CN112608931B - Deep-sea fungus FS140 anti-gliotoxin self-protection gene GliM and application thereof - Google Patents
Deep-sea fungus FS140 anti-gliotoxin self-protection gene GliM and application thereofDownload PDFInfo
- Publication number
- CN112608931B CN112608931BCN202011550170.9ACN202011550170ACN112608931BCN 112608931 BCN112608931 BCN 112608931BCN 202011550170 ACN202011550170 ACN 202011550170ACN 112608931 BCN112608931 BCN 112608931B
- Authority
- CN
- China
- Prior art keywords
- glim
- gliotoxin
- self
- gene
- deep
- Prior art date
- Legal status (The legal status is an assumption and is not a legal conclusion. Google has not performed a legal analysis and makes no representation as to the accuracy of the status listed.)
- Active
Links
Images
Classifications
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N9/00—Enzymes; Proenzymes; Compositions thereof; Processes for preparing, activating, inhibiting, separating or purifying enzymes
- C12N9/10—Transferases (2.)
- C12N9/1003—Transferases (2.) transferring one-carbon groups (2.1)
- C12N9/1007—Methyltransferases (general) (2.1.1.)
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/63—Introduction of foreign genetic material using vectors; Vectors; Use of hosts therefor; Regulation of expression
- C12N15/79—Vectors or expression systems specially adapted for eukaryotic hosts
- C12N15/80—Vectors or expression systems specially adapted for eukaryotic hosts for fungi
- C12N15/81—Vectors or expression systems specially adapted for eukaryotic hosts for fungi for yeasts
Landscapes
- Life Sciences & Earth Sciences (AREA)
- Health & Medical Sciences (AREA)
- Genetics & Genomics (AREA)
- Chemical & Material Sciences (AREA)
- Engineering & Computer Science (AREA)
- Zoology (AREA)
- Organic Chemistry (AREA)
- Bioinformatics & Cheminformatics (AREA)
- Wood Science & Technology (AREA)
- General Engineering & Computer Science (AREA)
- Biomedical Technology (AREA)
- Biotechnology (AREA)
- Microbiology (AREA)
- Biochemistry (AREA)
- Molecular Biology (AREA)
- General Health & Medical Sciences (AREA)
- Mycology (AREA)
- Medicinal Chemistry (AREA)
- Physics & Mathematics (AREA)
- Biophysics (AREA)
- Plant Pathology (AREA)
- Micro-Organisms Or Cultivation Processes Thereof (AREA)
- Measuring Or Testing Involving Enzymes Or Micro-Organisms (AREA)
Abstract
Description
Translated fromChinese技术领域technical field
本发明属于基因工程领域,具体涉及一种深海真菌Geosmithia pallida FS140抗胶霉毒素自我保护基因GliM及其应用。The invention belongs to the field of genetic engineering, in particular to a deep-sea fungus Geosmithia pallida FS140 anti-gliotoxin self-protection gene GliM and its application.
背景技术Background technique
前期本课题组从深海真菌Geosmithia pallida FS140中分离得到了20多个胶霉毒素类化合物,包括结构罕见的胶霉毒素类二聚体化合物,其中大部分胶霉毒素类化合物具有明显的抗肿瘤活性。In the early stage, our research group isolated more than 20 gliotoxin-like compounds from the deep-sea fungus Geosmithia pallida FS140, including gliotoxin-like dimer compounds with rare structures, most of which have obvious antitumor activity. .
胶霉毒素(gliotoxin,GT)是一类二酮哌嗪类化合物(epipolythiodioxopiperazine,ETP),ETP作为重要的毒力因子,其通过不同途经对各种细胞产生特异性毒性,在侵袭性曲霉病中发挥重要协同作用。ETP发挥毒副作用主要通过二硫键,巯基与靶蛋白的交联进而灭活该蛋白活性,并能通过氧化还原循环产生具有毒害的活性氧(reactiveoxygenspecies,ROS),ROS生成机制被认为是GT产生细胞毒性的一种机制。目前对GT的研究局限于作为ETP家族成员之一或烟曲霉毒素代谢物,因此胶霉毒素的很多毒性(如免疫抑制毒性)研究只集中于其作为初次级代谢产物而发挥毒性,而其本身毒性如细胞毒性、DNA损伤以及对宿主的其他毒性机制尚无定论,有待进一步研究。Gliotoxin (GT) is a class of diketopiperazine compounds (epipolythiodioxopiperazine, ETP), ETP is an important virulence factor, which produces specific toxicity to various cells through different pathways. play an important synergistic role. ETP exerts toxic side effects mainly through disulfide bonds, cross-linking of sulfhydryl groups and target proteins to inactivate the protein activity, and can generate toxic reactive oxygen species (ROS) through redox cycles. The mechanism of ROS generation is considered to be GT production. a mechanism of cytotoxicity. At present, the research on GT is limited to being a member of the ETP family or a metabolite of fumonisin, so many studies on the toxicity of gliotoxin (such as immunosuppressive toxicity) only focus on its toxicity as a primary metabolite, while its own Toxicity such as cytotoxicity, DNA damage, and other mechanisms of toxicity to the host are inconclusive and await further study.
胶霉毒素对宿主的毒性主要包括:诱导细胞凋亡;导致氧化还原反应失衡;抑制蛋白酶体活性,导致NF-κB活性被抑制,降低免疫活性等。它不仅对动植物细胞,而且对宿主菌也有毒性(Kamei K,Watanabe A.Aspergillus mycotoxins and their effect on thehost.Medical Mycology,2005,43:S95–S99.)。The toxicity of gliotoxin to the host mainly includes: inducing apoptosis; leading to the imbalance of redox reaction; inhibiting the activity of proteasome, leading to the inhibition of NF-κB activity and reducing the immune activity, etc. It is not only toxic to animal and plant cells, but also to host bacteria (Kamei K, Watanabe A. Aspergillus mycotoxins and their effect on the host. Medical Mycology, 2005, 43: S95–S99.).
发明内容SUMMARY OF THE INVENTION
本发明的第一个目的是提供一种深海真菌Geosmithia pallida FS140抗胶霉毒素自我保护基因GliM,所述的深海真菌Geosmithia pallida FS140抗胶霉毒素自我保护基因GliM,其核苷酸序列如SEQ ID NO.1所示。The first object of the present invention is to provide a deep-sea fungus Geosmithia pallida FS140 anti-gliotoxin self-protection gene GliM, the deep-sea fungus Geosmithia pallida FS140 anti-gliotoxin self-protection gene GliM, its nucleotide sequence is such as SEQ ID NO.1 is shown.
本发明的深海真菌Geosmithia pallida FS140抗胶霉毒素自我保护基因GliM通过以下方法获得的:通过转录组测序结果预测编码抗胶霉毒素自我保护基因GliM的序列,在其上下游设计特异性引物,其引物序列为GliM-F:5'-ATGGAAGCCAACAACACCGAC-3';GliM-R:5'-CTACTTCTTCAGCCGTAATTCCAA-3',以由深海真菌FS140转录组反转录而得的cDNA文库为模板,通过PCR扩增获得产物并纯化回收片段,获得抗胶霉毒素自我保护基因GliM,其核苷酸序列如SEQ ID NO.1所示。The deep-sea fungus Geosmithia pallida FS140 anti-gliotoxin self-protection gene GliM of the present invention is obtained by the following method: predicting the sequence encoding the anti-gliotoxin self-protection gene GliM from the results of transcriptome sequencing, and designing specific primers upstream and downstream thereof, which The primer sequences are GliM-F: 5'-ATGGAAGCCAACAACACCGAC-3'; GliM-R: 5'-CTACTTCTTCAGCCGTAATTCCAA-3', using the cDNA library reverse transcribed from the deep-sea fungus FS140 transcriptome as a template, and obtained by PCR amplification The product is purified and the recovered fragment is obtained to obtain the anti-gliotoxin self-protection gene GliM, the nucleotide sequence of which is shown in SEQ ID NO.1.
本发明利用同源重组法将GliM基因插入到酵母载体YEp352-TEF1-CYC1的表达盒内部。首先设计含有同源臂的GliM基因的上下游引物,其引物序列为YEp352-GliM-F:5'-ATAGCAATCTAATCTAAGTCTAGAATGGAAGCCAACAACACCGAC-3';YEp352-GliM-R:5'-TACATGATGCGGCCCGTCGACCTACTTCTTCAGCCGTAATTCCAA-3'(下划线序列为同源臂片段),通过PCR扩增获得产物并纯化回收片段。对已构建的YEp352-TEF1-CYC1载体采用内切酶Sal I和Xba I双酶切,然后使用ClonExpress II One Step Cloning Kit C112(Vazyme)将片段和酶切载体重组连接并转化至大肠杆菌感受态细胞,涂布于氨苄青霉素抗性平板筛选出阳性克隆。经过此轮分子克隆,目的基因GliM(其核苷酸序列如SEQ ID NO.1所示)插入到启动子TEF1和终止子CYC1之间,构建得到YEp352-TEF1-GliM载体,将其电转入酿酒酵母BJ5464-D细胞中,利用尿嘧啶缺陷型的SD培养基平板进行筛选和验证。与转入YEp352-TEF1-CYC1质粒(阴性对照)的酿酒酵母BJ5464-D相比,含有重组载体YEp352-TEF1-GliM的酿酒酵母生长速度明显加快,相同培养时间内菌落密度更高,证明功能基因GliM能有效协助酿酒酵母抵抗外源胶霉毒素,为在酿酒酵母内重构胶霉毒素生物合成通路奠定基础。In the present invention, the homologous recombination method is used to insert the GliM gene into the expression cassette of the yeast vector YEp352-TEF1-CYC1. First, the upstream and downstream primers of the GliM gene containing homology arms were designed, and the primer sequences were YEp352-GliM -F: 5'-ATAGCAATCTAATCTAAGTCTAGAATGGAAGCCAACAACACCGAC -3'; The underlined sequence is the homology arm fragment), the product was obtained by PCR amplification and the fragment was purified and recovered. The constructed YEp352-TEF1-CYC1 vector was double digested with endonuclease Sal I and Xba I, and then the fragment and the digested vector were recombined and transformed into E. coli competent using ClonExpress II One Step Cloning Kit C112 (Vazyme). The cells were plated on ampicillin-resistant plates to select positive clones. After this round of molecular cloning, the target gene GliM (the nucleotide sequence of which is shown in SEQ ID NO. 1) was inserted between the promoter TEF1 and the terminator CYC1 to construct the YEp352-TEF1-GliM vector, which was electroporated into Saccharomyces cerevisiae BJ5464-D cells were screened and validated using uracil-deficient SD medium plates. Compared with Saccharomyces cerevisiae BJ5464-D transferred into YEp352-TEF1-CYC1 plasmid (negative control), the growth rate of Saccharomyces cerevisiae containing the recombinant vector YEp352-TEF1-GliM was significantly faster, and the colony density was higher in the same culture time, which proved that the functional gene GliM can effectively assist Saccharomyces cerevisiae to resist exogenous gliotoxin, laying the foundation for the reconstruction of gliotoxin biosynthesis pathway in Saccharomyces cerevisiae.
本发明的第二个目的是提供一种表达载体,含有上述的深海真菌Geosmithiapallida FS140抗胶霉毒素自我保护基因GliM。The second object of the present invention is to provide an expression vector containing the above-mentioned deep-sea fungus Geosmithiapallida FS140 anti-gliotoxin self-protection gene GliM.
本发明的第三个目的是提供一种宿主细胞,含有上述的表达载体。The third object of the present invention is to provide a host cell containing the above-mentioned expression vector.
所述的宿主细胞优选为酿酒酵母Saccharomyces cerevisiae BJ5464。The host cell is preferably Saccharomyces cerevisiae BJ5464.
本发明的第四个目的是提供上述的深海真菌Geosmithia pallida FS140抗胶霉毒素自我保护基因GliM在协助宿主细胞抵抗胶霉毒素中的应用。The fourth object of the present invention is to provide the application of the above-mentioned deep-sea fungus Geosmithia pallida FS140 anti-gliotoxin self-protection gene GliM in assisting host cells to resist gliotoxin.
所述的宿主细胞优选为深海真菌Geosmithia pallida FS140抗胶霉毒素或酿酒酵母Saccha romyces cerevisiae BJ5464。The host cell is preferably a deep-sea fungus Geosmithia pallida FS140 anti-gliotoxin or Saccha romyces cerevisiae BJ5464.
本发明的第五个目的是提供一种表达盒,该表达盒含有上述的深海真菌Geosmithia pallida FS140抗胶霉毒素自我保护基因GliM。The fifth object of the present invention is to provide an expression cassette containing the above-mentioned deep-sea fungus Geosmithia pallida FS140 anti-gliotoxin self-protection gene GliM.
与现有技术相比,本发明具有以下有益效果:Compared with the prior art, the present invention has the following beneficial effects:
本发明所涉及的深海真菌Geosmithia pallida FS140分离自南海沉积物,本课题组前期对该菌株进行了转录组测序并对胶霉毒素生物合成相关基因进行了注释。鉴于目前关于深海真菌Geosmithia pallida FS140抗胶霉毒素自我保护基因的研究较少。因此本发明从深海真菌FS140的cDNA文库中获得了抗胶霉毒素自我保护基因GliM序列,并成功导入到酿酒酵母S.cerevisiae BJ5464中进行了抗毒功能验证,从而为后期提高酿酒酵母抗胶霉毒素的能力,提升胶霉毒素的异源表达水平并获得新型胶霉毒素奠定分子生物学基础。The deep-sea fungus Geosmithia pallida FS140 involved in the present invention was isolated from the sediments of the South China Sea. The research group performed transcriptome sequencing of the strain in the early stage and annotated the genes related to the biosynthesis of gliotoxin. In view of the fact that there are few studies on the self-protection genes of the deep-sea fungus Geosmithia pallida FS140 against gliotoxin. Therefore, the present invention obtained the anti-gluetoxin self-protection gene GliM sequence from the cDNA library of the deep-sea fungus FS140, and successfully imported it into S. cerevisiae BJ5464 to verify the anti-toxin function, so as to improve the anti-glue mold resistance of Saccharomyces cerevisiae in the later stage. The ability of the toxin to enhance the heterologous expression level of gliotoxin and obtain a new type of gliotoxin lays the foundation for molecular biology.
本发明的深海真菌Geosmithia pallida FS140,其公开于文献:Zhang-Hua Sun,Jiangyong Gu,Wei Ye,Liang-Xi Wen,Qi-Bin Lin,Sai-Ni Li,Yu-Chan Chen,Hao-HuaLi,Wei-Min Zhang.Geospallins A–C:New Thiodiketopiperazines with InhibitoryActivity against Angiotensin-Converting Enzyme from a Deep-Sea-Derived FungusGeosmithia pallida FS140.Marine Drugs,2018,16(12),464.https://doi.org/10.3390/md16120464。该菌种本申请人也持有,保证自发明的申请日起20年内向公众提供。The deep-sea fungus Geosmithia pallida FS140 of the present invention is disclosed in the literature: Zhang-Hua Sun, Jiangyong Gu, Wei Ye, Liang-Xi Wen, Qi-Bin Lin, Sai-Ni Li, Yu-Chan Chen, Hao-HuaLi, Wei -Min Zhang.Geospallins A–C:New Thiodiketopiperazines with InhibitoryActivity against Angiotensin-Converting Enzyme from a Deep-Sea-Derived FungusGeosmithia pallida FS140.Marine Drugs,2018,16(12),464.https://doi.org/10.3390 /md16120464. This strain is also held by the applicant and is guaranteed to be available to the public within 20 years from the filing date of the invention.
附图说明Description of drawings
图1为实验所使用胶霉毒素7-deoxy-6,7-didehydrogliotoxin(FS140-12-2)的结构式。Figure 1 shows the structural formula of the gliotoxin 7-deoxy-6,7-didehydrogliotoxin (FS140-12-2) used in the experiment.
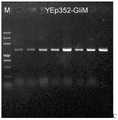
图2为深海真菌FS140 GliM基因序列的获得:以FS140 cDNA文库为模板,基因GliM扩增产物的电泳图;Figure 2 is the acquisition of the deep-sea fungus FS140 GliM gene sequence: using the FS140 cDNA library as a template, the electrophoresis diagram of the gene GliM amplification product;
图3为重组载体YEp352-TEF1-GliM的构建;其中A为YEp352-TEF1-CYC1载体图谱;B为YEp352-TEF1-GliM载体图谱;C为基因GliM的菌落PCR扩增产物的电泳图;Figure 3 is the construction of the recombinant vector YEp352-TEF1-GliM; wherein A is the map of the YEp352-TEF1-CYC1 vector; B is the map of the YEp352-TEF1-GliM vector; C is the electrophoresis map of the colony PCR amplification product of the gene GliM;
图4为三种酿酒酵母在YPD平板和YPD-FS140-12-2平板(2.5μM)培养30h的效果图。A、酿酒酵母BJ5464-D(YEp352-TEF1-CYC1);B、酿酒酵母BJ5464;C,酿酒酵母BJ5464-D(YEp352-TEF1-GliM)。10-2、10-3、10-4分别代表OD600约为0.01、0.001、0.0001的5μL菌液样品。Figure 4 is a graph showing the effect of three kinds of Saccharomyces cerevisiae cultured on YPD plate and YPD-FS140-12-2 plate (2.5 μM) for 30 h. A, Saccharomyces cerevisiae BJ5464-D (YEp352-TEF1-CYC1); B, Saccharomyces cerevisiae BJ5464; C, Saccharomyces cerevisiae BJ5464-D (YEp352-TEF1-GliM). 10-2 , 10-3 , and 10-4 represent 5 μL bacterial liquid samples with OD600 of about 0.01, 0.001, and 0.0001, respectively.
具体实施方式Detailed ways
以下实施例是对本发明的进一步说明,而不是对本发明的限制。The following examples are further illustrations of the present invention, rather than limitations of the present invention.
本实施例中所用的SD固体培养基的配方为:每升含有葡萄糖20g、Do supplement0.62g(-Leu/-Trp/-Ura,Clontech)、无氨基酵母氮源YNB 6.7g(普博欣)、亮氨酸0.06g、色氨酸0.04g和琼脂粉20g,余量为蒸馏水,其配制方法是将各成分混合均匀,灭菌制得。胶霉毒素FS140-12-2为本课题组分离自深海真菌Geosmithia pallida FS140。The formulation of SD solid medium used in this example is as follows: each liter contains 20 g of glucose, 0.62 g of Do supplement (-Leu/-Trp/-Ura, Clontech), and 6.7 g of amino-free yeast nitrogen source YNB (Pu Boxin) , leucine 0.06g, tryptophan 0.04g and agar powder 20g, the balance is distilled water, the preparation method is to mix the components evenly and sterilize to prepare. The gliotoxin FS140-12-2 was isolated from the deep-sea fungus Geosmithia pallida FS140 by our research group.
本实施例中所用的YPD固体培养基的配方为:每升含有酵母粉10g、蛋白胨20g、葡萄糖20g和琼脂粉20g,余量为蒸馏水,其配制方法是将各成分混合均匀,灭菌制得。The formula of the YPD solid medium used in this example is: each liter contains 10 g of yeast powder, 20 g of peptone, 20 g of glucose and 20 g of agar powder, and the balance is distilled water. .
实施例1深海真菌Geosmithia pallida FS140抗胶霉毒素自我保护基因序列的获得Example 1 Acquisition of the deep-sea fungus Geosmithia pallida FS140 anti-gliotoxin self-protection gene sequence
基因GliM的扩增:将深海真菌Geosmithia pallida FS140接种于YPD培养基平板,于37℃培养72h,挑取新鲜的菌丝体,利用真菌RNA提取试剂盒提取RNA,再用All-in-one RTMaster Kit逆转录获得cDNA。根据转录组测序结果预测编码抗单端孢霉烯自我保护基因GliM序列,在其上下游设计特异性引物,其引物序列为GliM-F:5'-ATGGAAGCCAACAACACCGAC-3';GliM-R:5'-CTACTTCTTCAGCCGTAATTCCAA-3',以cDNA文库为模板扩增,获得PCR产物(图1)。回收产物并用pEASY-T1试剂盒进行TA克隆,转化至大肠杆菌感受态细胞,涂布于氨苄青霉素抗性平板筛选出阳性克隆,以通用引物M13-F(5′-GTAAAACGACGGCCAGT-3′)和M13-R(5′-CAGGAAACAGCTATGAC-3′)进行菌液PCR验证阳性克隆并测序(图2),获得目的基因GliM序列(其核苷酸序列如SEQ ID NO.1所示,atggaagccaacaacaccgaccaaatccccaagcgcagacacaccgatgacctgtacgagctctcggttaatatctccagcgcggtcgagaccttcctcggaaggctggacgctgtaggagccccacgacccaccctggataacccgttcccggagcttatccacgacgaaggggcccagattgcccggatgaagattctccgcctatgcgagaggctcatggcgctggtgcagggccccgtccagtggctcatgtttcagaatatgtgcttcgtcgaaccagcttgtattggggcgatggcggaaatggggatccatgagattgtggctcctggtccggaaccgacgtctctagaccagattgtggaggctaccggtgcctctaaggatattctgaagcgggttatgcgagtctgtacccagagactggtctttgatgagattgcgccggagcaattcatccacaatggtgtctccttgcaatttcttgctcctcctgtacaggcgctgattagccatgcctgcgacgatggactgcgcattgcgtcccgtttctccgactctctgaagaaaaccaacttcaaaggcagcgataaaccagaagaaacggccttcagtctcgcctttggaacggataagggcctcttcgactacttctacagcgatgacatcgctcgcggacaacgcttcgccctcggtatggcaggtaatgatatcgtcagatcccggaccgaggatatgttcccattcgacaccctcccacagggcgcgaagttggtggacgttggaggcggtcgaggccatgtcgacgtgcgcattgccgagagggtccccggcttgaactttgtcgtgcaggacgacgtatccgttctcgaagccggacaggcagagggtgttcctgcggccgttaagggtcggatcgagttcatgccgcatgacttttttaaggagcagcctgtcaagggggcggatgcgtatctcctgcgattcattatgcatgatcacccggatagtgtctgtgcaaagatcctgtcccatattgtggacgcgatggaccccgagaagtctcggatcctgattgacgatgcggtcgtccccagcttcttgggtccggagagttcgcgattcttcaatttcctggatctgtatatgctgttctcactgaatgggaaggagaggactctggagatgtggaatcatctgttccagatggtcagtccgaacttggtgttggagaagatttggaaaatgcctggcagtgggcctgagtcggggaatgtgttggaattacggctgaagaagtag)。Amplification of gene GliM: Inoculate the deep-sea fungus Geosmithia pallida FS140 on YPD medium plate, culture at 37°C for 72h, pick fresh mycelium, extract RNA using fungal RNA extraction kit, and then use All-in-one RTMaster Kit reverse transcription to obtain cDNA. According to the transcriptome sequencing results, the GliM sequence encoding the anti-trichothecenes self-protection gene was predicted, and specific primers were designed upstream and downstream. The primer sequences were GliM-F: 5'-ATGGAAGCCAACAACACCGAC-3'; GliM-R: 5' -CTACTTCTTCAGCCGTAATTCCAA-3', using the cDNA library as a template to amplify to obtain a PCR product (Fig. 1). The product was recovered and TA cloned with pEASY-T1 kit, transformed into E. coli competent cells, coated on ampicillin-resistant plates to screen out positive clones, and universal primers M13-F (5′-GTAAAACGACGGCCAGT-3′) and M13 -R (5'-CAGGAAACAGCTATGAC-3') was used for bacterial liquid PCR to verify the positive clone and sequenced (Fig. 2) to obtain the target gene GliM sequence (its nucleotide sequence is shown in SEQ ID NO.1, gagagggtccccggcttgaactttgtcgtgcaggacgacgtatccgttctcgaagccggacaggcagagggtgttcctgcggccgttaagggtcggatcgagttcatgccgcatgacttttttaaggagcagcctgtcaagggggcggatgcgtatctcctgcgattcattatgcatgatcacccggatagtgtctgtgcaaagatcctgtcccatattgtggacgcgatggaccccgagaagtctcggatcctgattgacgatgcggtcgtccccagcttcttgggtccggagagttcgcgattcttcaatttcctggatctgtatatgctgttctcactgaatgggaaggagaggactctggagatgtggaatcatctgttccagatggtcagtccgaacttggtgttggagaagatttggaaaatgcctggcagtgggcctgagtcggggaatgtgttggaattacggctgaagaagtag)。
实施例2抗胶霉毒素自我保护基因GliM的功能验证Example 2 Functional verification of anti-gliotoxin self-protection gene GliM
利用同源重组法将基因GliM插入到酵母载体YEp352-TEF1-CYC1中(YEp352-TEF1-CYC1为早期构建质粒,携带有组成型启动子TEF1和终止子CYC1,载体图谱见图3A,为现有技术中的已知产品:Xiaodan Ouyang,Yaping Cha,Wen Li,Chaoyi Zhu,Muzi Zhu,ShuangLi,Min Zhuo,Shaobin Huang and Jianjun Li.Stepwise engineering ofSaccharomyces cerevisiae to produce(+)-valencene and its relatedsesquiterpenes,RSC Adv.,2019,9,30171,DOI:10.1039/c9ra05558d)。首先设计针对基因GliM(SEQ ID NO.1)扩增的上下游引物YEp352-GliM-F和YEp352-GliM-R,其引物序列为YEp352-GliM-F:5'-ATAGCAATCTAATCTAAGTCTAGAATGGAAGCCAACAACACCGAC-3';YEp352-GliM-R:5'-TACATGATGCGGCCCGTCGACCTACTTCTTCAGCCGTAATTCCAA-3'(下划线序列为同源臂片段),通过PCR扩增获得产物。对载体YEp352-TEF1-CYC1采用Sal I和Xba I双酶切并回收产物,然后使用ClonExpress II One Step Cloning Kit C112(Vazyme)将两个产物重组连接并转化至DH5α中筛选阳性克隆。采用引物YEp352-GliM-F和YEp352-GliM-R进行菌落PCR验证,结果表明基因GliM成功插入YEp352-TEF1-CYC1载体中(图3C),并通过测序予以确认,得到YEp352-TEF1-GliM载体(载体图谱见图3B)。The gene GliM was inserted into the yeast vector YEp352-TEF1-CYC1 by homologous recombination (YEp352-TEF1-CYC1 is an early construction plasmid, carrying the constitutive promoter TEF1 and terminator CYC1, the vector map is shown in Figure 3A, which is the existing Known products in technology: Xiaodan Ouyang, Yaping Cha, Wen Li, Chaoyi Zhu, Muzi Zhu, ShuangLi, Min Zhuo, Shaobin Huang and Jianjun Li. Stepwise engineering of Saccharomyces cerevisiae to produce(+)-valencene and its relatedsesquiterpenes, RSC Adv ., 2019, 9, 30171, DOI: 10.1039/c9ra05558d). First, the upstream and downstream primers YEp352-GliM-F and YEp352-GliM-R were designed for the amplification of the gene GliM (SEQ ID NO. 1). The primer sequences are YEp352-GliM-F: 5'-ATAGCAATCTAATCTAAGTCTAGA ATGGAAGCCAACAACACCGAC-3'; YEp352 -GliM-R:5'-TACATGATGCGGCCCGTCGAC CTACTTCTTCAGCCGTAATTCCAA-3' (the underlined sequence is a homology arm fragment), the product was obtained by PCR amplification. The vector YEp352-TEF1-CYC1 was double digested with Sal I and Xba I and the product was recovered, and then the two products were recombined and ligated with ClonExpress II One Step Cloning Kit C112 (Vazyme) and transformed into DH5α to screen for positive clones. Colony PCR was performed with primers YEp352-GliM-F and YEp352-GliM-R, and the results showed that the gene GliM was successfully inserted into the YEp352-TEF1-CYC1 vector (Figure 3C), which was confirmed by sequencing to obtain the YEp352-TEF1-GliM vector ( The vector map is shown in Figure 3B).
制备毒素敏感型酿酒酵母Saccharomyces cerevisiae BJ5464-D(relevantgenotype:Δpdr5Δpdr10Δpdr15)的感受态细胞(为现有技术中的已知产品,该菌株对毒性化合物更加敏感:Wolfgang Schweiger,Jayanand Boddu,Sanghyun Shin,BrigittePoppenberger,Franz Berthiller,Marc Lemmens,Gary J.Muehlbauer,and GerhardAdam.Validation of a Candidate Deoxynivalenol-Inactivating UDP-Glucosyltransferase from Barley by Heterologous Expression in Yeast,MPMI,2010,Vol.23,No.7,DOI:10.1094/MPMI-23-7-0977)。将YEp352-TEF1-GliM质粒载体以及YEp352-TEF1-CYC1质粒载体(阴性对照)分别电转入酿酒酵母BJ5464-D细胞中(1500V,5ms),均匀涂布于尿嘧啶缺陷型的SD平板中,在30℃培养2d,利用菌落PCR筛选阳性克隆,获得分别含有YEp352-TEF1-GliM质粒以及YEp352-TEF1-CYC1质粒的酿酒酵母BJ5464-D细胞。Preparation of competent cells of toxin-sensitive Saccharomyces cerevisiae BJ5464-D (relevantgenotype: Δpdr5Δpdr10Δpdr15) (a known product in the prior art, this strain is more sensitive to toxic compounds: Wolfgang Schweiger, Jayanand Boddu, Sanghyun Shin, Brigitte Poppenberger, Franz Berthiller, Marc Lemmens, Gary J. Muehlbauer, and Gerhard Adam. Validation of a Candidate Deoxynivalenol-Inactivating UDP-Glucosyltransferase from Barley by Heterologous Expression in Yeast, MPMI, 2010, Vol.23, No.7, DOI:10.1094/MPMI- 23-7-0977). The YEp352-TEF1-GliM plasmid vector and the YEp352-TEF1-CYC1 plasmid vector (negative control) were electroporated into Saccharomyces cerevisiae BJ5464-D cells (1500V, 5ms), and evenly spread on uracil-deficient SD plates, The cells were cultured at 30°C for 2 days, and positive clones were screened by colony PCR to obtain Saccharomyces cerevisiae BJ5464-D cells containing YEp352-TEF1-GliM plasmid and YEp352-TEF1-CYC1 plasmid respectively.
分别将酿酒酵母BJ5464-D(YEp352-TEF1-CYC1)、酿酒酵母BJ5464-D(YEp352-TEF1-GliM)接种于相应缺陷型的SD培养基中,在30℃培养2d。用分光光度计测量各菌液OD600,将各菌液用无菌水稀释到OD600≈1.0作为原液,再以100μL的原液加900μL的无菌水的方式稀释成10-1,以同样的方式稀释成10-2、10-3、10-4。各取5μL不同菌株的10-2、10-3、10-4的稀释液分别在YPD平板和YPD-FS140-12-2平板(含有2.5μM FS140-12-2胶霉毒素,图1)点板,在30℃培养并实时观察,以酿酒酵母BJ5464作为阳性对照。培养36h的平板结果显示(图4),酿酒酵母BJ5464-D(YEp352-TEF1-CYC1)、酿酒酵母BJ5464-D(YEp352-TEF1-GliM)在不添加任何毒素的YPD平板上生长状况近乎一致,但在含有2.5μM FS140-12-2胶霉毒素的YPD平板上,阴性对照BJ5464-D(YEp352-TEF1-CYC1)明显生长受阻,几乎都不能长。而导入GliM功能基因的酿酒酵母则生长良好,其不同稀释度下的菌体密度与正常酿酒酵母相当,说明来源于深海真菌Geosmithia pallida FS140的GliM功能基因部分或全部恢复了酿酒酵母BJ5464-D对外源添加毒素的耐受性,有效帮助酿酒酵母在含有毒素的环境下正常生长。Saccharomyces cerevisiae BJ5464-D (YEp352-TEF1-CYC1) and Saccharomyces cerevisiae BJ5464-D (YEp352-TEF1-GliM) were respectively inoculated into the corresponding deficient SD medium and cultured at 30°C for 2 days. Measure the OD600 of each bacterial solution with a spectrophotometer, dilute each bacterial solution with sterile water to OD600 ≈ 1.0 as a stock solution, and then add 100 μL of the stock solution to 900 μL of sterile water to dilute to 10-1 , in the same manner The method is diluted to 10-2 , 10-3 , 10-4 . Take 5 μL of the dilutions of 10-2 , 10-3 , and 10-4 of different strains on YPD plate and YPD-FS140-12-2 plate (containing 2.5μM FS140-12-2 gliotoxin, Figure 1) respectively. Plates were incubated at 30°C and observed in real time, with Saccharomyces cerevisiae BJ5464 as a positive control. The results of the plate cultured for 36h (Figure 4) showed that the growth conditions of Saccharomyces cerevisiae BJ5464-D (YEp352-TEF1-CYC1) and Saccharomyces cerevisiae BJ5464-D (YEp352-TEF1-GliM) on the YPD plate without any toxin were almost the same, However, on the YPD plate containing 2.5 μM FS140-12-2 gliotoxin, the negative control BJ5464-D (YEp352-TEF1-CYC1) obviously inhibited the growth and could hardly grow. The Saccharomyces cerevisiae introduced with the GliM functional gene grows well, and the cell density at different dilutions is comparable to that of normal Saccharomyces cerevisiae, indicating that the GliM functional gene derived from the deep-sea fungus Geosmithia pallida FS140 partially or fully restored the Saccharomyces cerevisiae BJ5464-D to the outside world. Source added toxin tolerance, effectively help Saccharomyces cerevisiae to grow normally in an environment containing toxins.
以上仅是本发明的优选实施方式,应当指出的是,上述优选实施方式不应视为对本发明的限制,本发明的保护范围应当以权利要求所限定的范围为准。对于本技术领域的普通技术人员来说,在不脱离本发明的精神和范围内,还可以做出若干改进和润饰,这些改进和润饰也应视为本发明的保护范围。The above are only the preferred embodiments of the present invention. It should be noted that the above preferred embodiments should not be regarded as limitations of the present invention, and the protection scope of the present invention should be based on the scope defined by the claims. For those skilled in the art, without departing from the spirit and scope of the present invention, several improvements and modifications can also be made, and these improvements and modifications should also be regarded as the protection scope of the present invention.
序列表sequence listing
<110> 广东省微生物研究所(广东省微生物分析检测中心)<110> Guangdong Institute of Microbiology (Guangdong Microbiological Analysis and Testing Center)
<120> 一种深海真菌FS140抗胶霉毒素自我保护基因GliM及其应用<120> A deep-sea fungus FS140 anti-gliotoxin self-protection gene GliM and its application
<160> 1<160> 1
<170> SIPOSequenceListing 1.0<170> SIPOSequenceListing 1.0
<210> 1<210> 1
<211> 1305<211> 1305
<212> DNA<212> DNA
<213> 深海真菌FS140(Geosmithia pallida)<213> Deep-sea fungi FS140 (Geosmithia pallida)
<400> 1<400> 1
atggaagcca acaacaccga ccaaatcccc aagcgcagac acaccgatga cctgtacgag 60atggaagcca acaacaccga ccaaatcccc aagcgcagac acaccgatga cctgtacgag 60
ctctcggtta atatctccag cgcggtcgag accttcctcg gaaggctgga cgctgtagga 120ctctcggtta atatctccag cgcggtcgag accttcctcg gaaggctgga cgctgtagga 120
gccccacgac ccaccctgga taacccgttc ccggagctta tccacgacga aggggcccag 180gccccacgac ccaccctgga taacccgttc ccggagctta tccacgacga aggggcccag 180
attgcccgga tgaagattct ccgcctatgc gagaggctca tggcgctggt gcagggcccc 240attgcccgga tgaagattct ccgcctatgc gagaggctca tggcgctggt gcagggcccc 240
gtccagtggc tcatgtttca gaatatgtgc ttcgtcgaac cagcttgtat tggggcgatg 300gtccagtggc tcatgtttca gaatatgtgc ttcgtcgaac cagcttgtat tggggcgatg 300
gcggaaatgg ggatccatga gattgtggct cctggtccgg aaccgacgtc tctagaccag 360gcggaaatgg ggatccatga gattgtggct cctggtccgg aaccgacgtc tctagaccag 360
attgtggagg ctaccggtgc ctctaaggat attctgaagc gggttatgcg agtctgtacc 420attgtggagg ctaccggtgc ctctaaggat attctgaagc gggttatgcg agtctgtacc 420
cagagactgg tctttgatga gattgcgccg gagcaattca tccacaatgg tgtctccttg 480cagagactgg tctttgatga gattgcgccg gagcaattca tccacaatgg tgtctccttg 480
caatttcttg ctcctcctgt acaggcgctg attagccatg cctgcgacga tggactgcgc 540caatttcttg ctcctcctgt acaggcgctg attagccatg cctgcgacga tggactgcgc 540
attgcgtccc gtttctccga ctctctgaag aaaaccaact tcaaaggcag cgataaacca 600attgcgtccc gtttctccga ctctctgaag aaaaccaact tcaaaggcag cgataaacca 600
gaagaaacgg ccttcagtct cgcctttgga acggataagg gcctcttcga ctacttctac 660gaagaaacgg ccttcagtct cgcctttgga acggataagg gcctcttcga ctacttctac 660
agcgatgaca tcgctcgcgg acaacgcttc gccctcggta tggcaggtaa tgatatcgtc 720agcgatgaca tcgctcgcgg acaacgcttc gccctcggta tggcaggtaa tgatatcgtc 720
agatcccgga ccgaggatat gttcccattc gacaccctcc cacagggcgc gaagttggtg 780agatcccgga ccgaggatat gttcccattc gacaccctcc cacagggcgc gaagttggtg 780
gacgttggag gcggtcgagg ccatgtcgac gtgcgcattg ccgagagggt ccccggcttg 840gacgttggag gcggtcgagg ccatgtcgac gtgcgcattg ccgagagggt ccccggcttg 840
aactttgtcg tgcaggacga cgtatccgtt ctcgaagccg gacaggcaga gggtgttcct 900aactttgtcg tgcaggacga cgtatccgtt ctcgaagccg gacaggcaga gggtgttcct 900
gcggccgtta agggtcggat cgagttcatg ccgcatgact tttttaagga gcagcctgtc 960gcggccgtta agggtcggat cgagttcatg ccgcatgact tttttaagga gcagcctgtc 960
aagggggcgg atgcgtatct cctgcgattc attatgcatg atcacccgga tagtgtctgt 1020aagggggcgg atgcgtatct cctgcgattc attatgcatg atcacccgga tagtgtctgt 1020
gcaaagatcc tgtcccatat tgtggacgcg atggaccccg agaagtctcg gatcctgatt 1080gcaaagatcc tgtcccatat tgtggacgcg atggaccccg agaagtctcg gatcctgatt 1080
gacgatgcgg tcgtccccag cttcttgggt ccggagagtt cgcgattctt caatttcctg 1140gacgatgcgg tcgtccccag cttcttgggt ccggagagtt cgcgattctt caatttcctg 1140
gatctgtata tgctgttctc actgaatggg aaggagagga ctctggagat gtggaatcat 1200gatctgtata tgctgttctc actgaatggg aaggagga ctctggagat gtggaatcat 1200
ctgttccaga tggtcagtcc gaacttggtg ttggagaaga tttggaaaat gcctggcagt 1260ctgttccaga tggtcagtcc gaacttggtg ttggagaaga tttggaaaat gcctggcagt 1260
gggcctgagt cggggaatgt gttggaatta cggctgaaga agtag 1305gggcctgagt cggggaatgt gttggaatta cggctgaaga agtag 1305
Claims (5)
Priority Applications (1)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| CN202011550170.9ACN112608931B (en) | 2020-12-24 | 2020-12-24 | Deep-sea fungus FS140 anti-gliotoxin self-protection gene GliM and application thereof |
Applications Claiming Priority (1)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| CN202011550170.9ACN112608931B (en) | 2020-12-24 | 2020-12-24 | Deep-sea fungus FS140 anti-gliotoxin self-protection gene GliM and application thereof |
Publications (2)
| Publication Number | Publication Date |
|---|---|
| CN112608931A CN112608931A (en) | 2021-04-06 |
| CN112608931Btrue CN112608931B (en) | 2022-09-02 |
Family
ID=75244653
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| CN202011550170.9AActiveCN112608931B (en) | 2020-12-24 | 2020-12-24 | Deep-sea fungus FS140 anti-gliotoxin self-protection gene GliM and application thereof |
Country Status (1)
| Country | Link |
|---|---|
| CN (1) | CN112608931B (en) |
Families Citing this family (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN110950940B (en)* | 2019-10-29 | 2023-03-31 | 广东省微生物研究所(广东省微生物分析检测中心) | Novel transcription regulation factor DcGliZ from deep-sea fungi and application thereof |
| CN116286897B (en)* | 2023-03-29 | 2024-05-03 | 佛山科学技术学院 | A Tri3 gene encoding acyltransferase and its application |
Citations (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN107858353A (en)* | 2017-11-10 | 2018-03-30 | 广东省微生物研究所(广东省微生物分析检测中心) | A kind of angstrom moral bacterium FS110 glutathione sulfurtransferase gene GliG promoters and its application |
| CN110950940A (en)* | 2019-10-29 | 2020-04-03 | 广东省微生物研究所(广东省微生物分析检测中心) | Novel transcription regulation factor DcGliZ from deep-sea fungi and application thereof |
Family Cites Families (6)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| JP2006249019A (en)* | 2005-03-11 | 2006-09-21 | Gunma Univ | A novel use of gliotoxin as an active ingredient for antimalarial drugs, and an antimalarial drug containing gliotoxin as an active ingredient. |
| WO2007128555A1 (en)* | 2006-05-08 | 2007-11-15 | MAX-PLANCK-Gesellschaft zur Förderung der Wissenschaften e.V. | Inhibition of gliotoxin |
| CN102839130B (en)* | 2012-08-24 | 2013-11-20 | 黄河三角洲京博化工研究院有限公司 | Bacterial strain for producing gliotoxin and method for producing gliotoxin by adopting bacterial strain |
| CN110904125B (en)* | 2019-11-26 | 2021-05-11 | 广东省微生物研究所(广东省微生物分析检测中心) | An anti-trichothecenes self-protection gene mfs1 of A553 from Rhizoctonia var. and its application |
| CN111057713A (en)* | 2019-12-18 | 2020-04-24 | 广东省微生物研究所(广东省微生物分析检测中心) | A CRISPR/Cas9 vector suitable for Escherichia coli FS110 and its construction method and application |
| CN111073902B (en)* | 2019-12-30 | 2022-04-26 | 广东省微生物研究所(广东省微生物分析检测中心) | CRISPR/dCas9 vector for enhancing the expression level of gliotoxin biosynthesis gene and its construction method and application |
- 2020
- 2020-12-24CNCN202011550170.9Apatent/CN112608931B/enactiveActive
Patent Citations (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN107858353A (en)* | 2017-11-10 | 2018-03-30 | 广东省微生物研究所(广东省微生物分析检测中心) | A kind of angstrom moral bacterium FS110 glutathione sulfurtransferase gene GliG promoters and its application |
| CN110950940A (en)* | 2019-10-29 | 2020-04-03 | 广东省微生物研究所(广东省微生物分析检测中心) | Novel transcription regulation factor DcGliZ from deep-sea fungi and application thereof |
Also Published As
| Publication number | Publication date |
|---|---|
| CN112608931A (en) | 2021-04-06 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| CN110904125B (en) | An anti-trichothecenes self-protection gene mfs1 of A553 from Rhizoctonia var. and its application | |
| CN106754960B (en) | One NLR genoid NLR1-V and its expression vector and application | |
| CN109735537B (en) | Myrothecium roridum A553 trichothecene synthase gene Tri5 promoter and application thereof | |
| Kusuya et al. | Transcription factor Afmac1 controls copper import machinery in Aspergillus fumigatus | |
| CN112608931B (en) | Deep-sea fungus FS140 anti-gliotoxin self-protection gene GliM and application thereof | |
| CN107858353B (en) | Edwardsiella FS110 glutathione S-transferase gene GliG promoter and application thereof | |
| Ökmen et al. | Functional analysis of the conserved transcriptional regulator CfWor1 in C ladosporium fulvum reveals diverse roles in the virulence of plant pathogenic fungi | |
| CN111748562B (en) | A gene encoding protein of rice sheath blight Atg 22 and its target fragment Rsatg22 and its application | |
| Izumitsu et al. | The MAPKK kinase ChSte11 regulates sexual/asexual development, melanization, pathogenicity, and adaptation to oxidative stress in Cochliobolus heterostrophus | |
| CN112680469B (en) | Application of gliotoxin self-protection gene GliK in assisting host cells in resisting gliotoxin | |
| CN111019945B (en) | A kind of Tri12 promoter of trichothecenes synthase gene A553 of Rhizoctonia var. and its application | |
| Barhoom et al. | Functional characterization of CgCTR2, a putative vacuole copper transporter that is involved in germination and pathogenicity in Colletotrichum gloeosporioides | |
| CN106916837A (en) | Hyperosmosis glycerine protein kinase gene RkHog1 and its recombinant expression carrier | |
| CN112626039B (en) | An oxidoreductase GliT and its application in resisting mycotoxins | |
| CN112646811B (en) | CRISPR/Cas9 system suitable for multi-gene knockout of yeast and construction method and application thereof | |
| CN112695044B (en) | A deep-sea fungus FS140 anti-gliotoxin self-protection gene mfs-get and its application | |
| CN112592954B (en) | Application of Gene GliT as Screening Marker Gene in Resistance Screening | |
| CN116064266B (en) | Recombinant saccharomyces cerevisiae with enhanced salt stress resistance, and construction method and application thereof | |
| CN117925430A (en) | Oxidation stress resistant saccharomyces cerevisiae engineering strain, construction method and application thereof | |
| CN103103208B (en) | Haynaldia villosa disulfide isomerase gene and application thereof | |
| Shao et al. | Effects of an alternative oxidase gene on conidia viability under external stresses in Monascus ruber M7 | |
| CN112725362B (en) | Myrothecium roridum A553 trichothecene-resistant self-protection gene GNAT11 and application thereof | |
| CN116286897B (en) | A Tri3 gene encoding acyltransferase and its application | |
| CN112795567B (en) | A deep-sea fungus FS140 O-methyltransferase gene GliM promoter and its application | |
| CN108659109B (en) | A Wheat Powdery Mildew Resistance-Related Protein TaSTKR1 and Its Encoding Gene and Application |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| PB01 | Publication | ||
| PB01 | Publication | ||
| SE01 | Entry into force of request for substantive examination | ||
| SE01 | Entry into force of request for substantive examination | ||
| GR01 | Patent grant | ||
| GR01 | Patent grant | ||
| CP01 | Change in the name or title of a patent holder | ||
| CP01 | Change in the name or title of a patent holder | Address after:510070 No.56 courtyard, No.100 Xianlie Middle Road, Yuexiu District, Guangzhou City, Guangdong Province Patentee after:Institute of Microbiology, Guangdong Academy of Sciences Address before:510070 No.56 courtyard, No.100 Xianlie Middle Road, Yuexiu District, Guangzhou City, Guangdong Province Patentee before:GUANGDONG INSTITUTE OF MICROBIOLOGY (GUANGDONG DETECTION CENTER OF MICROBIOLOGY) |