CN111612745A - Image straightening method, system, storage medium and device for curved chromosomes based on BagPix2Pix self-learning model - Google Patents
Image straightening method, system, storage medium and device for curved chromosomes based on BagPix2Pix self-learning modelDownload PDFInfo
- Publication number
- CN111612745A CN111612745ACN202010360518.1ACN202010360518ACN111612745ACN 111612745 ACN111612745 ACN 111612745ACN 202010360518 ACN202010360518 ACN 202010360518ACN 111612745 ACN111612745 ACN 111612745A
- Authority
- CN
- China
- Prior art keywords
- chromosome
- map
- image
- skeleton
- straightening
- Prior art date
- Legal status (The legal status is an assumption and is not a legal conclusion. Google has not performed a legal analysis and makes no representation as to the accuracy of the status listed.)
- Pending
Links
Images
Classifications
- G—PHYSICS
- G06—COMPUTING OR CALCULATING; COUNTING
- G06T—IMAGE DATA PROCESSING OR GENERATION, IN GENERAL
- G06T7/00—Image analysis
- G06T7/0002—Inspection of images, e.g. flaw detection
- G06T7/0012—Biomedical image inspection
- G—PHYSICS
- G06—COMPUTING OR CALCULATING; COUNTING
- G06T—IMAGE DATA PROCESSING OR GENERATION, IN GENERAL
- G06T5/00—Image enhancement or restoration
- G06T5/80—Geometric correction
- G—PHYSICS
- G06—COMPUTING OR CALCULATING; COUNTING
- G06T—IMAGE DATA PROCESSING OR GENERATION, IN GENERAL
- G06T7/00—Image analysis
- G06T7/10—Segmentation; Edge detection
- G06T7/12—Edge-based segmentation
- G—PHYSICS
- G06—COMPUTING OR CALCULATING; COUNTING
- G06T—IMAGE DATA PROCESSING OR GENERATION, IN GENERAL
- G06T7/00—Image analysis
- G06T7/60—Analysis of geometric attributes
- G06T7/62—Analysis of geometric attributes of area, perimeter, diameter or volume
- G—PHYSICS
- G06—COMPUTING OR CALCULATING; COUNTING
- G06T—IMAGE DATA PROCESSING OR GENERATION, IN GENERAL
- G06T2207/00—Indexing scheme for image analysis or image enhancement
- G06T2207/10—Image acquisition modality
- G06T2207/10056—Microscopic image
- G06T2207/10061—Microscopic image from scanning electron microscope
- G—PHYSICS
- G06—COMPUTING OR CALCULATING; COUNTING
- G06T—IMAGE DATA PROCESSING OR GENERATION, IN GENERAL
- G06T2207/00—Indexing scheme for image analysis or image enhancement
- G06T2207/10—Image acquisition modality
- G06T2207/10064—Fluorescence image
- G—PHYSICS
- G06—COMPUTING OR CALCULATING; COUNTING
- G06T—IMAGE DATA PROCESSING OR GENERATION, IN GENERAL
- G06T2207/00—Indexing scheme for image analysis or image enhancement
- G06T2207/20—Special algorithmic details
- G06T2207/20081—Training; Learning
- G—PHYSICS
- G06—COMPUTING OR CALCULATING; COUNTING
- G06T—IMAGE DATA PROCESSING OR GENERATION, IN GENERAL
- G06T2207/00—Indexing scheme for image analysis or image enhancement
- G06T2207/20—Special algorithmic details
- G06T2207/20084—Artificial neural networks [ANN]
- G—PHYSICS
- G06—COMPUTING OR CALCULATING; COUNTING
- G06T—IMAGE DATA PROCESSING OR GENERATION, IN GENERAL
- G06T2207/00—Indexing scheme for image analysis or image enhancement
- G06T2207/30—Subject of image; Context of image processing
- G06T2207/30004—Biomedical image processing
Landscapes
- Engineering & Computer Science (AREA)
- Physics & Mathematics (AREA)
- Theoretical Computer Science (AREA)
- General Physics & Mathematics (AREA)
- Computer Vision & Pattern Recognition (AREA)
- Health & Medical Sciences (AREA)
- Geometry (AREA)
- General Health & Medical Sciences (AREA)
- Medical Informatics (AREA)
- Nuclear Medicine, Radiotherapy & Molecular Imaging (AREA)
- Radiology & Medical Imaging (AREA)
- Quality & Reliability (AREA)
- Image Analysis (AREA)
Abstract
Translated fromChineseDescription
Translated fromChinese技术领域technical field
本发明设计图像处理技术领域,具体涉及一种基于BagPix2Pix自学习模型的弯曲染色体图像拉直方法、系统、存储介质及装置。The present invention is designed in the technical field of image processing, in particular to a method, system, storage medium and device for straightening a curved chromosome image based on the BagPix2Pix self-learning model.
背景技术Background technique
随着电子计算机技术和人工智能领域的发展,通过计算机算法实现图像的识别和分割已经可以初步实现,且医学上一般使用图像分析技术用于医疗影像中物体的检测、分离和提取。但是当被检测物体处于不同的形态时,例如弯曲和重叠,图形学算法往往不能像素级别地改变物体的形态。With the development of electronic computer technology and artificial intelligence, the recognition and segmentation of images through computer algorithms can be initially realized, and image analysis technology is generally used in medicine for the detection, separation and extraction of objects in medical images. But when the detected object is in different shapes, such as bending and overlapping, graphics algorithms often cannot change the shape of the object at the pixel level.
人类的基因信息是被承载在染色体上的,因而染色体核型分析是细胞遗传学研究的基本方法,是研究染色体形态和结构与其功能的联系,探究染色体异常和遗传缺陷的关联的重要手段。但是由于染色体自身的性质,染色体经常处于弯曲的形态。相较于处于竖直形态,弯曲形态的染色体往往给染色体鉴定,分类和功能研究带来阻碍,且人工分析耗时耗力巨大。现有的染色体拉直技术则基于切割和拼接等图形学的算法,其缺点是不能柔性的对图像进行像素级别的拉直,且会产生明显的切割痕迹和拼接处的空白。Human genetic information is carried on chromosomes, so chromosome karyotype analysis is the basic method of cytogenetics research, and it is an important means to study the relationship between chromosome morphology and structure and its function, and to explore the relationship between chromosomal abnormalities and genetic defects. But due to the nature of chromosomes themselves, chromosomes are often in a curved shape. Compared with vertical chromosomes, bent chromosomes often hinder chromosome identification, classification and functional studies, and manual analysis is time-consuming and labor-intensive. The existing chromosome straightening technology is based on graphics algorithms such as cutting and splicing. The disadvantage is that it cannot flexibly straighten the image at the pixel level, and will produce obvious cutting marks and blanks at the splicing.
例如文献《一种关于高度弯曲染色体图的拉直方法》Roshtkhari,M.J. andSetarehdan,S.K.,2008.A novel algorithm for straightening highly curved imagesof human chromosome.Pattern recognition letters,29(9),pp.1208——1217.,揭示了一种染色体拉直的手段,采用定位最大弯曲处,并在最大弯曲处切割后对两端进行旋转和拼接。这样的方式适用于染色体只存在一个较大弯曲,而在不同实验和精细度中拍摄的染色体照片经常会出现三个乃至更多的弯曲,所以这个算法经常出现拉直的错误,且切割再拼接的空白也会使染色体信息不连续,远远不能达到使用要求。For example, the document "A Straightening Method for Highly Curved Chromosome Maps" Roshtkhari, M.J. and Setarehdan, S.K., 2008. A novel algorithm for straightening highly curved images of human chromosome. Pattern recognition letters, 29(9), pp.1208-1217 ., revealed a means of chromosome straightening by locating the maximal bend, and rotating and splicing the ends after cutting at the maximal bend. This method is suitable for chromosomes with only one large curvature, and chromosome photos taken in different experiments and finesse often have three or more curvatures, so this algorithm often has straightening errors, and cutting and splicing The blank will also make the chromosome information discontinuous, which is far from meeting the requirements for use.
发明内容SUMMARY OF THE INVENTION
本发明的目的之一就是为了解决现有技术中存在的上述问题,提供一种基于BagPix2Pix自学习模型的人类弯曲染色体的拉直方法,利用已学习到从骨架生成相应染色体图的模式的弯曲染色体图像拉直模型,实现通过竖直骨架生成相应的柔性拉直染色体的预测图。One of the objectives of the present invention is to solve the above-mentioned problems in the prior art, to provide a method for straightening human bent chromosomes based on the BagPix2Pix self-learning model, using the bent chromosomes that have learned the pattern of generating the corresponding chromosome map from the skeleton The image straightening model realizes the generation of the corresponding prediction map of the flexible straightened chromosomes through the vertical skeleton.
本发明的目的通过以下技术方案来实现:The object of the present invention is achieved through the following technical solutions:
基于BagPix2Pix自学习模型的弯曲染色体图像拉直方法,包括如下步骤,The image straightening method for curved chromosomes based on the BagPix2Pix self-learning model includes the following steps:
S1,接收弯曲染色体原图,处理得到标记图;S1, receive the original image of the curved chromosome, and process it to obtain the marked image;
S2,根据S1得到的标记图,生成染色体骨架锚点图;S2, according to the marker map obtained in S1, generate a chromosome skeleton anchor map;
S3,根据染色体骨架锚点图,生成染色体拉直骨架图;S3, according to the chromosome skeleton anchor point map, generate a chromosome straightening skeleton map;
S4,将染色体拉直骨架图输入训练收敛的、接收骨架图可输出匹配的预测图的弯曲染色体图像拉直模型中,生成弯曲染色体拉直图像。S4, the chromosome straightening skeleton map is input into the curved chromosome image straightening model converged by training, receiving the skeleton map and outputting a matching prediction map, to generate a curved chromosome straightening image.
优选的,所述的基于BagPix2Pix自学习模型的弯曲染色体图像拉直方法中,所述S1包括:Preferably, in the method for straightening curved chromosome images based on the BagPix2Pix self-learning model, the S1 includes:
S11,获取显微镜拍摄的弯曲染色体图像,并转化为灰度图;S11, acquiring the image of the curved chromosomes photographed by the microscope, and converting it into a grayscale image;
S12,对S11得到的灰度图进行标记,将图中染色体标记为1值像素,杂质和背景部分标记为0值像素,获得标记图。S12, the grayscale image obtained in S11 is marked, and the chromosomes in the image are marked as 1-value pixels, and the impurities and background parts are marked as 0-value pixels to obtain a marked image.
优选的,所述的基于BagPix2Pix自学习模型的弯曲染色体图像拉直方法中,所述S2包括如下步骤:Preferably, in the method for straightening curved chromosome images based on the BagPix2Pix self-learning model, the S2 includes the following steps:
S21,记录S12中标记图的每一行的首个1值像素和末位1值像素;S21, record the first 1-value pixel and the last 1-value pixel of each row of the marked image in S12;
S22,求出每一行的首个1值像素和末位1值像素的中间点的坐标(X,Y);S22, obtain the coordinates (X, Y) of the middle point of the first 1-value pixel and the last 1-value pixel of each row;
S23,将所有求出的Y值平均分成n等分形成n个均分点;S23, all the obtained Y values are equally divided into n equal divisions to form n equal division points;
S24,将等分后,除第一个均分点和最后一个均分点之外的均分点定为弯曲染色体的骨架锚点;S24, after dividing, the dividing points except the first dividing point and the last dividing point are determined as the backbone anchor points of the curved chromosome;
S25,新建一张a×a大小的黑色背景图片,在此图片上定位所述S24 步骤确定的骨架锚点,得到染色体骨架锚点图。S25, create a black background picture with a size of a×a, locate the skeleton anchor point determined in the step S24 on this picture, and obtain a chromosome skeleton anchor point map.
优选的,所述的基于BagPix2Pix自学习模型的弯曲染色体图像拉直方法中,所述S2中,若染色体弯曲幅度过大或骨架锚点定位在染色体部分之外,则人工标记目标数量个点作为骨架锚点。Preferably, in the method for straightening bent chromosome images based on the BagPix2Pix self-learning model, in S2, if the chromosome bending amplitude is too large or the skeleton anchor point is located outside the chromosome part, manually mark the target number of points as Skeleton anchor point.
优选的,所述的基于BagPix2Pix自学习模型的弯曲染色体图像拉直方法中,所述S3包括Preferably, in the method for straightening curved chromosome images based on the BagPix2Pix self-learning model, the S3 includes:
S31,创建a×a大小的黑色背景图片;S31, create a black background image of size a×a;
S32,计算出染色体骨架锚点图中每两个相邻骨架锚点之间的距离;S32, calculate the distance between every two adjacent skeleton anchor points in the chromosome skeleton anchor point map;
S33,在S31的黑色背景图片中,通过多条分别与一对相邻骨架锚点之间的距离相等且位置居中的线段依次连接构成染色体拉直骨架图。S33 , in the black background picture of S31 , a chromosome straightening skeleton diagram is formed by sequentially connecting a plurality of line segments that are respectively at the same distance from a pair of adjacent skeleton anchor points and are located in the center.
优选的,所述的基于BagPix2Pix自学习模型的弯曲染色体图像拉直方法中,每条所述线段的宽度为10像素,且多条线段的像素值渐大或渐小。Preferably, in the method for straightening a curved chromosome image based on the BagPix2Pix self-learning model, the width of each line segment is 10 pixels, and the pixel values of the plurality of line segments gradually increase or decrease.
优选的,所述的基于BagPix2Pix自学习模型的弯曲染色体图像拉直方法中,所述S4中,所述弯曲染色体图像拉直模型基于对抗训练方法学习从染色体骨架图生成相应的染色体图的模式,其包括一个生成器和一个判别器,且生成器包括一条编码路径和一条解码路径。Preferably, in the method for straightening the image of the curved chromosome based on the BagPix2Pix self-learning model, in the S4, the straightening model for the curved chromosome image learns the pattern of generating the corresponding chromosome map from the chromosome skeleton map based on the adversarial training method, It includes a generator and a discriminator, and the generator includes an encoding path and a decoding path.
本发明的另一目的是提供一种实现上述方法的基于BagPix2Pix自学习模型的弯曲染色体图像拉直系统,包括Another object of the present invention is to provide an image straightening system based on the BagPix2Pix self-learning model for realizing the above method, comprising:
预处理单元,用于接收弯曲染色体原图,处理得到标记图;The preprocessing unit is used to receive the original image of the curved chromosome, and process it to obtain the marked image;
弯曲骨架图生成单元,用于根据标记图,生成染色体骨架锚点图;The curved skeleton map generation unit is used to generate the chromosome skeleton anchor point map according to the marker map;
拉直骨架图生成单元,用于根据染色体骨架锚点图,生成染色体拉直骨架图;The straightening skeleton map generation unit is used to generate the chromosome straightening skeleton map according to the chromosome skeleton anchor point map;
拉直染色体图生成单元,用于将染色体拉直骨架图输入训练收敛的、接收骨架图可输出匹配的预测图的弯曲染色体图像拉直模型中,生成弯曲染色体拉直图像。The straight chromosome map generating unit is used for inputting the chromosome straightening skeleton map into the curved chromosome image straightening model converged by training, receiving the skeleton map and outputting a matching prediction map, and generating the curved chromosome straightening image.
本发明的又一目的是提供一种可读存储介质,存储有实现上述任一所述的基于BagPix2Pix自学习模型的弯曲染色体图像拉直方法的程序。Another object of the present invention is to provide a readable storage medium storing a program for implementing any of the above-described methods for straightening images of curved chromosomes based on the BagPix2Pix self-learning model.
本发明的又一目的是提供一种图像处理装置,包括存储器、处理器及通信总线;所述通信总线用于实现处理器和存储器之间的连接通信;所述处理器用于执行存储器中存储的软件程序以实现上述任一所述的基于 BagPix2Pix自学习模型的弯曲染色体图像拉直方法。Another object of the present invention is to provide an image processing device, including a memory, a processor and a communication bus; the communication bus is used to implement connection and communication between the processor and the memory; the processor is used to execute data stored in the memory. A software program to implement any of the above-mentioned methods for straightening images of curved chromosomes based on the BagPix2Pix self-learning model.
本发明技术方案的优点主要体现在:The advantages of the technical solution of the present invention are mainly reflected in:
本方案能够准确地通过单一染色体图构建增广数据集,并通过拉直骨架来精确生成拉直的染色体图,并保留染色体细节信息,以此高质量的完成人类弯曲染色体柔性拉直目标,与现有的基于图像学的方法相比,此方法的结果不包含明显的断裂和切面,极大的提升了拉直后染色体图像的质量和有效特征的完整性和连续性,且不受弯曲个数和弯曲部位的影响,准确性高,实用性强。This solution can accurately construct an augmented data set through a single chromosome map, and accurately generate a straightened chromosome map by straightening the skeleton, and retain the chromosome detail information, so as to complete the flexible straightening goal of human bending chromosomes with high quality. Compared with the existing image-based methods, the results of this method do not contain obvious breaks and sections, which greatly improves the quality of the straightened chromosome image and the integrity and continuity of the effective features, and is not subject to bending. The influence of the number and the bending part, the accuracy is high, and the practicability is strong.
本方案基于深度学习模型,能在保留染色体细节信息的前提下提取染色体图像特征,并通过对抗性训练的方法自学习图像局部与全局特征和由骨架图生成相应染色体图的生成方式,以此来对染色体进行柔性的拉直。Based on the deep learning model, this scheme can extract chromosome image features on the premise of retaining the detailed information of chromosomes, and self-learn the local and global features of the image through the method of adversarial training and the generation method of generating the corresponding chromosome map from the skeleton map. Flexible straightening of chromosomes.
拉直的染色体相比于原始弯曲的染色体,能为神经网络提供更高的分类准确率,并协助计算机构建更准确的分类模型和帮助医护人员更便捷的观察区分染色体中的异常和病变区域。Compared with the original bent chromosomes, the straightened chromosomes can provide a higher classification accuracy for the neural network, and help the computer to build a more accurate classification model and help medical staff to observe and distinguish abnormal and diseased regions in the chromosomes more conveniently.
附图说明Description of drawings
图1为背景技术中描述的图形学算法生成的染色体拉直图;1 is a chromosome straightening diagram generated by the graphics algorithm described in the background art;
图2为本发明的流程示意图;Fig. 2 is the schematic flow chart of the present invention;
图3是本发明的S12中选取的细胞中期显微镜下经荧光染色的染色体的标记图像;Fig. 3 is the labeling image of the chromosome that is fluorescently stained under the microscope of the cell mid-phase selected in S12 of the present invention;
图4为S13中经标记图像过滤出的染色体图像;Fig. 4 is the chromosome image filtered out by marked image in S13;
图5为S2得到的染色体弯曲骨架图;Fig. 5 is the chromosome bending skeleton diagram obtained by S2;
图6为S3得到的染色体拉直骨架图;Fig. 6 is the chromosome straightening skeleton diagram obtained by S3;
图7为S4得到的弯曲染色体拉直图像;Figure 7 is the straightened image of bent chromosomes obtained by S4;
图8为S40得到的增广后的染色体缩放图;Fig. 8 is the enlarged chromosome zoom diagram obtained by S40;
图9为S40得到的增广后的染色体弯曲骨架图;Fig. 9 is the expanded chromosome bending skeleton diagram obtained by S40;
图10为本方案的系统的第一实施例的示意图;FIG. 10 is a schematic diagram of the first embodiment of the system of this scheme;
图11为本方案的系统的第二实施例的示意图。FIG. 11 is a schematic diagram of a second embodiment of the system of the present scheme.
具体实施方式Detailed ways
本发明的目的、优点和特点,将通过下面优选实施例的非限制性说明进行图示和解释。这些实施例仅是应用本发明技术方案的典型范例,凡采取等同替换或者等效变换而形成的技术方案,均落在本发明要求保护的范围之内。The objects, advantages and features of the present invention will be illustrated and explained by the following non-limiting description of the preferred embodiments. These embodiments are only typical examples of applying the technical solutions of the present invention, and all technical solutions formed by taking equivalent replacements or equivalent transformations fall within the scope of protection of the present invention.
在方案的描述中,需要说明的是,术语“中心”、“上”、“下”、“左”、“右”、“前”、“后”、“竖直”、“水平”、“内”、“外”等指示的方位或位置关系为基于附图所示的方位或位置关系,仅是为了便于描述和简化描述,而不是指示或暗示所指的装置或元件必须具有特定的方位、以特定的方位构造和操作,因此不能理解为对本发明的限制。此外,术语“第一”、“第二”、“第三”仅用于描述目的,而不能理解为指示或暗示相对重要性。并且,在方案的描述中,以操作人员为参照,靠近操作者的方向为近端,远离操作者的方向为远端。In the description of the scheme, it should be noted that the terms "center", "upper", "lower", "left", "right", "front", "rear", "vertical", "horizontal", " The orientation or positional relationship indicated by "inside", "outside", etc. is based on the orientation or positional relationship shown in the drawings, which is only for convenience and simplification of description, rather than indicating or implying that the indicated device or element must have a specific orientation , constructed and operated in a specific orientation, and therefore should not be construed as limiting the invention. Furthermore, the terms "first", "second", and "third" are used for descriptive purposes only and should not be construed to indicate or imply relative importance. In addition, in the description of the solution, with reference to the operator, the direction close to the operator is the proximal end, and the direction away from the operator is the distal end.
下面结合附图对本发明揭示的基于BagPix2Pix自学习模型的弯曲染色体图像拉直方法进行阐述,如附图2所示,其包括如下步骤,The method for straightening a curved chromosome image based on the BagPix2Pix self-learning model disclosed in the present invention will be described below with reference to the accompanying drawings. As shown in FIG. 2 , it includes the following steps:
S1,接收弯曲染色体原图,处理得到标记图;S1, receive the original image of the curved chromosome, and process it to obtain the marked image;
S2,根据S1得到的标记图,生成染色体骨架锚点图;S2, according to the marker map obtained in S1, generate a chromosome skeleton anchor map;
S3,根据染色体骨架锚点图,生成染色体拉直骨架图;S3, according to the chromosome skeleton anchor point map, generate a chromosome straightening skeleton map;
S4,将染色体拉直骨架图输入训练收敛的、接收骨架图可输出匹配的预测图的弯曲染色体图像拉直模型中,生成弯曲染色体拉直图像。S4, the chromosome straightening skeleton map is input into the curved chromosome image straightening model converged by training, receiving the skeleton map and outputting a matching prediction map, to generate a curved chromosome straightening image.
这一方法是基于已经形成了弯曲染色体图像拉直模型,将采集的某一弯曲染色体图像经过一定处理后得到该弯曲染色体的拉直骨架图,然后将该拉直骨架图输入弯曲染色体图像拉直模型,该模型能够通过其生成器将构建的相同长度的拉直骨架,还原成为拉直的染色体,得到弯曲染色体拉直图。This method is based on a curved chromosome image straightening model that has been formed. After a certain curved chromosome image is collected, a straightened skeleton image of the curved chromosome is obtained, and then the straightened skeleton image is input into the curved chromosome image to straighten The model can restore the straightened skeleton of the same length constructed by its generator into straightened chromosomes, and obtain a straightened diagram of curved chromosomes.
具体的,所述S1的处理过程如下:Specifically, the processing process of the S1 is as follows:
S11,获取显微镜拍摄的弯曲染色体图像,并转化为灰度图。为了获得真实的染色体图像,需要对细胞中期的染色体进行荧光标记,通过显微镜在合适的倍数下选择染色体进行拍摄,并将图像导入计算机,计算机接收显微镜拍摄的弯曲染色体图像后转换为灰度图。如果该染色体和其他染色体发生接触或重叠现象,将其按照原本的边缘形状进行切割和提取即可。S11, the curved chromosome image captured by the microscope is acquired and converted into a grayscale image. In order to obtain a real chromosome image, it is necessary to fluorescently label the chromosomes in the metaphase of the cell, select the chromosomes under the appropriate magnification through the microscope, and import the image into the computer. The computer receives the curved chromosome image captured by the microscope and converts it into a grayscale image. If the chromosome contacts or overlaps with other chromosomes, it can be cut and extracted according to the original edge shape.
S12,为了去除边缘像素的污染,对S11得到的灰度图中的染色体、杂质和背景部分进行标记,将图中染色体标记为1值像素,将杂质和背景部分标记为0值像素,获得标记图,如附图3所示,其中杂质包括但不限于细胞质,细胞核等细胞器。S12, in order to remove the contamination of the edge pixels, the chromosomes, impurities and background parts in the grayscale image obtained in S11 are marked, the chromosomes in the image are marked as 1-value pixels, and the impurities and background parts are marked as 0-value pixels to obtain the mark As shown in Figure 3, the impurities include but are not limited to cytoplasm, nucleus and other organelles.
进一步,所述S12步骤之后还可以具有S13,按照S12的标记图来进行染色体图像的提取,即对标记图进行反色处理,将标记图中1值像素(染色体)保持原色,将0值像素(杂质和背景部分)转变为黑色,从而去除杂质,提取得到仅包含染色体部分和单一色背景(黑色背景)的染色体图,如附图4所示。该步骤可以用于后续形成训练模型所需的缩放图。Further, after the step S12, there may be a step S13 to extract the chromosome image according to the marker map in S12, that is, perform inverse color processing on the marker map, keep the 1-value pixels (chromosomes) in the marker map as primary colors, and use the 0-value pixels in the marker map. (impurities and background parts) are converted to black, thereby removing impurities, and extracting a chromosome map containing only chromosome parts and a single-color background (black background), as shown in FIG. 4 . This step can be used to subsequently form the scaled map needed to train the model.
在得到标记图后,需要构建弯曲染色体的弯曲骨架图,具体包括如下步骤:After obtaining the labeled map, it is necessary to construct a curved skeleton map of curved chromosomes, which specifically includes the following steps:
S21,记录S12得到的标记图的每一行的首个1值像素和末位1值像素 (即从一张图像的第一行开始至最后一行,分别记录每一行的第一个1值像素和最后一个1值像素)。S21, record the first 1-value pixel and the last 1-value pixel of each row of the marked image obtained in S12 (that is, from the first row to the last row of an image, record the first 1-value pixel and the last 1-valued pixel).
S22,求出每一行的首个1值像素和末位1值像素的中间点的坐标(X,Y),即得到多个中间点。S22, obtain the coordinates (X, Y) of the middle point of the first 1-value pixel and the last 1-value pixel of each row, that is, to obtain a plurality of middle points.
S23,将所有求出的中间点的坐标的Y值平均分成n等分,例如均分为 12等分,得到12个均分点。S23: Divide all the obtained Y values of the coordinates of the intermediate points into n equal parts, for example, into 12 equal parts, to obtain 12 equal parts.
S24,将等分后的,除第一个均分点和最后一个均分点之外的均分点定为弯曲染色体的骨架锚点,即将12个点中的第一个均分点和最后一个均分点去除,保留它们之间的10个均分点作为骨架锚点。S24, after the equalization, the equalization points except the first equalization point and the last equalization point are determined as the skeleton anchor points of the curved chromosome, that is, the first equalization point and the last equalization point among the 12 points One split point is removed, and the 10 split points between them are retained as skeleton anchor points.
S25,新建一张a×a大小的黑色(像素值为0)背景图片,在此图片上定位所述S24步骤确定的10个骨架锚点,得到染色体骨架锚点图。S25, create a black (pixel value is 0) background image of size a×a, locate the 10 skeleton anchor points determined in the step S24 on this image, and obtain a chromosome skeleton anchor point map.
所述S2还可以包括S26,用一定宽度的线段将S25中的骨架锚点依次连接起来,得到染色体弯曲骨架图,优选采用宽度为10像素的线段将骨架锚点连接,最终形成具有9条线段的染色体弯曲骨架图,如附图5所示,并且9条所述线段的像素值逐渐提高或逐渐降低,优选为从上至下依次升高,即9条线段的像素值依次为46,69,92,115,138,161,184,207,从而可以用来在下述的模型训练时来记录染色体的弯曲和方向信息。The S2 may also include S26, connecting the skeleton anchor points in S25 with a certain width of line segments in turn to obtain a chromosome bending skeleton diagram, preferably using a line segment with a width of 10 pixels to connect the skeleton anchor points, and finally form a line segment with 9 lines. The chromosome bending skeleton diagram is shown in Figure 5, and the pixel values of the 9 line segments gradually increase or decrease, preferably from top to bottom, that is, the pixel values of the 9 line segments are 46, 69 , 92, 115, 138, 161, 184, 207, which can be used to record chromosome bending and orientation information during model training as described below.
若所选染色体的弯曲幅度过大或骨架锚点定位在染色体之外,则将该染色体弯曲骨架图舍弃,并可通过人工标记的方式标记出骨架锚点,再重新构件染色体弯曲骨架图。If the bending amplitude of the selected chromosome is too large or the skeleton anchor point is located outside the chromosome, the chromosome bending skeleton map is discarded, and the skeleton anchor point can be marked manually, and then the chromosome bending skeleton map can be rebuilt.
构建染色体骨架锚点图后,可以依此构建染色体拉直骨架图,具体过程如下:After the chromosome skeleton anchor point map is constructed, the chromosome straightening skeleton map can be constructed accordingly. The specific process is as follows:
所述S3包括The S3 includes
S31,创建a×a大小的黑色背景图片;S31, create a black background image of size a×a;
S32,计算出染色体骨架锚点图中没相邻两个骨架锚点之间的距离,以上述的10个骨架锚点为例,最终得到9个距离值。S32, calculate the distance between two skeleton anchor points that are not adjacent in the chromosome skeleton anchor point map, taking the above 10 skeleton anchor points as an example, and finally obtain 9 distance values.
S33,在S31的黑色背景图片中,通过多条分别与一对相邻骨架锚点之间的距离相等且位置居中的线段依次连接构成染色体拉直骨架图,即采用9 条与9个距离值一一对应的线段依次连接构成拉直骨架图,并且染色体拉直骨架图的每条线段的宽度与所述染色体弯曲骨架图的线段的宽度相同,为10像素,并且,染色体拉直骨架图的多条线段的像素值同样可以递减或递增,优选为与染色体拉直骨架图的每条线段的像素值一一对应,如附图6所示。S33, in the black background picture of S31, a chromosome straightening skeleton diagram is formed by sequentially connecting a plurality of line segments with the same distance between a pair of adjacent skeleton anchor points and centered positions, that is, 9 lines and 9 distance values are used. One-to-one corresponding line segments are sequentially connected to form a straight skeleton diagram, and the width of each line segment of the chromosome straightened skeleton diagram is the same as the width of the line segment of the chromosome bent skeleton diagram, which is 10 pixels, and the width of each line segment of the chromosome straightened skeleton diagram is 10 pixels. The pixel values of the plurality of line segments may also decrease or increase, and preferably correspond to the pixel values of each line segment of the chromosome straightening skeleton diagram one-to-one, as shown in FIG. 6 .
得到染色体拉直骨架图后,即可将其输入已训练收敛的弯曲染色体图像拉直模型,对于新输入的拉直骨架,通过收敛模型的生成器预测并生成相应的媲美真实染色体的预测图片,即得到弯曲染色体拉直图,如附图7 所示。After obtaining the chromosome straightening skeleton map, it can be input into the trained convergent curved chromosome image straightening model. For the newly input straightening skeleton, the generator of the convergent model predicts and generates a corresponding prediction image comparable to the real chromosome. That is, the straightening diagram of the bent chromosome is obtained, as shown in FIG. 7 .
从附图7中,我们可以看到深度学习的模型的拉直效果和局部细节非常接近如附图4所述的染色体图。与现有的基于图像学的方法得到的如附图1所示的拉直图,本方法的结果不包含明显的断裂和切面,极大的提升了拉直后染色体图像的质量和有效特征的完整性和连续性,且不受弯曲个数和弯曲部位的影响。From Fig. 7, we can see that the straightening effect and local details of the deep learning model are very close to the chromosome map as described in Fig. 4. Compared with the straightened image shown in Figure 1 obtained by the existing image-based method, the result of this method does not contain obvious breaks and sections, which greatly improves the quality of the straightened chromosome image and the efficiency of effective features. Integrity and continuity, and are not affected by the number of bends and the location of bends.
本方案进一步揭示了一种生成上述弯曲染色体图像拉直模型及其生成方法,所述弯曲染色体图像拉直模型基于BagPix2Pix自学习模型,采用深度学习中的卷积神经网络,对经过弹性形变和随机旋转等增广后的染色体骨架图和对应的染色体缩放图的高纬度特征进行提取,并通过对抗训练的方法使模型学习到从骨架生成相应染色体图的模式,从而实现通过染色体竖直骨架生成相应的柔性拉直染色体的预测图。This solution further discloses a method for generating the above-mentioned curved chromosome image straightening model and its generation method. The curved chromosome image straightening model is based on the BagPix2Pix self-learning model and adopts the convolutional neural network in deep learning. The high-dimensional features of the chromosome skeleton map after rotation and the corresponding chromosome zoom map are extracted, and the model learns the pattern of generating the corresponding chromosome map from the skeleton through the method of adversarial training, so as to realize the generation of the corresponding chromosome map through the vertical skeleton of the chromosome. Prediction plot of flexible straightened chromosomes.
下面具体阐述所述弯曲染色体图像拉直模型生成方法,包括如下步骤:The method for generating the straightening model of the curved chromosome image is specifically described below, including the following steps:
S10,获取一组(例如k张)弯曲染色体原图,分别对每张弯曲染色体原图进行处理得到k张去除杂质的染色体图,其具体过程如下:S10, obtain a set of (for example, k) original images of curved chromosomes, and process each original image of bent chromosomes to obtain k images of chromosomes with impurities removed. The specific process is as follows:
S101,获取k张显微镜拍摄的弯曲染色体图像,并转化为灰度图。为了获得真实的染色体图像,需要对细胞中期的染色体进行荧光标记,通过显微镜在合适的倍数下选择染色体进行拍摄,并将图像导入计算机,计算机接收显微镜拍摄的弯曲染色体图像后转换为灰度图。如果该染色体和其他染色体发生接触或重叠现象,将其按照原本的边缘形状进行切割和提取即可。S101, acquiring k images of curved chromosomes photographed by a microscope, and converting them into grayscale images. In order to obtain a real chromosome image, it is necessary to fluorescently label the chromosomes in the metaphase of the cell, select the chromosomes under the appropriate magnification through the microscope, and import the image into the computer. The computer receives the curved chromosome image captured by the microscope and converts it into a grayscale image. If the chromosome contacts or overlaps with other chromosomes, it can be cut and extracted according to the original edge shape.
S102,为了去除边缘像素的污染,对S101得到的k张灰度图分别进行标记,将每张图中染色体标记为1值像素,将杂质和背景部分标记为0值像素,获得k张标记图(标记图可作为下述染色体弯曲骨架图生成时的染色体图),其中杂质包括但不限于细胞质,细胞核等细胞器。S102, in order to remove the pollution of the edge pixels, the k grayscale images obtained in S101 are marked respectively, the chromosomes in each image are marked as 1-value pixels, and the impurities and background parts are marked as 0-value pixels to obtain k marked images (marking The map can be used as the chromosome map when generating the following chromosome bending skeleton map), in which impurities include but are not limited to cytoplasm, nucleus and other organelles.
S103,按照S102得到的标记图来进行染色体图像的提取,即将标记图中染色体部分保持原色,将杂质和背景部分转变为黑色,从而去除杂质,得到仅包含染色体部分和单一色背景(黑色)的染色体图(只含有黑色背景和反色),得到k张染色体图。S103, extract the chromosome image according to the marker image obtained in S102, that is, keep the primary color of the chromosome part in the marker image, convert the impurity and the background part to black, thereby remove the impurity, and obtain an image containing only the chromosome part and a single-color background (black). Chromosome map (only contains black background and inverse color), get k chromosome maps.
S20,根据每张染色体图,生成k张染色体缩放图,生成每张染色体缩放图的具体过程如下:S20, according to each chromosome map, generate k chromosome zoom maps, and the specific process of generating each chromosome zoom map is as follows:
S201,计算S103得到的染色体图中染色体部分(非黑色区域)的长和宽及判断长、宽与设定值a的大小,此处,所述设定值a可以是256或512,若染色体部分的长和宽均小于设定值a,则执行S202;若染色体部分的长或宽任一大于a,则执行S203。S201, calculate the length and width of the chromosome part (non-black area) in the chromosome diagram obtained in S103 and determine the size of the length and width and the set value a, here, the set value a can be 256 or 512, if the chromosome If the length and width of the part are both smaller than the set value a, execute S202; if either the length or width of the chromosome part is greater than a, execute S203.
S202,以染色体部分为中心,将此染色体图的黑色区域扩充至a×a像素大小,得到a×a的图像。S202 , taking the chromosome part as the center, expand the black area of the chromosome map to a pixel size of a×a to obtain an a×a image.
S203,将长、宽中较大的一个缩放至a,将另一个按比例缩放,并将不足a长度的部分填充为黑色,形成a×a的图像。S203: Scale the larger one of the length and the width to a, scale the other proportionally, and fill the part less than the length of a with black to form an a×a image.
S30,根据染色体图,生成k张染色体弯曲骨架图,生成每张染色体骨架图的具体过程如下:S30, according to the chromosome map, generate k chromosomal curved skeleton maps, and the specific process of generating each chromosome skeleton map is as follows:
S301,记录染色体图的每一行中属于染色体部分的首个像素及最后一个像素;由于在上述s102中已对染色体、杂质及背景部分进行标记,可以方便地进行染色体部分的区分,因此此处可以记录S102得到的标记图的每一行的首个1值像素和末位1值像素,即从一张标记图的第一行开始至最后一行,分别记录每一行的第一个1值像素和最后一个1值像素的位置。S301, record the first pixel and the last pixel belonging to the chromosome part in each row of the chromosome map; since the chromosome, impurities and background parts have been marked in the above s102, the chromosome part can be easily distinguished, so here it can be Record the first 1-value pixel and the last 1-value pixel of each row of the marker image obtained in S102, that is, from the first row to the last row of a marker image, record the first 1-value pixel and the last 1-value pixel of each row respectively. The location of a 1-valued pixel.
S302,求出S301中记录的染色体图的每一行的属于染色体部分的首个像素及最后一个像素的中间点的坐标(X,Y),当记录的是标记图中记录的首个1值像素和末位1至像素时,即求出每一行的首个1值像素和末位1 值像素的中间点的坐标(X,Y),最终得到多个中间点。S302, obtain the coordinates (X, Y) of the middle point of the first pixel and the last pixel belonging to the chromosome part in each row of the chromosome map recorded in S301, when the record is the first 1-valued pixel recorded in the marker map When the last 1-value pixel is added to the pixel, the coordinates (X, Y) of the middle point between the first 1-value pixel and the last 1-value pixel of each row are obtained, and finally multiple middle points are obtained.
S303,将所有求出的中间点的坐标的Y值平均分成n等分,例如均分为12等分,得到12个均分点。S303: Divide all the obtained Y values of the coordinates of the intermediate point into n equal parts, for example, into 12 equal parts, to obtain 12 equal parts.
S304,将等分后的,除第一个均分点和最后一个均分点之外的均分点定为弯曲染色体的骨架锚点,即将12个点中的第一个均分点和最后一个均分点去除后的10个均分点定为骨架锚点。S304, after the equalization, the equalization points except the first equalization point and the last equalization point are determined as the skeleton anchor points of the curved chromosome, that is, the first equalization point and the last equalization point among the 12 points The 10 equalization points after one equalization point is removed are designated as skeleton anchor points.
S305,新建一张a×a大小的黑色(像素值为0)背景图片,在此图片上定位所述S304步骤确定的10个骨架锚点。S305 , create a black background image of size a×a (pixel value is 0), and locate the 10 skeleton anchor points determined in the step S304 on the image.
S306,用一定宽度的线段将S305中的骨架锚点依次连接起来,得到染色体弯曲骨架图,优选采用宽度为10像素的线段将10个骨架锚点依次连接,最终形成具有9条线段的染色体弯曲骨架图,并且9条所述线段的像素值逐渐提高或逐渐降低,优选为从上至下依次升高,更优选9条线段的像素值依次为46,69,92,115,138,161,184,207。S306, connect the skeleton anchor points in S305 with line segments of a certain width in turn to obtain a chromosome bending skeleton diagram, preferably using a line segment with a width of 10 pixels to connect 10 skeleton anchor points in turn, and finally form a chromosome bending with 9 line segments A skeleton diagram, and the pixel values of the 9 line segments gradually increase or decrease, preferably from top to bottom, more preferably, the pixel values of the 9 line segments are 46, 69, 92, 115, 138, 161, 184, 207 in sequence.
但是,若所选染色体的弯曲幅度过大或骨架锚点定位在染色体之外,则将该染色体弯曲骨架图舍弃,并可通过人工标记的方式标记出骨架锚点,然后重新构建染色体弯曲骨架图。However, if the bending amplitude of the selected chromosome is too large or the skeleton anchor point is located outside the chromosome, the chromosome bending skeleton map will be discarded, and the skeleton anchor point can be marked manually, and then the chromosome bending skeleton map can be reconstructed. .
通过S20和S30,得到K对染色体缩放图和染色体弯曲骨架图,即一个染色体缩放图和与其对应同一个弯曲染色体的染色体弯曲骨架图为一对。Through S20 and S30, K pairs of chromosome scaling diagrams and chromosome curved skeleton diagrams are obtained, that is, a chromosome scaling diagram and the chromosome curved skeleton diagram corresponding to the same curved chromosome are a pair.
S40,对K对染色体缩放图和染色体弯曲骨架图进行数据增广,一对染色体缩放图和染色体弯曲骨架图的数据增广过程如下:S40, perform data augmentation on the K-pair chromosome zoom map and the chromosome curved skeleton map. The data augmentation process for a pair of chromosome zoom maps and the chromosome curved skeleton map is as follows:
S401,采用弹性形变法和随机旋转法进行染色体缩放图和染色体弯曲骨架图数据增广;对于a=512的图像,参数选择为sigma=25,points=4;对于a=256的图像,参数选择为sigma=18,points=3;并且随机旋转的角度范围在-45°~45°之间。S401, using elastic deformation method and random rotation method to augment chromosome zoom map and chromosome bending skeleton map; for the image with a=512, the parameter selection is sigma=25, points=4; for the image with a=256, the parameter selection is sigma=18, points=3; and the random rotation angle ranges from -45° to 45°.
其中,带有宽度的染色体弯曲骨架可以携带弹性形变和随机旋转的数据增广信息,用于之后的BagPix2Pix自学习模型的训练。Among them, the chromosome bending skeleton with width can carry the data augmentation information of elastic deformation and random rotation, which is used for the training of the BagPix2Pix self-learning model.
S402,重复S401,形成m对增广后的染色体缩放图(如附图8所示)和和染色体弯曲骨架图(如附图9所示),构成一套针对该染色体的训练数据,例如m为1000,即重复S401步骤1000次,生成1000对增广后的染色体缩放图和染色体弯曲骨架图。S402, repeating S401, to form m pairs of augmented chromosome zoom maps (as shown in FIG. 8) and chromosome bending skeleton maps (as shown in FIG. 9) to form a set of training data for the chromosome, for example m is 1000, that is, repeating step S401 1000 times to generate 1000 pairs of augmented chromosome zoom maps and chromosome bending skeleton maps.
通过S40步骤,最终得到k套用于模型训练的过程数据,每套过程数据包括1000对增广后的染色体缩放图和染色体弯曲骨架图。Through step S40, k sets of process data for model training are finally obtained, and each set of process data includes 1000 pairs of augmented chromosome zoom maps and chromosome bending skeleton maps.
S50,对S40增广后的染色体缩放图和染色体弯曲骨架图进行归一化处理得到训练数据集,具体包括:S50, normalizing the chromosome zoom map and the chromosome bending skeleton map augmented by S40 to obtain a training data set, which specifically includes:
S501,对S40得到的每套增广后的染色体缩放图和染色体弯曲骨架图进行归一化处理。S501, normalize each set of augmented chromosome zoom map and chromosome curved skeleton map obtained in S40.
S502,将经过归一化处理的每套的m对数据随机分成两组,一组作为训练集,一组作为验证集,以1000对增广后的染色体缩放图和染色体弯曲骨架图数据为例,训练集采用800对数据,验证集采用200对数据。S502: Randomly divide m pairs of data of each set after normalization into two groups, one group is used as training set and the other group is used as validation set, taking 1000 pairs of augmented chromosome zoom map and chromosome bending skeleton map data as an example , the training set uses 800 pairs of data, and the validation set uses 200 pairs of data.
S60,将训练数据集输入弯曲染色体图像拉直模型进行训练至模型收敛,将训练数据集输入自学习模型后,通过对局部与全局信息的提取,对生成的染色体和真实染色体之间损失的计算,和自学习模型中生成器和判别器之间的对抗训练,使得收敛的模型可以仅通过骨架图生成媲美真实染色体的预测图片。在以642张经分别训练至收敛的模型拉直的染色体图片为数据集的分类任务中(因长度较短的染色体较不易弯曲,此次验证仅选取染色体编号1-7),以3:1的随机交叉验证的方式,在vgg16和densenet121 的模型训练下,相同染色体拉直后的验证集图片的分类表现,平均准确率分别达到了93.28%和86.41%,对比原始弯曲染色体图片的分类表现,准确率分别平均提高了1.40%和2.82%。以此可以初步说明,经过拉直后的染色体能使分类神经网络达到更好的训练结果。S60, input the training data set into the curved chromosome image straightening model for training until the model converges, and after the training data set is input into the self-learning model, through the extraction of local and global information, calculate the loss between the generated chromosome and the real chromosome , and the adversarial training between the generator and the discriminator in the self-learning model, so that the converged model can generate prediction pictures comparable to real chromosomes only through the skeleton map. In the classification task with 642 chromosome images straightened by the models trained to convergence as the dataset (because the chromosomes with shorter lengths are more difficult to bend, only chromosome numbers 1-7 are selected for this verification), with a ratio of 3:1 The method of random cross-validation, under the model training of vgg16 and densenet121, the classification performance of the validation set images after the same chromosome straightening, the average accuracy rate reached 93.28% and 86.41%, respectively. Compared with the classification performance of the original bent chromosome images, The accuracy is improved by an average of 1.40% and 2.82%, respectively. This can preliminarily show that the straightened chromosomes can make the classification neural network achieve better training results.
BagPix2Pix自学习模型(弯曲染色体图像拉直模型)训练具体过程均如下:The specific training process of the BagPix2Pix self-learning model (bending chromosome image straightening model) is as follows:
S601,使用BagPix2Pix自学习模型中的卷积核根据步长在输入的图片上逐行滑动提取图片的局部与全局特征,用以保留局部与全局特征中的细节和空间信息。S601, use the convolution kernel in the BagPix2Pix self-learning model to slide line by line on the input picture according to the step size to extract the local and global features of the picture, so as to retain the details and spatial information in the local and global features.
S602,使用包括但不限于批归一化函数对S601生成的特征映射图进行批归一化操作,以提升训练效果。S602, use a batch normalization function including but not limited to perform a batch normalization operation on the feature map generated in S601, so as to improve the training effect.
S603,使用包括但不限于LeakyReLU和ReLU激活函数对S602中提取的特征进行非线性映射来增强对特征的提取效果。S603, using activation functions including but not limited to LeakyReLU and ReLU to perform nonlinear mapping on the features extracted in S602 to enhance the feature extraction effect.
S604,使用包括但不限于Dropout函数对S603中提取的特征添加随机噪声。S604, adding random noise to the features extracted in S603 using functions including but not limited to Dropout.
S605,使用包括但不限于零填充函数的方法,将要串联起来的特征图保持同一纬度。S605, using a method including but not limited to a zero-padding function, keep the feature maps to be concatenated in the same latitude.
S606,使用上采样的方式对输入的特征映射图进行深度的加深。S606, using an up-sampling manner to deepen the depth of the input feature map.
S607,使用反卷积的方式还原图像分辨率和细节特征,并与同维度下卷积核提取的图像特征进行串联,使得图像细节特征进行最大程度的保留,使从骨架图生成的染色体图与真实图片的差距减小。S607, use deconvolution to restore the image resolution and detail features, and concatenate them with the image features extracted by the convolution kernel in the same dimension, so that the image detail features are preserved to the greatest extent, so that the chromosome map generated from the skeleton map is The gap of the real picture is reduced.
S608,模型生成器的最后一层卷积层后使用如下tanh函数,S608, the following tanh function is used after the last convolutional layer of the model generator,
通过非线性映射的方式,获得输入图经过生成器后生成的输出图。Through nonlinear mapping, the output image generated by the input image after passing through the generator is obtained.
S609,通过极小化生成器(G)和极大化判别器(D)的方法来进行对抗神经网络训练,以缩小损失函数,S609, perform adversarial neural network training by minimizing the generator (G) and maximizing the discriminator (D) to reduce the loss function,
其中,xB为输入的骨架图,yB为输入的染色体图,z为通过Dropout函数输入的随机噪声。Among them, xB is the input skeleton map, yB is the input chromosome map, and z is the random noise input through the Dropout function.
BagPix2Pix自学习模型会提取图片的高纬度的局部与全局特征,并通过对抗学习的方式训练从骨架生成至相应染色体之间的映射关系,从而具备由新输入的拉直骨架生成拉直染色体的预测图的能力。The BagPix2Pix self-learning model will extract the high-latitude local and global features of the picture, and train the mapping relationship from skeleton generation to the corresponding chromosomes through adversarial learning, so as to have the prediction of straightening chromosomes generated from the newly input straightening skeleton the ability to map.
所述BagPix2Pix自学习模型基于对抗训练的学习方式,该模型由一个生成器和一个判别器构成,且生成器由一条编码路径和一条解码路径构成。所述生成器和判别器通过S49中所述的损失函数进行对抗性训练。The BagPix2Pix self-learning model is based on the learning method of adversarial training. The model consists of a generator and a discriminator, and the generator consists of an encoding path and a decoding path. The generator and discriminator are adversarially trained with the loss function described in S49.
所述生成器的编码路径共包含17个卷积层、12个批归一化函数、16个 LeakyReLU激活函数和10个Dropout函数;解码路径包含14个反卷积层、 8个Dropout函数、14个批归一化函数,14个ReLU激活函数、5个零填充函数、1个卷积层、一个上采样层、和一个tanh激活函数。The encoding path of the generator contains a total of 17 convolutional layers, 12 batch normalization functions, 16 LeakyReLU activation functions and 10 Dropout functions; the decoding path contains 14 deconvolution layers, 8 Dropout functions, 14 A batch normalization function, 14 ReLU activation functions, 5 zero-padding functions, 1 convolutional layer, an upsampling layer, and a tanh activation function.
所述生成器的编码路径包含两条分支,分支一中不同层和函数的连接方式为:卷积层1——LeakyReLU函数——卷积层2——批归一化函数—— LeakyReLU函数——卷积层3——批归一化函数——LeakyReLU函数——卷积层4——批归一化函数——LeakyReLU函数——Dropout函数——卷积层 5——批归一化函数——LeakyReLU函数——Dropout函数——卷积层6 ——批归一化函数——LeakyReLU函数——Dropout函数——卷积层7——批归一化函数——LeakyReLU函数——Dropout函数——卷积层8——LeakyReLU函数——Dropout函数。The encoding path of the generator includes two branches, and the connection modes of different layers and functions in branch one are: convolution layer 1 - LeakyReLU function - convolution layer 2 - batch normalization function - LeakyReLU function - - Convolutional layer 3 - Batch normalization function - LeakyReLU function - Convolutional layer 4 - Batch normalization function - LeakyReLU function - Dropout function - Convolutional layer 5 - Batch normalization function - LeakyReLU function - Dropout function - Convolution layer 6 - Batch normalization function - LeakyReLU function - Dropout function - Convolution layer 7 - Batch normalization function - LeakyReLU function - Dropout function - Convolutional layer 8 - LeakyReLU function - Dropout function.
所述生成器分支一的解码路径不同层和函数的连接方式为:反卷积层 1——批归一化函数——ReLU函数——Dropout函数——反卷积层2——批归一化函数——ReLU函数——Dropout函数——反卷积层3——批归一化函数——ReLU函数——零填充函数——Dropout函数——反卷积层4——批归一化函数——ReLU函数——Dropout函数——反卷积层5——批归一化函数——ReLU函数——零填充函数——反卷积层6——批归一化函数——ReLU函数——反卷积层7——批归一化函数——ReLU函数。The connection modes of different layers and functions of the decoding path of the generator branch 1 are: deconvolution layer 1 - batch normalization function - ReLU function - Dropout function - deconvolution layer 2 - batch normalization Deconvolution function - ReLU function - Dropout function - Deconvolution layer 3 - Batch normalization function - ReLU function - Zero padding function - Dropout function - Deconvolution layer 4 - Batch normalization Function - ReLU function - Dropout function - Deconvolution layer 5 - Batch normalization function - ReLU function - Zero padding function - Deconvolution layer 6 - Batch normalization function - ReLU function - Deconvolution layer 7 - Batch normalization function - ReLU function.
所述分支一编码路径的卷积层1和卷积层8均采用3×3的卷积核,以步长为1,零填充为1的形式对输入特征进行信息提取;卷积层2、卷积层 4、和卷积层6均采用3×3卷积核,以步长为2,零填充为1的形式对输入特征进行信息提取;卷积层3、卷积层5、卷积层7均采用1×1的卷积核,以步长为1,零填充为0的形式对输入特征进行信息提取。The convolutional layer 1 and the convolutional layer 8 of the branch-one encoding path both use a 3×3 convolution kernel, and perform information extraction on the input features in the form of a stride of 1 and zero padding of 1; Convolutional layer 4 and convolutional layer 6 both use 3 × 3 convolution kernels to extract information from input features in the form of stride 2 and zero padding 1; convolutional layer 3, convolutional layer 5, convolutional layer Layer 7 uses a 1 × 1 convolution kernel to extract information from the input features with a stride of 1 and zero padding of 0.
所述分支一解码路径中反卷积层1采用3×3的反卷积核,以步长为1,零填充为1的形式对输入特征进行信息的还原;所述反卷积层3、反卷积层 5、反卷积层7均采用3×3的反卷积核,以步长为2,零填充为1的形式对输入特征进行信息的还原;所述反卷积层2、反卷积层4、反卷积层6均采用1×1的反卷积核,以步长为1,零填充为0的形式对输入特征进行信息还原;所述卷积层9采用3×3的卷积核,以步长为1,零填充为1的形式对输入特征映射图进行特征提取。In the branch-1 decoding path, the deconvolution layer 1 adopts a 3×3 deconvolution kernel, and restores the information of the input features in the form of a step size of 1 and a zero padding of 1; the deconvolution layer 3, The deconvolution layer 5 and the deconvolution layer 7 both use a 3×3 deconvolution kernel to restore the information of the input features in the form of a step size of 2 and zero padding of 1; the deconvolution layers 2, The deconvolution layer 4 and the deconvolution layer 6 both use a 1×1 deconvolution kernel to restore the input features with a stride of 1 and zero padding as 0; the convolution layer 9 uses a 3×1 deconvolution kernel. A convolution kernel of 3 to perform feature extraction on the input feature map in the form of stride 1 and zero padding 1.
所述生成器分支一编码路径中的卷积层8的输入特征图与输出特征图串联作为反卷积层1的输入;其输出的特征图经过批归一化函数、ReLU函数、 Dropout函数后与卷积层7的输入进行串联作为反卷积层2的输入;其输出的特征图经过批归一化函数函数、ReLU函数、Dropout函数后与卷积层6 的输入进行串联作为反卷积层3的输入;其输出的特征图经过批归一化函数、ReLU函数、Dropout函数后与卷积层5的输入进行串联作为反卷积层4 的输入;其输出的特征图经过批归一化函数、ReLU函数的输出与卷积层4 的输入进行串联作为反卷积层5的输入;其输出的特征图经过批归一化函数、ReLU函数后与卷积层3的输入进行串联作为反卷积层6的输入;其输出的特征图经过批归一化函数、ReLU函数后与卷积层2的输入进行串联作为反卷积层7的输入。The input feature map and the output feature map of the convolution layer 8 in the generator branch-encoding path are connected in series as the input of the deconvolution layer 1; It is concatenated with the input of convolution layer 7 as the input of deconvolution layer 2; the output feature map is concatenated with the input of convolution layer 6 after batch normalization function, ReLU function, and Dropout function as deconvolution. The input of layer 3; the output feature map is concatenated with the input of convolution layer 5 after batch normalization function, ReLU function, and Dropout function as the input of deconvolution layer 4; the output feature map is batch normalized The output of the normalization function and the ReLU function is concatenated with the input of the convolutional layer 4 as the input of the deconvolution layer 5; the output feature map is concatenated with the input of the convolutional layer 3 after the batch normalization function and the ReLU function. The input of deconvolution layer 6; the output feature map is concatenated with the input of convolution layer 2 after batch normalization function and ReLU function as the input of deconvolution layer 7.
所述生成器的编码路径分支二中不同层和函数的连接方式为:卷积层 9——LeakyReLU函数——卷积层10——批归一化函数——LeakyReLU函数——卷积层11——批归一化函数——LeakyReLU函数——卷积层12——批归一化函数——LeakyReLU函数——Dropout函数——卷积层13——批归一化函数——LeakyReLU函数——Dropout函数——卷积层14——批归一化函数——LeakyReLU函数——Dropout函数——卷积层15——批归一化函数——LeakyReLU函数——Dropout函数——卷积层16——LeakyReLU函数——Dropout函数。The connection modes of different layers and functions in the coding path branch two of the generator are: convolution layer 9 - LeakyReLU function - convolution layer 10 - batch normalization function - LeakyReLU function - convolution layer 11 - Batch Normalization Function - LeakyReLU Function - Convolutional Layer 12 - Batch Normalization Function - LeakyReLU Function - Dropout Function - Convolutional Layer 13 - Batch Normalization Function - LeakyReLU Function - - Dropout function - Convolutional layer 14 - Batch normalization function - LeakyReLU function - Dropout function - Convolution layer 15 - Batch normalization function - LeakyReLU function - Dropout function - Convolutional layer 16 - LeakyReLU function - Dropout function.
所述生成器分支二的解码路径不同层和函数的连接方式为:反卷积层 8——批归一化函数——ReLU函数——Dropout函数——反卷积层9——批归一化函数——ReLU函数——Dropout函数——反卷积层10——批归一化函数——ReLU函数——零填充函数——Dropout函数——反卷积层11——批归一化函数——ReLU函数——Dropout函数——反卷积层12——批归一化函数——ReLU函数——零填充函数——反卷积层13——批归一化函数——ReLU函数——反卷积层14——批归一化函数——ReLU函数——上采样层——与分支一解码路径得到的特征图串联——零填充函数——卷积层 17——tanh函数。The connection modes of different layers and functions of the decoding path of the generator branch 2 are: deconvolution layer 8 - batch normalization function - ReLU function - Dropout function - deconvolution layer 9 - batch normalization Normalization function - ReLU function - Dropout function - Deconvolution layer 10 - Batch normalization function - ReLU function - Zero padding function - Dropout function - Deconvolution layer 11 - Batch normalization Function - ReLU function - Dropout function - Deconvolution layer 12 - Batch normalization function - ReLU function - Zero padding function - Deconvolution layer 13 - Batch normalization function - ReLU function ——Deconvolution layer 14——Batch normalization function——ReLU function——Upsampling layer——Concatenation with feature map obtained by branch-decoding path——Zero padding function——Convolution layer 17——tanh function .
所述分支二编码路径的卷积层9至卷积层16均采用4×4的卷积核,以步长为2,零填充为1的形式对输入特征进行信息提取;The convolutional layers 9 to 16 of the branch two encoding path all use a 4×4 convolution kernel, and perform information extraction on the input features in the form of a stride of 2 and zero padding of 1;
所述分支二解码路径中反卷积层8至反卷积层14采用4×4的反卷积核,以步长为2,零填充为1的形式对输入特征进行信息的还原;所述卷积层 17采用4×4的卷积核,以步长为1,零填充为1的形式对输入特征映射图进行特征提取;所述上采样层以倍数为2对输入的特征图进行深度的加深;所述tanh函数将输入的特征图进行生成器最后的非线性映射操作。The deconvolution layer 8 to the deconvolution layer 14 in the branch two decoding path use a 4×4 deconvolution kernel to restore the information of the input features in the form of a stride of 2 and zero padding of 1; the The convolutional layer 17 uses a 4×4 convolution kernel to perform feature extraction on the input feature map in the form of a stride of 1 and zero padding of 1; the upsampling layer performs depth on the input feature map with a multiple of 2. The deepening of ; the tanh function performs the final nonlinear mapping operation of the generator on the input feature map.
所述生成器分支二编码路径中的卷积层16的输入特征图与输出特征图串联作为反卷积层8的输入;其输出的特征图经过批归一化函数、ReLU函数、Dropout函数后与卷积层15的输入进行串联作为反卷积层9的输入;其输出的特征图经过批归一化函数函数、ReLU函数、Dropout函数后与卷积层14的输入进行串联作为反卷积层10的输入;其输出的特征图经过批归一化函数、ReLU函数、Dropout函数后与卷积层13的输入进行串联作为反卷积层11的输入;其输出的特征图经过批归一化函数、ReLU函数的输出与卷积层12的输入进行串联作为反卷积层12的输入;其输出的特征图经过批归一化函数、ReLU函数后与卷积层11的输入进行串联作为反卷积层 13的输入;其输出的特征图经过批归一化函数、ReLU函数后与卷积层10 的输入进行串联作为反卷积层14的输入。The input feature map and the output feature map of the convolutional layer 16 in the second encoding path of the generator branch are connected in series as the input of the deconvolution layer 8; It is concatenated with the input of the convolution layer 15 as the input of the deconvolution layer 9; the output feature map is concatenated with the input of the convolution layer 14 as a deconvolution after the batch normalization function, ReLU function, and Dropout function. The input of layer 10; the output feature map is concatenated with the input of convolution layer 13 after batch normalization function, ReLU function and Dropout function as the input of deconvolution layer 11; the output feature map is batch normalized The output of the normalization function and the ReLU function is concatenated with the input of the convolution layer 12 as the input of the deconvolution layer 12; the output feature map is concatenated with the input of the convolution layer 11 after the batch normalization function and the ReLU function as the input. The input of the deconvolution layer 13; the output feature map is concatenated with the input of the convolution layer 10 after the batch normalization function and the ReLU function as the input of the deconvolution layer 14.
所述生成器分支一经卷积层和反卷积层后所得的特征映射图深度依次为,64,128,256,512,512,512,512,512,512,512,512,512,256, 128,64,128;分支二经卷积层和反卷积层后所得的特征映射图深度依次为64,128,256,512,512,512,512,512,512,512,512,512,256, 128,64,128;分支一与分支二的输出特征图串联后所得的特征映射图依次为256,1。The depths of the feature maps obtained by the generator branch after the convolution layer and the deconvolution layer are successively: 64, 128, 256, 512, 512, 512, 512, 512, 512, 512, 512, 512, 256, 128, 64, 128; the depths of feature maps obtained by branch two after the convolution layer and deconvolution layer are 64, 128, 256, 512, 512, 512, 512, 512, 512, 512, 512, 512, 256, 128, 64, 128; the feature maps obtained by concatenating the output feature maps of branch one and branch two are 256, 1 in turn.
所述判别器包含6个卷积层,5个批归一化函数和5个LeakyReLU激活函数;所述判别器不同层和函数的连接方式为:卷积层1——批归一化函数——LeakyReLU函数——卷积层2——批归一化函数——LeakyReLU函数——卷积层3——批归一化函数——LeakyReLU函数——卷积层4——批归一化函数——LeakyReLU函数——卷积层5——批归一化函数—— LeakyReLU函数——卷积层6。The discriminator includes 6 convolution layers, 5 batch normalization functions and 5 LeakyReLU activation functions; the connection modes of different layers and functions of the discriminator are: convolution layer 1—batch normalization function— - LeakyReLU function - convolution layer 2 - batch normalization function - LeakyReLU function - convolution layer 3 - batch normalization function - LeakyReLU function - convolution layer 4 - batch normalization function - LeakyReLU function - convolution layer 5 - batch normalization function - LeakyReLU function - convolution layer 6.
所述判别器的卷积层1和卷积层5均采用3×3的卷积核,以步长为1,零填充为1的形式对输入特征进行信息提取;卷积层2、卷积层3、和卷积层4均采用3×3卷积核,以步长为2,零填充为1的形式对输入特征进行信息提取;卷积层6采用1×1的卷积核,以步长为1,零填充为0的形式对输入特征进行信息提取;The convolutional layer 1 and the convolutional layer 5 of the discriminator both use a 3×3 convolution kernel, and perform information extraction on the input features in the form of a stride of 1 and zero padding of 1; Layer 3 and convolution layer 4 both use 3 × 3 convolution kernels to extract information from input features with a stride of 2 and zero padding of 1; convolution layer 6 uses a 1 × 1 convolution kernel to The step size is 1 and the zero padding is 0 to extract information from the input features;
所述判别器经卷积层后所得的特征映射图深度以此为,64,128,256, 512,1。The depth of the feature map obtained by the discriminator after the convolutional layer is 64, 128, 256, 512, 1.
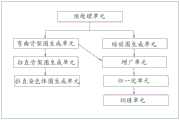
本方案还揭示了一种基于BagPix2Pix自学习模型的弯曲染色体图像拉直系统,如附图10所示,包括This scheme also discloses a curved chromosome image straightening system based on the BagPix2Pix self-learning model, as shown in Figure 10, including
预处理单元,用于接收弯曲染色体原图,处理得到标记图及染色体图;The preprocessing unit is used to receive the original image of the curved chromosome, and process it to obtain the marker map and the chromosome map;
弯曲骨架图生成单元,至少用于根据标记图,生成染色体骨架锚点图,其还可以根据染色体骨架锚点图生成染色体弯曲骨架图;The curved skeleton map generation unit is at least used to generate a chromosome skeleton anchor point map according to the marker map, and it can also generate a chromosome curved skeleton map according to the chromosome skeleton anchor point map;
拉直骨架图生成单元,用于根据染色体弯曲骨架图,生成染色体拉直骨架图;The straightening skeleton map generation unit is used to generate the chromosome straightening skeleton map according to the chromosome bending skeleton map;
拉直染色体图生成单元,用于将染色体拉直骨架图输入训练收敛的、接收骨架图可输出匹配的预测图的弯曲染色体图像拉直模型中,生成弯曲染色体拉直图像。The straight chromosome map generating unit is used for inputting the chromosome straightening skeleton map into the curved chromosome image straightening model converged by training, receiving the skeleton map and outputting a matching prediction map, and generating the curved chromosome straightening image.
如附图11所示,所述的基于BagPix2Pix自学习模型的弯曲染色体图像拉直系统还包括:As shown in FIG. 11 , the described system for straightening images of curved chromosomes based on the BagPix2Pix self-learning model further includes:
缩放图生成单元,用于根据染色体图生成染色体缩放图;The zoom map generation unit is used to generate the chromosome zoom map according to the chromosome map;
增广单元,用于进行染色体缩放图和染色体弯曲骨架图的数据增广;Augmentation unit for data augmentation of chromosome zoom map and chromosome curved skeleton map;
归一化单元,用于对增广单元得到的增广图进行预处理得到训练数据集;The normalization unit is used to preprocess the augmented graph obtained by the augmentation unit to obtain a training data set;
训练单元,用于将训练数据集输入弯曲染色体图像拉直模型进行训练至模型收敛得到弯曲染色体图像拉直模型。The training unit is used for inputting the training data set into the curved chromosome image straightening model for training until the model converges to obtain the curved chromosome image straightening model.
本方案仅以揭示了一种可读存储介质,存储有实现上述任一所述的基于BagPix2Pix自学习模型的弯曲染色体图像拉直方法的程序。This solution merely discloses a readable storage medium, which stores a program for realizing any one of the above-mentioned methods for straightening curved chromosome images based on the BagPix2Pix self-learning model.
本方案还揭示了一种图像处理装置,包括存储器、处理器及通信总线;所述通信总线用于实现处理器和存储器之间的连接通信;所述处理器用于执行存储器中存储的软件程序以实现上述任一所述的基于BagPix2Pix自学习模型的弯曲染色体图像拉直方法。The solution also discloses an image processing device, including a memory, a processor and a communication bus; the communication bus is used to implement connection and communication between the processor and the memory; the processor is used to execute a software program stored in the memory to Implement any of the above-mentioned methods for straightening curved chromosome images based on the BagPix2Pix self-learning model.
本发明尚有多种实施方式,凡采用等同变换或者等效变换而形成的所有技术方案,均落在本发明的保护范围之内。The present invention still has multiple embodiments, and all technical solutions formed by using equivalent transformations or equivalent transformations fall within the protection scope of the present invention.
Claims (10)
Priority Applications (1)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| CN202010360518.1ACN111612745A (en) | 2020-04-30 | 2020-04-30 | Image straightening method, system, storage medium and device for curved chromosomes based on BagPix2Pix self-learning model |
Applications Claiming Priority (1)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| CN202010360518.1ACN111612745A (en) | 2020-04-30 | 2020-04-30 | Image straightening method, system, storage medium and device for curved chromosomes based on BagPix2Pix self-learning model |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| CN111612745Atrue CN111612745A (en) | 2020-09-01 |
Family
ID=72199714
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| CN202010360518.1APendingCN111612745A (en) | 2020-04-30 | 2020-04-30 | Image straightening method, system, storage medium and device for curved chromosomes based on BagPix2Pix self-learning model |
Country Status (1)
| Country | Link |
|---|---|
| CN (1) | CN111612745A (en) |
Cited By (1)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN114820596A (en)* | 2022-06-23 | 2022-07-29 | 西湖大学 | Curved chromosome image straightening method based on combined model |
Citations (3)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US20140016843A1 (en)* | 2012-06-19 | 2014-01-16 | Health Discovery Corporation | Computer-assisted karyotyping |
| CN109146838A (en)* | 2018-06-20 | 2019-01-04 | 湖南自兴智慧医疗科技有限公司 | A kind of aobvious band adhering chromosome dividing method of the G merged based on geometrical characteristic with region |
| CN110533684A (en)* | 2019-08-22 | 2019-12-03 | 杭州德适生物科技有限公司 | Chromosome karyotype image cutting method |
- 2020
- 2020-04-30CNCN202010360518.1Apatent/CN111612745A/enactivePending
Patent Citations (3)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US20140016843A1 (en)* | 2012-06-19 | 2014-01-16 | Health Discovery Corporation | Computer-assisted karyotyping |
| CN109146838A (en)* | 2018-06-20 | 2019-01-04 | 湖南自兴智慧医疗科技有限公司 | A kind of aobvious band adhering chromosome dividing method of the G merged based on geometrical characteristic with region |
| CN110533684A (en)* | 2019-08-22 | 2019-12-03 | 杭州德适生物科技有限公司 | Chromosome karyotype image cutting method |
Cited By (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN114820596A (en)* | 2022-06-23 | 2022-07-29 | 西湖大学 | Curved chromosome image straightening method based on combined model |
| CN114820596B (en)* | 2022-06-23 | 2022-10-11 | 西湖大学 | Image Straightening Method for Curved Chromosomes Based on Joint Model |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| Melekhov et al. | Dgc-net: Dense geometric correspondence network | |
| US12106482B2 (en) | Learning-based active surface model for medical image segmentation | |
| CN110599528A (en) | Unsupervised three-dimensional medical image registration method and system based on neural network | |
| CN108596833A (en) | Super-resolution image reconstruction method, device, equipment and readable storage medium storing program for executing | |
| CN110334645B (en) | Moon impact pit identification method based on deep learning | |
| CN105981051A (en) | Hierarchical interlinked multi-scale convolutional network for image parsing | |
| CN115147426B (en) | Model training and image segmentation method and system based on semi-supervised learning | |
| CN114358388A (en) | A kind of pedicle screw placement operation path planning method, device and equipment | |
| CN113706562B (en) | Image segmentation method, device and system and cell segmentation method | |
| CN114663880B (en) | Three-dimensional object detection method based on multi-level cross-modal self-attention mechanism | |
| CN111553422A (en) | Automatic identification and recovery method and system for surgical instruments | |
| CN110991258A (en) | A face fusion feature extraction method and system | |
| CN114708237A (en) | Detection algorithm for hair health condition | |
| CN106203269A (en) | A kind of based on can the human face super-resolution processing method of deformation localized mass and system | |
| CN111612744A (en) | Curved chromosome image straightening model generation method, model application, system, readable storage medium and computer equipment | |
| Zarei et al. | PlantSegNet: 3D point cloud instance segmentation of nearby plant organs with identical semantics | |
| CN115035089B (en) | Brain anatomical structure localization method for two-dimensional brain image data | |
| Aonty et al. | Multi-person pose estimation using group-based convolutional neural network model | |
| CN119027357B (en) | Method, system, device and medium for recovering key information deformation of 3D document | |
| CN104268550A (en) | Feature extraction method and device | |
| CN111612745A (en) | Image straightening method, system, storage medium and device for curved chromosomes based on BagPix2Pix self-learning model | |
| CN113688842B (en) | Local image feature extraction method based on decoupling | |
| Somasundaram et al. | Straightening of highly curved human chromosome for cytogenetic analysis | |
| CN103679764A (en) | Image generation method and device | |
| CN109993695B (en) | Image fragment splicing method and system for irregular graphic annotation |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| PB01 | Publication | ||
| PB01 | Publication | ||
| SE01 | Entry into force of request for substantive examination | ||
| SE01 | Entry into force of request for substantive examination | ||
| RJ01 | Rejection of invention patent application after publication | ||
| RJ01 | Rejection of invention patent application after publication | Application publication date:20200901 |