmax is not satisfied between the two sets of genetic parts G2 and G5, and they are not connected (C=0). Another example is that the shared repeat sequence aboutLmax is not satisfied between the two groups of genetic parts G1 and G5, and they are not connected (C=0). If the longest repeat sequence is a key factor in the stability of a genetic system, this factor is reflected in the C value, which is the basis for the design of the star-linked repeat graph described below.
参见图3,创建重复图MAP,将每个遗传部分用一个节点表示,当任意一对节点表征的两组遗传部分之间存在超过最大共享重复长度(Lmax)的共享重复序列时,定义此对节点具有连接边。连接边的定义与前述C值密切相关。Referring to Figure 3, create a repeat map MAP, representing each genetic part with a node, when there is a shared repeat sequence exceeding the maximum shared repeat length (Lmax ) between the two sets of genetic parts represented by any pair of nodes, define this Has connecting edges to nodes. The definition of the connecting edge is closely related to the aforementioned C value.
参见图3,可以借助图2的相关描述来解释:图2的遗传部分G1切换到用当前重复图中的带有圆圈的顶点(含G1的顶点)来表示、图2的遗传部分G2切换到用当前重复图中的带有圆圈的顶点(含G2的顶点)来表示、图2的遗传部分G5切换到用当前重复图中的带有圆圈的顶点(含G5的顶点)来表示。图3的遗传部分G3-G4等虽然并未在展示重复序列的图2中赘述,但它们和G1-G2及G5基本类似。Referring to Fig. 3, it can be explained with the help of the relevant description of Fig. 2: the genetic part G1 of Fig. 2 is switched to be represented by the vertices with circles (vertices containing G1) in the current repetition graph, the genetic part G2 of Fig. 2 is switched to It is represented by the circled vertices (including G2 vertices) in the current repetition graph, and the genetic part G5 of Figure 2 is switched to be represented by the circled vertices (including G5 vertices) in the current repetition graph. Although the genetic parts G3-G4 and the like in FIG. 3 are not described in detail in FIG. 2 showing the repeated sequences, they are basically similar to G1-G2 and G5.
参见图3,暂以有限的节点G1-G5(实际应用中节点数量远超示范的五个)来表示局部重复图MAP。节点G1-G2表征的两组遗传部分之间存在超过最大共享重复长度的共享重复序列时则它们具有连接边C12。节点G1-G5表征的两组遗传部分之间不存在超过最大共享重复长度的共享重复序列时则它们无连接边。节点G2-G5表征的两组遗传部分之间不存在超过最大共享重复长度的共享重复序列时则它们无连接边。Referring to FIG. 3 , a limited number of nodes G1-G5 (in practical applications, the number of nodes far exceeds the five in the demonstration) is temporarily used to represent the local repetition graph MAP. When there is a shared repeat sequence exceeding the maximum shared repeat length between the two sets of genetic parts represented by nodes G1-G2, they have a connecting edge C12. Nodes G1-G5 have no connected edges when there is no shared repeat sequence exceeding the maximum shared repeat length between the two sets of genetic parts represented by nodes G1-G5. Nodes G2-G5 have no connecting edges when there is no shared repeat sequence exceeding the maximum shared repeat length between the two sets of genetic parts represented by nodes G2-G5.
参见图3,特殊情况是,具有内部重复序列的遗传部分可以由单个节点表示,因为某个遗传部分不仅仅与外部的其他遗传部分存在重复序列,在其内部同样也存在着某一些重复的序列而且同样影响着遗传系统的稳定性。Referring to Figure 3, a special case is that a genetic part with an internal repeat sequence can be represented by a single node, because a genetic part not only has repeat sequences with other genetic parts outside, but also has some repeat sequences inside it It also affects the stability of the genetic system.
参见图3,类似的,节点G1-G4具有连接边C14、节点G3-G4具有连接边C34以及重复图中节点G2-G3具有连接边C23、节点G3-G5具有连接边C35。基于简洁起见,图中更多的顶点和连接边并没有一一展示,顶点的多寡取决于DNA库的大小而连接边取决于遗传部分之间是否存在超过Lmax的共享重复序列。至此,重复图MAP得以创建。Referring to Figure 3, similarly, nodes G1-G4 have a connecting edge C14, nodes G3-G4 have a connecting edge C34, and nodes G2-G3 have a connecting edge C23, and nodes G3-G5 have a connecting edge C35 in the repeating graph. For the sake of brevity, more vertices and connecting edges are not shown in the figure. The number of vertices depends on the size of the DNA pool and the connecting edges depend on whether there are shared repeats between the genetic parts that exceedLmax . At this point, the repetition map MAP is created.
参见图3,创建重复图MAP的目的之一是寻找重复图的最小节点覆盖状态。最小节点覆盖状态表达了贡献至少一个共享重复的遗传部分的最小数目。倘若依据最小节点覆盖状态来删除不稳定的DNA,那么可以得到稳定的DNA库。Referring to Figure 3, one of the purposes of creating a repeated graph MAP is to find the minimum node coverage state of the repeated graph. The minimum node coverage state expresses the minimum number of genetic parts that contribute to at least one shared repeat. If the unstable DNA is deleted according to the minimum node coverage state, then a stable DNA pool can be obtained.
参见图4,全局重复图MAP分布有完整的多节点和多连接边。假设图中的虚线框定的众多节点或顶点(位于MMIN内部)视为某一类不受欢迎的节点,不受欢迎的理由就在于其可能是同源重组以及顺带产生遗传不稳定性的根源,例如20bp的重复就让DNA组装的初步成功率降到40%。虚线框MMIN内部覆盖的节点如果被删除,在DNA片段的合成和组装过程当中产生的不必要的副产物近乎会大幅度降低,阻止下一代测序(NGS)读数的正确对齐这种现象要少得多,修复错误的能力也大为增强。Referring to Figure 4, the global repeating graph MAP is distributed with complete multi-nodes and multi-connected edges. Assuming that many nodes or vertices (located inside MMIN ) framed by dashed lines in the graph are regarded as a certain type of unwelcome nodes, the reason for unpopularity is that it may be the source of homologous recombination and incidental genetic instability , for example, a 20bp repeat reduces the initial success rate of DNA assembly to 40%. If the nodes covered inside the dotted box MMIN were deleted, unwanted by-products generated during the synthesis and assembly of DNA fragments would be substantially reduced, preventing the correct alignment of next-generation sequencing (NGS) reads. Much more, and the ability to fix bugs is greatly enhanced.
参见图4,然而删除虚线框MMIN内部覆盖的节点,只是试图删除不稳定DNA和得到稳定的DNA库的一种意愿,这种意愿却不能阻止稳定DNA被删除、不稳定DNA被保留的这种现象的发生。所以虚线框MMIN随机删除节点的方式并不可取,尤其是造成不稳定因素的节点是随机分布的,很难聚拢成如图所示的一簇,因此合理的节点删除机制在此情况下显得尤为重要。利用基于伊辛机的量子退火法寻找重复图MAP的最小节点覆盖状态和最大限度的减少指定类别的如不受欢迎的节点是较佳的选择,至此,伊辛机量子退火和基因稳定性筛选通过重复图MAP建立了内在联系。Referring to Figure 4, however, deleting the nodes covered inside the dotted box MMIN is just a willingness to delete unstable DNA and obtain a stable DNA pool, but this willingness cannot prevent stable DNA from being deleted and unstable DNA being retained. occurrence of a phenomenon. Therefore, it is not advisable to delete nodes randomly in the dotted box MMIN , especially the nodes that cause instability are randomly distributed, and it is difficult to gather into a cluster as shown in the figure. Therefore, a reasonable node deletion mechanism appears in this case. especially important. Using the Ising machine-based quantum annealing method to find the minimum node coverage state of the repeated graph MAP and to minimize the unpopular nodes of the specified category is a better choice. So far, the Ising machine quantum annealing and gene stability screening Intrinsic connections are established by repeating the graph MAP.
参见图4,试图用基于伊辛机(IsingMachine)的量子退火法寻找重复图MAP的最小节点覆盖状态,量子退火与重复图相契合之处应当满足以下两个条件:首先保障每条连接边至少与一个指定类别的节点相连,再者是最大限度的减少指定类别的节点。在可选实施例中要求每条连接边至少与一个指定类别的节点相连,用于限定具有共享重复序列的一组节点恰好与具有共享重复序列的另一组节点强制性耦合对接,同时这两组节点因为具有共同的公共共享节点而产生了邻接效应,直至所有节点全部被吸纳在重复图中。在可选实施例中要求最大限度的减少指定类别的节点,其中指定类别的节点的减少并非是无限制的减少而是要达到重复图的最小节点覆盖状态,也就是说,在最小节点覆盖状态之下需得到贡献了至少一个共享重复序列的一系列遗传部分的最小数目。图4的虚线框MMIN随机或依经验删除具有重复序列的节点的方式将切换到图5的伊辛哈密顿量构建上。Referring to Figure 4, an attempt is made to find the minimum node coverage state of a repeating graph MAP by using the quantum annealing method based on IsingMachine. The quantum annealing and repeating graph should meet the following two conditions: First, ensure that each connecting edge has at least Connect to a node of a specified category, and minimize the number of nodes of the specified category. In an optional embodiment, each connecting edge is required to be connected to at least one node of a specified class, which is used to define that a group of nodes with a shared repeat sequence is just forcibly coupled and docked with another group of nodes with a shared repeat sequence, and at the same time these two Group nodes have an adjacency effect because they have a common common shared node until all nodes are absorbed into the repeating graph. In an alternative embodiment, it is required to minimize the number of nodes of the specified class, wherein the reduction of the nodes of the specified class is not an unlimited reduction but to reach the minimum node coverage state of the repeated graph, that is, in the minimum node coverage state Below is the minimum number of a series of genetic parts that contribute at least one shared repeat sequence. The dashed box MMIN of Fig. 4 switches to the Ising Hamiltonian construction of Fig. 5 by randomly or empirically deleting nodes with repeated sequences.
参见图5,构建伊辛哈密顿的一个分量HA,将节点映射到分量HA的自旋,指定类别的节点的自旋x取1以及非指定类别的节点的自旋x取0,以限定每条连接边至少与一个指定类别的节点相连:Referring to Figure 5, construct a componentHA of the Ising Hamiltonian, map the nodes to the spin of the componentHA , the spin x of the node of the specified class is 1 and the spin x of the node of the non-specified class is 0, to Restrict each connecting edge to at least one node of the specified class:
第u个自旋为xu及第v个自旋为xv,A为分量HA的预设强度系数。在重复图中可以理解成与第u个节点对应的自旋为xu及与第v个节点相对的自旋为xv。伊辛哈密顿量与重复图相结合时可理解成重复图中第u个节点对应于伊辛哈密顿量中第u个自旋xu以及同样重复图中第v个节点对应于伊辛哈密顿量中第v个自旋xv。节点数目E使得(u,v)∈E。The u-th spin is xu and the v-th spin is xv , andA is the preset intensity coefficient of the component HA. In the repetition graph, it can be understood that the spin corresponding to the u-th node is xu and the spin corresponding to the v-th node is xv . When the Ising Hamiltonian is combined with the repeating graph, it can be understood that the uth node in the repeating graph corresponds to the uth spin xu in the Ising Hamiltonian and the vth node in the same repeating graph corresponds to Isinghamian The vth spin xv in the quantum. The number of nodes E is such that (u,v)∈E.
参见图5,分量HA的意义在于:用基于伊辛机的量子退火寻找重复图MAP的最小节点覆盖状态时,足以保障每条连接边至少与一个指定类别的节点(例如自旋x取1的带有阴影颜色的节点)相连,自旋x取0的节点表征了非指定类别的节点。因此分量HA是保障每条连接边至少与一个指定类别的节点相连的可选实施例。指定类别的节点例如定义它的自旋取值为1,相对的自旋取值为0的节点定义为非指定类别的节点。伊辛哈密顿量输入伊辛机进行量子退火演化即可得到最小节点覆盖状态,在重复图中对应的是每个节点的自旋值的基态解,本实施例中带有分量HA的伊辛哈密顿量还可以进一步优化。Referring to Figure 5, the significance of the componentHA is: when using the Ising machine-based quantum annealing to find the minimum node coverage state of the repeated graph MAP, it is sufficient to ensure that each connecting edge is at least connected to a node of a specified class (for example, the spin x takes 1 The nodes with shaded colors) are connected, and the nodes whose spin x takes 0 represent the nodes of non-specified categories. The componentHA is therefore an optional embodiment that guarantees that each connecting edge is connected to at least one node of a given class. For example, a node of a specified category defines its spin value as 1, and a node with a relative spin value of 0 is defined as a node of a non-specified category. The Ising Hamiltonian is input into the Ising machine to perform quantum annealing evolution to obtain the minimum node coverage state. In the repetition graph, the corresponding ground state solution of the spin value of each node is the ground state solution with the componentHA in this embodiment. The Sing Hamiltonian can be further optimized.
参见图5,构建伊辛哈密顿的另一个分量HB,以最大限度的减少指定类别的节点:Referring to Figure 5, another component of the Ising Hamiltonian, HB , is constructed to minimize the number of nodes of a given class:
其中B为分量HB的预设强度系数。whereB is the preset intensity coefficient of the component HB.
参见图5,在优化后的实施例中给伊辛哈密顿的分量HA叠加上了分量HB。Referring to Fig. 5, in the optimized embodiment, the componentHB is superimposed on the componentHA of the Ising Hamiltonian.
参见图5,最小节点覆盖状态对应的伊辛哈密顿量HIsing为:Referring to Figure 5, the Ising Hamiltonian HIsing corresponding to the minimum node coverage state is:
伊辛哈密顿量HIsing为HIsing=HA+HB。The Ising Hamiltonian HIsing is HIsing =HA +HB .
由于本文与量子相关,关于量子机器和量子数据的相关内容如下文所述:Since this article is quantum related, the relevant content about quantum machines and quantum data is as follows:
本文所谓“量子机器”包含已知的量子计算设备和量子芯片等,也可以用量子硬件替代量子器件这类术语。典型的“量子机器”包括但不限制于:量子计算机、量子信息处理系统或量子密码系统、量子模拟器、处理量子数据的所有种类的装置、设备和机器。The term "quantum machine" in this paper includes known quantum computing devices and quantum chips, etc., and the term quantum device can also be replaced by quantum hardware. Typical "quantum machines" include, but are not limited to, quantum computers, quantum information processing systems or quantum cryptography systems, quantum simulators, all kinds of devices, devices, and machines that process quantum data.
关于量子退火的商业应用——最典型的就是量子退火机,例如加拿大的一家量子计算机专业公司D-Wave。D-Wave商业销售的量子计算机原理是用金属铌制成的微小电流环形成量子比特,实现量子退火现象,模仿量子计算中比特数据存储大量数值的效果。Regarding the commercial application of quantum annealing - the most typical one is the quantum annealing machine, such as D-Wave, a professional quantum computer company in Canada. The principle of the quantum computer commercially sold by D-Wave is to use tiny current loops made of metal niobium to form qubits to realize the phenomenon of quantum annealing, imitating the effect of bit data storing a large number of values in quantum computing.
值得注意的是,在商业应用落地上,量子退火法可以通过使用叠加状态搜索各种可能性来有效地解决优化问题,有效满足当前对于实际工作方案的提效与加速需求。It is worth noting that in commercial applications, the quantum annealing method can effectively solve optimization problems by using superposition states to search for various possibilities, effectively meeting the current requirements for efficiency improvement and acceleration of practical work solutions.
截至目前为止,量子退火已在物流、人工智能、材料科学、药物发现、网络安全及故障检测和财务建模等各个领域,构建了多款早期应用程序。目前常用的退火算法分模拟退火和量子退火两种,量子退火更胜一筹。退火本质上是将金属缓慢加热到一定温度并保持足够时间然后以适宜速度冷却的金属热处理工艺。以半导体芯片为例,在经过离子注入以后就需要退火处理,因为往半导体中注入杂质离子时,高能量的入射离子会与半导体晶格上的原子碰撞使晶格原子发生位移,进行退火可恢复晶体结构和消除缺陷。实际退火解决的是材料在研制过程中的硬件工艺不稳定问题,而模拟退火和量子退火则是解决组合优化等数学计算中的非优解问题。To date, quantum annealing has built several early-stage applications in fields such as logistics, artificial intelligence, materials science, drug discovery, cybersecurity and fault detection, and financial modeling. At present, the commonly used annealing algorithms are divided into two types: simulated annealing and quantum annealing, and quantum annealing is better. Annealing is essentially a metal heat treatment process in which the metal is slowly heated to a certain temperature for a sufficient time and then cooled at a suitable rate. Taking a semiconductor chip as an example, annealing is required after ion implantation, because when impurity ions are implanted into the semiconductor, the high-energy incident ions will collide with the atoms on the semiconductor lattice to displace the lattice atoms, which can be recovered by annealing. Crystal structure and elimination of defects. Practical annealing solves the problem of hardware process instability during material development, while simulated annealing and quantum annealing solve non-optimal solutions in mathematical calculations such as combinatorial optimization.
参见图5,将伊辛哈密顿量(HIsing)输入伊辛机,开始量子退火演化,演化结束后可以得到对应的解就是顶点覆盖的点的集合,最小顶点覆盖表示贡献至少一个共享重复的遗传部分的最小数目。量子退火可以通过例如超导电路、相干量子计算(CIM)实施激光脉冲等方式、以及基于模拟退火(SA)的相干量子计算,与数字电路例如现场可编程门阵列等一起实现的量子算法。Referring to Figure 5, the Ising Hamiltonian (HIsing ) is input into the Ising machine to start the quantum annealing evolution. After the evolution, the corresponding solution can be obtained as the set of vertex-covered points, and the minimum vertex-coverage represents the contribution of at least one shared repetition. Minimum number of genetic parts. Quantum annealing can be accomplished by means such as superconducting circuits, coherent quantum computing (CIM) implementing laser pulses, and coherent quantum computing based on simulated annealing (SA), quantum algorithms implemented with digital circuits such as field programmable gate arrays and the like.
参见图5,从重复图MAP中删除指定类别节点(如带有阴影颜色的节点)并保留余下的其他节点,籍此得到与余下节点(如没有带阴影颜色的节点)相对应的体现为稳定集合的非重复的遗传部分。Referring to Figure 5, delete the specified category node (such as a node with a shaded color) from the repeating graph MAP and keep the remaining other nodes, thereby obtaining a stable representation corresponding to the remaining nodes (such as a node without a shaded color). The non-repetitive genetic part of the collection.
参见图5,非重复遗传部分包括启动子、核糖体结合位点、终止子。其中启动子通常是指代RNA聚合酶识别、结合和开始转录的一段DNA序列。核糖体结合位点即核蛋白体结合位点而终止子是RNA聚合酶转录终止信号的DNA序列。通常在一个操纵元中至少在结构基因群最后一个基因的后面设置一个终止子。重复图MAP的意义在于剔除了不稳定的重复内容而保障了启动子、核糖体结合位点、终止子等部分的稳定性。Referring to Figure 5, the non-repetitive genetic portion includes a promoter, a ribosome binding site, and a terminator. The promoter generally refers to a DNA sequence that RNA polymerase recognizes, binds and initiates transcription. The ribosome binding site is the ribosome binding site and the terminator is the DNA sequence that signals the termination of transcription by RNA polymerase. A terminator is usually placed in an operon at least after the last gene in a structural gene group. The significance of the repeat map MAP is to eliminate the unstable repeat content and ensure the stability of the promoter, ribosome binding site, terminator and other parts.
参见图5,非重复遗传部分在DNA片段合成和组装过程中用于降低同源重组。Referring to Figure 5, non-repetitive genetic moieties are used to reduce homologous recombination during DNA fragment synthesis and assembly.
参见图6,基于伊辛机量子退火的基因稳定性筛选系统包括:分布有多个节点的重复图模块Mod(用于提供重复图MAP)和伊辛机。重复图模块Mod:需要将每个遗传部分用一个节点表示,当任意一对节点表征的两组遗传部分之间存在超过最大共享重复长度的共享重复序列时,定义该一对节点具有连接边。伊辛机:其利用量子退火法寻找所述重复图模块的最小节点覆盖状态(前文已阐释),保障每条连接边至少与一个指定类别的节点相连的前提下,最大限度的减少指定类别的节点。Referring to FIG. 6 , the gene stability screening system based on Ising machine quantum annealing includes: a repetitive graph module Mod (used to provide a repetitive graph MAP) distributed with multiple nodes and an Ising machine. Repetition graph module Mod: Each genetic part needs to be represented by a node. When there is a shared repeat sequence exceeding the maximum shared repeat length between the two sets of genetic parts represented by any pair of nodes, it is defined that the pair of nodes has connecting edges. Ising machine: It uses the quantum annealing method to find the minimum node coverage state of the repeating graph module (explained above), and on the premise that each connecting edge is connected to at least one node of a specified category, it minimizes the specified category of nodes. node.
参见图6,通过实验可以获得大量的基因集合,例如图示的广义NDA库。然后需要从广义NDA库中识别非重复的基因部分。将每个遗传部分由一个节点来表示,当两个遗传部分有一个大于Lmax(bp)的共享重复序列时,它们的节点通过一条边连接,创建一个前文所言的重复图MAP。具有内部重复序列的遗传部分由带环边的单个节点表示。Referring to Figure 6, large gene sets, such as the generalized NDA library illustrated, can be obtained experimentally. Non-repetitive gene parts then need to be identified from the generalized NDA library. Each genetic part is represented by a node. When two genetic parts have a shared repeat sequence greater thanLmax (bp), their nodes are connected by an edge, creating a repeating graph MAP as described above. Genetic parts with internal repeats are represented by single nodes with loop edges.
参见图6,在可选的实施例中,解决方案是使用基于伊辛机的量子退火算法找到重复图中的最小顶点覆盖,所谓的最小顶点覆盖的意思是在重复图中的每条边至少与一个阴影节点相连的情况下,找到重复图中最小数量的阴影节点。Referring to Figure 6, in an alternative embodiment, the solution is to use the Ising machine-based quantum annealing algorithm to find the minimum vertex cover in the repeated graph, the so-called minimum vertex cover means that each edge in the repeated graph has at least Find the minimum number of shaded nodes in the repeating graph when connected to one shaded node.
参见图6,在可选的实施例中,首先构建哈密顿量HA以保证每条边至少与一个阴影节点相连,自旋x取值为0和1,0表示节点无阴影,1表示节点有阴影。Referring to Fig. 6, in an optional embodiment, a HamiltonianHA is first constructed to ensure that each edge is connected to at least one shaded node, and the spin x values are 0 and 1, 0 means the node has no shadow, and 1 means the node There are shadows.
参见图6,在可选的实施例中,构建哈密顿量HB以使阴影节点的数量尽可能少。Referring to Figure 6, in an alternative embodiment, the HamiltonianHB is constructed to keep the number of shadow nodes as small as possible.
参见图6,在可选的实施例中,数学计算化简伊辛哈密顿量Hising可得到标准形式的伊辛哈密顿量例如:Referring to FIG. 6 , in an optional embodiment, the Ising Hamiltonian in standard form can be obtained by mathematically simplifying the Ising Hamiltonian Hising , for example:
将事先设计好的标准形式的参数Jij和hi输入伊辛机,开始量子退火演化。演化结束后得到对应的解(各个节点对应的自旋的取值)就是节点阴影覆盖的点的集合,最小节点覆盖表示贡献了至少一个共享重复的遗传部分的最小数目。对伊辛哈密顿量HIsing进行量子退火演化后得到的自旋的基态解定义成最小节点覆盖状态下的节点分布。The parameters Jij andhi in the standard form designed in advance are input into the Ising machine to start the quantum annealing evolution. After the evolution, the corresponding solution (the value of the spin corresponding to each node) is the set of points covered by the shadow of the node, and the minimum node coverage represents the minimum number of genetic parts that contribute to at least one shared repeat. The ground state solution of the spin obtained by quantum annealing evolution of the Ising Hamiltonian HIsing is defined as the node distribution in the minimum node coverage state.
参见图6,在可选的实施例中,从重复图中删除节点阴影覆盖的点的集合,创建一个断开的图(也即重复图MAP离散化),一旦这些遗传部分被移除,剩下的节点代表的就是非重复遗传部分的稳定集合,例如图示的稳定DNA库。Referring to Fig. 6, in an alternative embodiment, the set of points covered by node shadows is removed from the repeated graph, creating a disconnected graph (i.e. the repeated graph MAP discretization), once these genetic parts are removed, the remaining The nodes below represent stable collections of non-repetitive genetic parts, such as the stable DNA library shown.
参见图6,通过以上步骤就可以用伊辛机量子退火解决DNA稳定性的筛选。Referring to Fig. 6, the screening of DNA stability can be solved by Ising machine quantum annealing through the above steps.
参见图6,对比广义DNA库和稳定DNA库:广义DNA库应用于基因工程时它其中的重复序列很容易发生工程系统故障,单纯以人工的形式来按照经验编辑基因,绝大部分时候无法做到近乎100%的重复序列剔除完整率。图5的退火法决解的问题是:其等效于是非重复部分排除器,可快速生成数千个高度非重复的遗传部分(包括启动子、核糖体结合位点和终止子等),此时广义DNA库过滤到稳定DNA库,非重复性遗传部分大大减少同源重组从而保证了更大的遗传稳定性。See Figure 6 for a comparison between the generalized DNA library and the stable DNA library: when the generalized DNA library is applied to genetic engineering, the repetitive sequences in it are prone to engineering system failures, and it is impossible to edit genes in the form of manual editing based on experience most of the time. to nearly 100% repeat depletion completeness. The problem solved by the annealing method of Figure 5 is that it is equivalent to a non-repetitive part excluder, which can rapidly generate thousands of highly non-repetitive genetic parts (including promoters, ribosome binding sites and terminators, etc.), which When the generalized DNA library is filtered to the stable DNA library, the non-repetitive genetic part greatly reduces homologous recombination thus ensuring greater genetic stability.
参见图6,基于伊辛机量子退火的基因稳定性筛选系统包括两大部分。首先是经典的计算机这部分。计算机需要完成的任务是:构建重复图模块,从而可得到分布有多个节点的重复图模块,将每个遗传部分用一个节点表示,当任意一对节点表征的两组遗传部分之间存在超过最大共享重复长度的共享重复序列时,定义该一对节点具有连接边。Referring to Fig. 6, the gene stability screening system based on Ising machine quantum annealing includes two parts. The first is the classic computer part. The task that the computer needs to complete is: build a repeating graph module, so that a repeating graph module with multiple nodes can be obtained, and each genetic part is represented by a node. When the shared repeat sequence of the maximum shared repeat length, the pair of nodes is defined to have connecting edges.
参见图6,基于伊辛机量子退火的基因稳定性筛选系统还包括伊辛机。伊辛机利用量子退火法寻找重复图模块(基于经典计算机)的最小节点覆盖状态,在保障每条所述连接边至少与一个指定类别的节点相连的前提下,最大限度的减少指定类别的节点。兼顾了经典计算机的通用计算优势和量子退火的独有高算力。Referring to Fig. 6, the gene stability screening system based on Ising machine quantum annealing also includes Ising machine. The Ising machine uses the quantum annealing method to find the minimum node coverage state of the repeating graph module (based on classical computers), and minimizes the number of nodes of the specified category on the premise that each of the connecting edges is connected to at least one node of the specified category. . It takes into account the general computing advantages of classical computers and the unique high computing power of quantum annealing.
参见图6,基于伊辛机(IsingMachine)量子退火的基因稳定性筛选系统,从现有的基因工具箱中识别最大的非重复遗传部件子集,其兼容现有基因库的优势是能够快速的生成数千个乃至更多数量的高度非重复遗传部分,这类遗传部分又典型的包括启动子和核糖体结合位点以及终止子等遗传信息。非重复性的遗传部分会大大减少同源重组,从而在保证更大的遗传稳定性的前提下,实现了期望的基因稳定性筛选之目的。Referring to Figure 6, a gene stability screening system based on Ising Machine quantum annealing identifies the largest subset of non-repetitive genetic components from the existing gene toolbox. The advantage of its compatibility with existing gene libraries is that it can rapidly Generate thousands or more of highly non-repetitive genetic parts, which typically include genetic information such as promoters and ribosome binding sites and terminators. The non-repetitive genetic portion will greatly reduce homologous recombination, thereby achieving the desired purpose of genetic stability screening while ensuring greater genetic stability.
以上通过说明和附图的内容,给出了具体实施方式的特定结构的典型实施例,上述申请内容提出了现有的较佳实施例,但这些内容并不作为局限。对于本领域的技术人员而言在阅读上述说明后,各种变化和修正无疑将显而易见。因此,所附的权利要求书应当看作是涵盖本发明的真实意图和范围的全部变化和修正。在权利要求书范围之内的任何和所有等价的范围与内容,都应认为仍属本发明的意图和范围内。Typical examples of specific structures of specific implementations are given above through the content of the description and the accompanying drawings. The content of the above application proposes the existing preferred embodiments, but these contents are not intended to be limiting. Various changes and modifications will no doubt become apparent to those skilled in the art upon reading the above description. Therefore, the appended claims should be construed to cover all changes and modifications within the true intent and scope of this invention. Any and all equivalent ranges and content within the scope of the claims should still be considered to be within the intent and scope of the invention.
Claims (12)
Translated fromChinesePriority Applications (1)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| CN202210179637.6ACN114464250A (en) | 2022-02-25 | 2022-02-25 | Gene stability screening method and system based on Ito quantum annealing |
Applications Claiming Priority (1)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| CN202210179637.6ACN114464250A (en) | 2022-02-25 | 2022-02-25 | Gene stability screening method and system based on Ito quantum annealing |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| CN114464250Atrue CN114464250A (en) | 2022-05-10 |
Family
ID=81414695
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| CN202210179637.6APendingCN114464250A (en) | 2022-02-25 | 2022-02-25 | Gene stability screening method and system based on Ito quantum annealing |
Country Status (1)
| Country | Link |
|---|---|
| CN (1) | CN114464250A (en) |
Cited By (1)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO2023226310A1 (en)* | 2022-05-23 | 2023-11-30 | 华为云计算技术有限公司 | Molecule optimization method and apparatus |
Citations (3)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US20080052055A1 (en)* | 2006-07-28 | 2008-02-28 | Geordie Rose | Systems, methods and apparatus for protein folding simulation |
| US20180276550A1 (en)* | 2017-01-13 | 2018-09-27 | D-Wave Systems Inc. | Problem solving using quantum annealer, useful for example in sequencing, for instance nucleic acid sequencing |
| CN110890126A (en)* | 2018-09-11 | 2020-03-17 | 富士通株式会社 | Devices and Methods for Exploring Compounds |
- 2022
- 2022-02-25CNCN202210179637.6Apatent/CN114464250A/enactivePending
Patent Citations (3)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US20080052055A1 (en)* | 2006-07-28 | 2008-02-28 | Geordie Rose | Systems, methods and apparatus for protein folding simulation |
| US20180276550A1 (en)* | 2017-01-13 | 2018-09-27 | D-Wave Systems Inc. | Problem solving using quantum annealer, useful for example in sequencing, for instance nucleic acid sequencing |
| CN110890126A (en)* | 2018-09-11 | 2020-03-17 | 富士通株式会社 | Devices and Methods for Exploring Compounds |
Non-Patent Citations (2)
| Title |
|---|
| ANDREW LUCAS: "Ising formulations of many NP problems", 《FRONTIERS IN PHYSCIS》, 12 February 2014 (2014-02-12)* |
| AYAAN HOSSAIN, ET AL: "Automated design of thousands of nonrepetitive parts for engineering stable genetic systems", 《NATURE BIOTECHNOLOGY》, 13 July 2020 (2020-07-13), pages 1466 - 1475* |
Cited By (1)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO2023226310A1 (en)* | 2022-05-23 | 2023-11-30 | 华为云计算技术有限公司 | Molecule optimization method and apparatus |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| Murata et al. | Genetic algorithms for flowshop scheduling problems | |
| Rasmussen et al. | Unified modeling of gene duplication, loss, and coalescence using a locus tree | |
| Sevinç et al. | An evolutionary genetic algorithm for optimization of distributed database queries | |
| Li et al. | Maximal biclique subgraphs and closed pattern pairs of the adjacency matrix: A one-to-one correspondence and mining algorithms | |
| US6532453B1 (en) | Genetic programming problem solver with automatically defined stores loops and recursions | |
| Koza | Spontaneous emergence of self-replicating and evolutionarily self-improving computer programs | |
| Bonyadi et al. | Logic optimization for majority gate-based nanoelectronic circuits based on genetic algorithm | |
| Luo et al. | A hybrid algorithm combining genetic algorithm and variable neighborhood search for process sequencing optimization of large-size problem | |
| Tseng et al. | A genetic local search algorithm for minimizing total flowtime in the permutation flowshop scheduling problem | |
| Squires et al. | Stability of Boolean networks: The joint effects of topology and update rules | |
| US20200040329A1 (en) | Systems and methods for predicting repair outcomes in genetic engineering | |
| CN114464250A (en) | Gene stability screening method and system based on Ito quantum annealing | |
| Ghasemi et al. | A hybridizing-enhanced differential evolution for optimization | |
| Lin et al. | Mining high-utility sequential patterns from big datasets | |
| Zimmermann et al. | Set-based graph methods for fast equation sorting in large DAE systems | |
| Khanum et al. | Reflected adaptive differential evolution with two external archives for large-scale global optimization | |
| Ponnambalam et al. | Estimation of optimum genetic control parameters for job shop scheduling | |
| Hajeer et al. | Distributed genetic algorithm to big data clustering | |
| Oltean | Solving even-parity problems using traceless genetic programming | |
| Bentley et al. | Overview of a generic evolutionary design system | |
| Zaenudin et al. | A parallel algorithm to generate connected network motifs | |
| Agrawal et al. | Evolutionary programming for fast and robust point pattern matching | |
| Bhanderi et al. | Parallel frequent set mining using inverted matrix approach | |
| Rolland et al. | Heuristic solution procedures for the graph partitioning problem | |
| Shi et al. | Gene Sequence Assembly Algorithm Model Based on the DBG Strategy and Its Application |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| PB01 | Publication | ||
| PB01 | Publication | ||
| SE01 | Entry into force of request for substantive examination | ||
| SE01 | Entry into force of request for substantive examination | ||
| WD01 | Invention patent application deemed withdrawn after publication | Application publication date:20220510 | |
| WD01 | Invention patent application deemed withdrawn after publication |