CN111235288A - A microfluidic chip kit for rapid detection of pathogenic bacteria on wound surfaces - Google Patents
A microfluidic chip kit for rapid detection of pathogenic bacteria on wound surfacesDownload PDFInfo
- Publication number
- CN111235288A CN111235288ACN202010169329.6ACN202010169329ACN111235288ACN 111235288 ACN111235288 ACN 111235288ACN 202010169329 ACN202010169329 ACN 202010169329ACN 111235288 ACN111235288 ACN 111235288A
- Authority
- CN
- China
- Prior art keywords
- primer
- seq
- reverse
- loop
- bip
- Prior art date
- Legal status (The legal status is an assumption and is not a legal conclusion. Google has not performed a legal analysis and makes no representation as to the accuracy of the status listed.)
- Withdrawn
Links
- 238000001514detection methodMethods0.000titleclaimsabstractdescription47
- 244000052616bacterial pathogenSpecies0.000titleclaimsabstractdescription21
- 241000191967Staphylococcus aureusSpecies0.000claimsabstractdescription35
- 241000894006BacteriaSpecies0.000claimsabstractdescription22
- 241000588724Escherichia coliSpecies0.000claimsabstractdescription19
- 241000588747Klebsiella pneumoniaeSpecies0.000claimsabstractdescription19
- 241000589517Pseudomonas aeruginosaSpecies0.000claimsabstractdescription19
- 241000222126[Candida] glabrataSpecies0.000claimsabstractdescription19
- 208000032343candida glabrata infectionDiseases0.000claimsabstractdescription19
- 241000588626Acinetobacter baumanniiSpecies0.000claimsabstractdescription18
- 241000222122Candida albicansSpecies0.000claimsabstractdescription18
- RJQXTJLFIWVMTO-TYNCELHUSA-NMethicillinChemical compoundCOC1=CC=CC(OC)=C1C(=O)N[C@@H]1C(=O)N2[C@@H](C(O)=O)C(C)(C)S[C@@H]21RJQXTJLFIWVMTO-TYNCELHUSA-N0.000claimsabstractdescription18
- 229940095731candida albicansDrugs0.000claimsabstractdescription18
- 229960003085meticillinDrugs0.000claimsabstractdescription18
- 241000235395MucorSpecies0.000claimsabstractdescription17
- 208000027418Wounds and injuryDiseases0.000claimsabstractdescription14
- 206010048629Wound secretionDiseases0.000claimsabstractdescription9
- 238000006243chemical reactionMethods0.000claimsdescription27
- 239000000203mixtureSubstances0.000claimsdescription21
- 238000003199nucleic acid amplification methodMethods0.000claimsdescription20
- 238000003908quality control methodMethods0.000claimsdescription20
- 230000003321amplificationEffects0.000claimsdescription19
- 150000007523nucleic acidsChemical class0.000claimsdescription19
- 108020004707nucleic acidsProteins0.000claimsdescription17
- 102000039446nucleic acidsHuman genes0.000claimsdescription17
- 229910021642ultra pure waterInorganic materials0.000claimsdescription14
- 239000012498ultrapure waterSubstances0.000claimsdescription14
- 229920000936AgarosePolymers0.000claimsdescription13
- 238000012360testing methodMethods0.000claimsdescription7
- 239000012634fragmentSubstances0.000claimsdescription6
- 239000007788liquidSubstances0.000claimsdescription5
- 238000002156mixingMethods0.000claimsdescription5
- 240000004808Saccharomyces cerevisiaeSpecies0.000claimsdescription4
- 238000011901isothermal amplificationMethods0.000claimsdescription4
- 238000012545processingMethods0.000claimsdescription3
- 239000012295chemical reaction liquidSubstances0.000claims1
- 108090000623proteins and genesProteins0.000abstractdescription22
- 108020004414DNAProteins0.000description65
- 206010052428WoundDiseases0.000description12
- 238000000034methodMethods0.000description9
- 239000011541reaction mixtureSubstances0.000description9
- 238000007397LAMP assayMethods0.000description8
- 239000000243solutionSubstances0.000description8
- 238000005516engineering processMethods0.000description6
- 239000006228supernatantSubstances0.000description6
- 230000001580bacterial effectEffects0.000description5
- 238000006073displacement reactionMethods0.000description4
- 102000016928DNA-directed DNA polymeraseHuman genes0.000description3
- 108010014303DNA-directed DNA polymeraseProteins0.000description3
- WSFSSNUMVMOOMR-UHFFFAOYSA-NFormaldehydeChemical compoundO=CWSFSSNUMVMOOMR-UHFFFAOYSA-N0.000description3
- 238000002347injectionMethods0.000description3
- 239000007924injectionSubstances0.000description3
- 238000012986modificationMethods0.000description3
- 230000004048modificationEffects0.000description3
- 239000013642negative controlSubstances0.000description3
- 230000035945sensitivityEffects0.000description3
- 241000894007speciesSpecies0.000description3
- 239000000126substanceSubstances0.000description3
- 108091028043Nucleic acid sequenceProteins0.000description2
- 239000011324beadSubstances0.000description2
- 239000003153chemical reaction reagentSubstances0.000description2
- 230000000295complement effectEffects0.000description2
- 238000012136culture methodMethods0.000description2
- 239000003814drugSubstances0.000description2
- 239000011521glassSubstances0.000description2
- 208000015181infectious diseaseDiseases0.000description2
- 239000002184metalSubstances0.000description2
- 244000052769pathogenSpecies0.000description2
- 238000002360preparation methodMethods0.000description2
- FFRBMBIXVSCUFS-UHFFFAOYSA-N2,4-dinitro-1-naphtholChemical compoundC1=CC=C2C(O)=C([N+]([O-])=O)C=C([N+]([O-])=O)C2=C1FFRBMBIXVSCUFS-UHFFFAOYSA-N0.000description1
- 241000193830Bacillus <bacterium>Species0.000description1
- KWIUHFFTVRNATP-UHFFFAOYSA-NBetaineNatural productsC[N+](C)(C)CC([O-])=OKWIUHFFTVRNATP-UHFFFAOYSA-N0.000description1
- 241000222120Candida <Saccharomycetales>Species0.000description1
- 208000028399Critical IllnessDiseases0.000description1
- 230000004544DNA amplificationEffects0.000description1
- AHCYMLUZIRLXAA-SHYZEUOFSA-NDeoxyuridine 5'-triphosphateChemical compoundO1[C@H](COP(O)(=O)OP(O)(=O)OP(O)(O)=O)[C@@H](O)C[C@@H]1N1C(=O)NC(=O)C=C1AHCYMLUZIRLXAA-SHYZEUOFSA-N0.000description1
- 102000004190EnzymesHuman genes0.000description1
- 108090000790EnzymesProteins0.000description1
- 241000192125FirmicutesSpecies0.000description1
- 241000233866FungiSpecies0.000description1
- 201000008225Klebsiella pneumoniaDiseases0.000description1
- 241000589248LegionellaSpecies0.000description1
- 208000007764Legionnaires' DiseaseDiseases0.000description1
- KWIUHFFTVRNATP-UHFFFAOYSA-ON,N,N-trimethylglyciniumChemical compoundC[N+](C)(C)CC(O)=OKWIUHFFTVRNATP-UHFFFAOYSA-O0.000description1
- 206010035717Pneumonia klebsiellaDiseases0.000description1
- 101150071661SLC25A20 geneProteins0.000description1
- 206010040047SepsisDiseases0.000description1
- 241000191940StaphylococcusSpecies0.000description1
- 241000193998Streptococcus pneumoniaeSpecies0.000description1
- 206010053615Thermal burnDiseases0.000description1
- 206010048038Wound infectionDiseases0.000description1
- 206010058041Wound sepsisDiseases0.000description1
- 230000001154acute effectEffects0.000description1
- 239000003242anti bacterial agentSubstances0.000description1
- 229940088710antibiotic agentDrugs0.000description1
- 238000003491arrayMethods0.000description1
- 230000009286beneficial effectEffects0.000description1
- 229960003237betaineDrugs0.000description1
- 230000015572biosynthetic processEffects0.000description1
- 230000036770blood supplyEffects0.000description1
- 101150102633cact geneProteins0.000description1
- 238000004113cell cultureMethods0.000description1
- 238000005119centrifugationMethods0.000description1
- 230000001684chronic effectEffects0.000description1
- 238000010276constructionMethods0.000description1
- 238000007796conventional methodMethods0.000description1
- 238000005336crackingMethods0.000description1
- 238000012864cross contaminationMethods0.000description1
- 238000012258culturingMethods0.000description1
- 125000004122cyclic groupChemical group0.000description1
- SUYVUBYJARFZHO-RRKCRQDMSA-NdATPChemical compoundC1=NC=2C(N)=NC=NC=2N1[C@H]1C[C@H](O)[C@@H](COP(O)(=O)OP(O)(=O)OP(O)(O)=O)O1SUYVUBYJARFZHO-RRKCRQDMSA-N0.000description1
- SUYVUBYJARFZHO-UHFFFAOYSA-NdATPNatural productsC1=NC=2C(N)=NC=NC=2N1C1CC(O)C(COP(O)(=O)OP(O)(=O)OP(O)(O)=O)O1SUYVUBYJARFZHO-UHFFFAOYSA-N0.000description1
- RGWHQCVHVJXOKC-SHYZEUOFSA-JdCTP(4-)Chemical compoundO=C1N=C(N)C=CN1[C@@H]1O[C@H](COP([O-])(=O)OP([O-])(=O)OP([O-])([O-])=O)[C@@H](O)C1RGWHQCVHVJXOKC-SHYZEUOFSA-J0.000description1
- HAAZLUGHYHWQIW-KVQBGUIXSA-NdGTPChemical compoundC1=NC=2C(=O)NC(N)=NC=2N1[C@H]1C[C@H](O)[C@@H](COP(O)(=O)OP(O)(=O)OP(O)(O)=O)O1HAAZLUGHYHWQIW-KVQBGUIXSA-N0.000description1
- NHVNXKFIZYSCEB-XLPZGREQSA-NdTTPChemical compoundO=C1NC(=O)C(C)=CN1[C@@H]1O[C@H](COP(O)(=O)OP(O)(=O)OP(O)(O)=O)[C@@H](O)C1NHVNXKFIZYSCEB-XLPZGREQSA-N0.000description1
- 230000006378damageEffects0.000description1
- 230000034994deathEffects0.000description1
- 230000007547defectEffects0.000description1
- 230000007850degenerationEffects0.000description1
- 230000001934delayEffects0.000description1
- 238000004925denaturationMethods0.000description1
- 230000036425denaturationEffects0.000description1
- 238000010586diagramMethods0.000description1
- 229940079593drugDrugs0.000description1
- 230000000694effectsEffects0.000description1
- 230000007613environmental effectEffects0.000description1
- 238000002474experimental methodMethods0.000description1
- 238000000605extractionMethods0.000description1
- 238000001917fluorescence detectionMethods0.000description1
- 230000009545invasionEffects0.000description1
- 238000002955isolationMethods0.000description1
- 238000002032lab-on-a-chipMethods0.000description1
- 230000017074necrotic cell deathEffects0.000description1
- 244000039328opportunistic pathogenSpecies0.000description1
- 239000013641positive controlSubstances0.000description1
- 230000008569processEffects0.000description1
- 239000000047productSubstances0.000description1
- 238000004393prognosisMethods0.000description1
- 102000004169proteins and genesHuman genes0.000description1
- 238000003753real-time PCRMethods0.000description1
- 230000010076replicationEffects0.000description1
- 238000005070samplingMethods0.000description1
- 238000000926separation methodMethods0.000description1
- 230000008591skin barrier functionEffects0.000description1
- 229940031000streptococcus pneumoniaeDrugs0.000description1
- 238000006467substitution reactionMethods0.000description1
- 230000004083survival effectEffects0.000description1
- 238000003786synthesis reactionMethods0.000description1
- XLYOFNOQVPJJNP-UHFFFAOYSA-NwaterSubstancesOXLYOFNOQVPJJNP-UHFFFAOYSA-N0.000description1
Images
Classifications
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q1/00—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions
- C12Q1/68—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions involving nucleic acids
- C12Q1/6876—Nucleic acid products used in the analysis of nucleic acids, e.g. primers or probes
- C12Q1/6888—Nucleic acid products used in the analysis of nucleic acids, e.g. primers or probes for detection or identification of organisms
- C12Q1/689—Nucleic acid products used in the analysis of nucleic acids, e.g. primers or probes for detection or identification of organisms for bacteria
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q1/00—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions
- C12Q1/68—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions involving nucleic acids
- C12Q1/6844—Nucleic acid amplification reactions
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q1/00—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions
- C12Q1/68—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions involving nucleic acids
- C12Q1/6876—Nucleic acid products used in the analysis of nucleic acids, e.g. primers or probes
- C12Q1/6888—Nucleic acid products used in the analysis of nucleic acids, e.g. primers or probes for detection or identification of organisms
- C12Q1/6895—Nucleic acid products used in the analysis of nucleic acids, e.g. primers or probes for detection or identification of organisms for plants, fungi or algae
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q2600/00—Oligonucleotides characterized by their use
- C12Q2600/166—Oligonucleotides used as internal standards, controls or normalisation probes
Landscapes
- Chemical & Material Sciences (AREA)
- Life Sciences & Earth Sciences (AREA)
- Organic Chemistry (AREA)
- Proteomics, Peptides & Aminoacids (AREA)
- Engineering & Computer Science (AREA)
- Analytical Chemistry (AREA)
- Health & Medical Sciences (AREA)
- Zoology (AREA)
- Wood Science & Technology (AREA)
- Biotechnology (AREA)
- Microbiology (AREA)
- Biochemistry (AREA)
- Biophysics (AREA)
- Molecular Biology (AREA)
- Physics & Mathematics (AREA)
- Genetics & Genomics (AREA)
- General Health & Medical Sciences (AREA)
- Immunology (AREA)
- Bioinformatics & Cheminformatics (AREA)
- General Engineering & Computer Science (AREA)
- Botany (AREA)
- Mycology (AREA)
- Chemical Kinetics & Catalysis (AREA)
- Measuring Or Testing Involving Enzymes Or Micro-Organisms (AREA)
Abstract
Description
Translated fromChinese技术领域technical field
本发明涉及医药生物技术领域,特别涉及基因检测技术领域;具体涉及一种能够快速检测创面常见致病菌的微流控芯片快速检测方法。The invention relates to the technical field of medicine and biology, in particular to the technical field of gene detection, and in particular to a microfluidic chip rapid detection method capable of rapidly detecting common pathogenic bacteria on wound surfaces.
背景技术Background technique
急慢性创面因皮肤屏障的破坏加上局部血运障碍、变性坏死覆盖、蛋白渗出等给予细菌提供了良好的生存环境,有利于机会致病菌的侵入和繁殖。临床上若不能早期处理创面上定植的细菌,在良好的“生存”环境下,致病菌大量繁殖最终可进展为创面脓毒症。尤其在大面积烧伤患者中,创面感染引发的脓毒症是导致其死亡的最主要原因之一。因此如果能早期快速识别创面中致病菌种类,针对不同种类的致病菌做出及时有效的处理,会对患者的预后有重要帮助。Acute and chronic wounds provide a good living environment for bacteria due to the destruction of the skin barrier, local blood supply obstruction, degeneration and necrosis coverage, and protein exudation, which is conducive to the invasion and reproduction of opportunistic pathogens. In clinical practice, if the bacteria colonized on the wound surface cannot be treated early, in a good "survival" environment, the large number of pathogenic bacteria can eventually progress to wound sepsis. Especially in patients with extensive burns, sepsis caused by wound infection is one of the leading causes of death. Therefore, if the types of pathogenic bacteria in the wound can be quickly identified early, and timely and effective treatment of different types of pathogenic bacteria can be made, it will be of great help to the prognosis of patients.
目前临床上对创面病原学的检测鉴定的方法主要是细菌培养法。主要步骤包括创面分泌物的取样、细菌的分离和培养、细菌的鉴定、以及细菌的药敏试验。该方法目前存在以下缺陷:1.此方法对检验科实验操作环境及操作员的操作技术有较高的要求,实验室工作人员在分离培养病原菌时存在着高风险被感染的可能,在乡镇医院、社区医疗单位尚不能大规模开展。2.对于批量鉴定时,操作过程中存在交叉污染,所得到的最终结果不一定能反应出创面的真实感染情况。3.难以检出需要特殊培养条件的细菌,如军团菌、L-型细菌、肺炎链球菌等。4.整个培养法检测时间较长,至少用时2-3天,对于临床急危重患者的救治,不能及时快速的回报结果大大延误了救治。综上所述,临床上需要一种快速实验环境要求低、准确,操作简单的创面分泌物致病菌检测试剂盒。At present, the clinical detection and identification of the etiology of wounds is mainly the bacterial culture method. The main steps include the sampling of wound secretions, the isolation and culture of bacteria, the identification of bacteria, and the drug susceptibility test of bacteria. This method currently has the following defects: 1. This method has high requirements on the laboratory experimental operating environment and the operator's operating skills, and laboratory staff are at high risk of infection when isolating and culturing pathogenic bacteria. , Community medical units can not be carried out on a large scale. 2. For batch identification, there is cross-contamination during the operation, and the final result obtained may not reflect the real infection of the wound. 3. It is difficult to detect bacteria that require special culture conditions, such as Legionella, L-type bacteria, Streptococcus pneumoniae, etc. 4. The detection time of the whole culture method is long, at least 2-3 days. For the treatment of clinically critically ill patients, the failure to report the results in a timely and rapid manner greatly delays the treatment. To sum up, there is a clinical need for a rapid test kit for detection of pathogenic bacteria in wound secretions with low requirements, accuracy, and simple operation.
对于细菌基因学的检测,常规的q-PCR具有操作的繁琐性,对实验操作人员的技术有较高的要求,在大的临床检验中心可以运用,尚不能在基层医疗机构大量推广。恒温核酸扩增分析仪是一种集快速核酸扩增、实时荧光检测、智能数据处理和自动结果判读于一体的PCR仪,能够对特定基因靶标进行快速检测。恒温核酸扩增分析仪具有仪器小巧,质量轻便,操作简便,检测快速等优点。环介导等温扩增技术(Loop-mediated isothermalamplification,LAMP)是利用一种具有链置换活性和瀑布式核酸扩增功能的Bst DNA聚合酶,在等温条件下进行核酸的变性和自动循环的链置换核酸扩增反应。LAMP针对靶序列的6个区域设计4条特异引物,利用具备链置换功能的DNA聚合酶在恒定温度下不断复制扩增DNA。为了提高反应效率,可在反应体系中添加两条环引物,使之分别与茎环结构结合,启动链置换合成,循环复制。LAMP具有简便、快速、灵敏、特异的优势。For the detection of bacterial genetics, conventional q-PCR is cumbersome to operate, and has high requirements for the skills of experimental operators. It can be used in large clinical testing centers, but cannot be widely promoted in primary medical institutions. The constant temperature nucleic acid amplification analyzer is a PCR instrument that integrates rapid nucleic acid amplification, real-time fluorescence detection, intelligent data processing and automatic result interpretation, which can rapidly detect specific gene targets. The constant temperature nucleic acid amplification analyzer has the advantages of compact instrument, light weight, easy operation and rapid detection. Loop-mediated isothermal amplification (LAMP) uses a Bst DNA polymerase with strand displacement activity and waterfall nucleic acid amplification function to perform nucleic acid denaturation and automatic cycle strand displacement under isothermal conditions. Nucleic acid amplification reaction. LAMP designs 4 specific primers for 6 regions of the target sequence, and uses a DNA polymerase with strand displacement function to continuously replicate and amplify DNA at a constant temperature. In order to improve the reaction efficiency, two loop primers can be added to the reaction system to bind them to the stem-loop structure respectively to initiate strand displacement synthesis and cyclic replication. LAMP has the advantages of simplicity, rapidity, sensitivity and specificity.
微流控芯片技术(Microfluidic chip)又称芯片实验室(Lab-on-a-chip),它将化学中所涉及的样品预处理、反应、分离、检测,生命科学中的细胞培养、分选、裂解等基本操作单元集成到一块几平方厘米大小的芯片上,并以微通道网络贯穿各个实验环节,从而实现对整个实验系统的灵活操控,承载传统化学或生物实验室的各项功能。微流控芯片采用微通道将多种功能单元集成一个整体,在减少样本用量和试剂消耗量的基础上能够自动化检测样品,具有经济、环保等优点。Microfluidic chip technology (Microfluidic chip), also known as Lab-on-a-chip, integrates sample pretreatment, reaction, separation and detection involved in chemistry, cell culture and sorting in life sciences Basic operation units such as , cracking, etc. are integrated into a chip with a size of several square centimeters, and run through each experimental link with a micro-channel network, so as to realize flexible control of the entire experimental system and carry various functions of traditional chemical or biological laboratories. The microfluidic chip uses microchannels to integrate various functional units into a whole, which can automatically detect samples on the basis of reducing sample consumption and reagent consumption, and has the advantages of economy and environmental protection.
本发明所开发的创面病原菌检测的试剂盒,采用环介导等温扩增法,用微流控芯片在恒温核酸扩增分析仪上进行。使用该方法对创面病原菌中特定的核酸进行检测,其具有特异性好,灵敏度高,操作简单,检测迅速等优点,能对特定病原菌进行检测,为临床抗生素的使用提供指导性意见。The kit for detection of pathogenic bacteria on wound surface developed by the present invention adopts a loop-mediated isothermal amplification method, and uses a microfluidic chip to carry out on a constant temperature nucleic acid amplification analyzer. Using this method to detect specific nucleic acid in pathogenic bacteria on wound surface has the advantages of good specificity, high sensitivity, simple operation, rapid detection, etc. It can detect specific pathogenic bacteria and provide guidance for the use of clinical antibiotics.
发明内容SUMMARY OF THE INVENTION
鉴于现有技术存在的问题,本发明的解决的技术问题是通过微流控芯片和环介导等温扩增技术快速检测创面分泌物中的致病菌,包括革兰阳性菌2种(耐甲氧西林金黄色葡萄球菌、金黄色葡萄球菌),革兰阴性菌(大肠埃希菌、肺炎克雷伯菌、铜绿假单胞菌、鲍曼不动杆菌)4种,真菌(白色念珠菌、光滑念珠菌、毛霉菌)3种,共计9种常见致病菌的快速检出。In view of the problems existing in the prior art, the technical problem solved by the present invention is to rapidly detect pathogenic bacteria in wound secretions by microfluidic chip and loop-mediated isothermal amplification technology, including 2 kinds of Gram-positive bacteria (formaldehyde-resistant bacteria). Oxycillin Staphylococcus aureus, Staphylococcus aureus), Gram-negative bacteria (Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii) 4 species, fungi (Candida albicans, Candida glabrata, Mucor) 3 kinds, a total of 9 kinds of common pathogens can be quickly detected.
具体的,本发明通过以下技术方案实现:Specifically, the present invention is realized through the following technical solutions:
快速检测创面病原菌的微流控芯片试剂盒包括对如下至少两种菌的快速检出组分,所述至少两种菌选自:耐甲氧西林金黄色葡萄球菌、金黄色葡萄球菌、大肠埃希菌、肺炎克雷伯菌、铜绿假单胞菌、鲍曼不动杆菌、白色念珠菌、光滑念珠菌、毛霉菌9种常见致病菌,快速检出组分为上述致病菌的特异性引物组;优选地,所述试剂盒包含至少3种菌的快速检出组分,更优选地,所述试剂盒9种菌的快速检出组分。The microfluidic chip kit for rapid detection of pathogenic bacteria on wound surfaces includes rapid detection components for at least two bacteria selected from the group consisting of: Methicillin-resistant Staphylococcus aureus, Staphylococcus aureus, Escherichia coli 9 kinds of common pathogenic bacteria, including Herba, Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, Candida albicans, Candida glabrata, and Mucor Preferably, the kit includes rapid detection components of at least 3 species of bacteria, more preferably, the rapid detection components of 9 species of bacteria in the kit.
所述试剂盒还包括核酸扩增反应液、阴性及阳性质控模板。The kit also includes nucleic acid amplification reaction solution, negative and positive quality control templates.
所述引物组包括耐甲氧西林金黄色葡萄球菌、金黄色葡萄球菌、大肠埃希菌、肺炎克雷伯菌、铜绿假单胞菌、鲍曼不动杆菌、白色念珠菌、光滑念珠菌、毛霉菌9种病原菌检测引物组,可用于扩增靶基因特异碱基序列,所述靶基因分别为:耐甲氧西林金黄色葡萄球菌、金黄色葡萄球菌、大肠埃希菌、肺炎克雷伯菌、铜绿假单胞菌、鲍曼不动杆菌、白色念珠菌、光滑念珠菌、毛霉菌,为所述引物与所述靶基因核酸序列的一部分或其互补连互补。所述引物组由耐甲氧西林金黄色葡萄球菌、金黄色葡萄球菌、大肠埃希菌、肺炎克雷伯菌、铜绿假单胞菌、鲍曼不动杆菌、白色念珠菌、光滑念珠菌、毛霉菌9种病原菌的各6条引物组成。The primer set includes Methicillin-resistant Staphylococcus aureus, Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, Candida albicans, Candida glabrata, The primer set for detection of 9 pathogenic bacteria of Mucor, can be used to amplify the specific base sequences of target genes. The target genes are: Methicillin-resistant Staphylococcus aureus, Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae bacteria, Pseudomonas aeruginosa, Acinetobacter baumannii, Candida albicans, Candida glabrata, Mucor, the primer is complementary to a part of the nucleic acid sequence of the target gene or its complementary link. The primer set is composed of methicillin-resistant Staphylococcus aureus, Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, Candida albicans, Candida glabrata, It consists of 6 primers for each of 9 pathogenic bacteria of Mucor.
所述引物组包含:The primer set includes:
一种耐甲氧西林金黄色葡萄球菌检测用引物组,由下列引物组成:A primer set for detection of methicillin-resistant Staphylococcus aureus consists of the following primers:
正向外引物F3-TTATGTATCAGGTACTTAT(Seq ID No.1)Forward primer F3-TTATGTATCAGGTACTTAT (Seq ID No.1)
反向外引物B3-TTTTGTTATTTAACCCAATCATTTA(Seq ID No.2)Reverse outer primer B3-TTTTGTTATTTAACCCAATCATTTA (Seq ID No.2)
正向内引物FIPForward Inward Primer FIP
ATTCTTCGTTACTCATTACATACATGTGAATTATTATAACTTGTATAAC(Seq ID No.3)ATTCTTCGTTACTCATTACATACATGTGAATTATTATAACTTGTATAAC(Seq ID No.3)
反向内引物BIPreverse inner primer BIP
AACCGAAGATAAAAAAGAACCTCTGAATATTTTTTGAGTTGAACCTGGTG(Seq ID No.4)AACCGAAGATAAAAAAGAACCTCTGAATTTTTTTGAGTTGAACCTGGTG(Seq ID No.4)
正向环引物LF-AATGGATAGACGTCATATGAAGGT(Seq ID No.5)Forward loop primer LF-AATGGATAGACGTCATATGAAGGT (Seq ID No.5)
反向环引物LB-CTCAACAAGTTCCAGATTACAACTT(Seq ID No.6)Reverse loop primer LB-CTCAACAAGTTCCAGATTACAACTT (Seq ID No.6)
用于扩增耐甲氧西林金黄色葡萄球菌特异性序列;Used to amplify methicillin-resistant Staphylococcus aureus-specific sequences;
一种金黄色葡萄球菌检测用引物组,由下列引物组成:A primer set for detection of Staphylococcus aureus, consisting of the following primers:
正向外引物F3-GTTACTTTACAGATATAATG(Seq ID No.7)Forward primer F3-GTTACTTTACAGATATAATG (Seq ID No.7)
反向外引物B3-GAAAAAGTGTACGAGTTCTTGA(Seq ID No.8)Reverse outer primer B3-GAAAAAGTGTACGAGTTCTTGA (Seq ID No.8)
正向内引物FIPForward Inward Primer FIP
GTTTCATAACCTTCATAAATATTTCCATACAGTCATTTCACTAA(Seq ID No.9)GTTTCATAACCTTCATAAATATTTCCATACAGTCATTTCACTAA(Seq ID No.9)
反向内引物BIPreverse inner primer BIP
GAGGTCATTTAATATTTATTACTTCGATCACTGGACCTAG(Seq ID No.10)GAGGTCATTTAATATTTATTACTTCGATCACTGGACCTAG(Seq ID No.10)
正向环引物LF-AACTCATAGTGTACAACA(Seq ID No.11)Forward loop primer LF-AACTCATAGTGTACAACA (Seq ID No.11)
反向环引物LB-GTACCTGTTATGAAAGTGTTCA(Seq ID No.12)Reverse loop primer LB-GTACCTGTTATGAAAGTGTTCA (Seq ID No.12)
用于扩增金黄色葡萄球菌特异性序列;Used to amplify Staphylococcus aureus-specific sequences;
一种大肠埃希菌检测用引物组,由下列引物组成:A primer set for detection of Escherichia coli consists of the following primers:
正向外引物F3-GTAATCGTGGTGATTGATGA(Seq ID No.13)Forward primer F3-GTAATCGTGGTGATTGATGA (Seq ID No.13)
反向外引物B3-GGTTCGTTGTAAATACTCCA(Seq ID No.14)Reverse outer primer B3-GGTTCGTTGTAAATACTCCA (Seq ID No.14)
正向内引物FIP-TCTTTCGTATTGTTTACCTACTTATGTCGTATTTAACCTC(Seq ID No.15)Forward Inner Primer FIP-TCTTTCGTATTGTTTACCTACTTATGTCGTATTTAACCTC(Seq ID No.15)
反向内引物BIPreverse inner primer BIP
TACATAGAAGAGTAAGTCAACGTTTTGGTTTTTGTCACTATATATC(Seq ID No.16)TACATAGAAGAGTAAGTCAACGTTTTGGTTTTTGTCACTATATATC(Seq ID No.16)
正向环引物LF-TTCGAAACCAATTACTAAAGA(Seq ID No.17)Forward loop primer LF-TTCGAAAACCAATTACTAAAGA (Seq ID No.17)
反向环引物LB-TATAACTTACAGTAGATT(Seq ID No.18)Reverse loop primer LB-TATAACTTACAGTAGATT (Seq ID No.18)
用于扩增大肠埃希菌特异性序列;Used to amplify Escherichia coli-specific sequences;
一种肺炎克雷伯菌检测用引物组,由下列引物组成:A primer set for Klebsiella pneumoniae detection, consisting of the following primers:
正向外引物F3-GTCTCTACAATAGGAAT(Seq ID No.19)Forward primer F3-GTCTCTACAATAGGAAT (Seq ID No.19)
反向外引物B3-ATGAGACCTACTGGGACA(Seq ID No.20)Reverse outer primer B3-ATGAGACCTACTGGGACA (Seq ID No.20)
正向内引物FIP-AACCTAAATGTTAAAATAAGTCAGGGAGACGACCGT(Seq ID No.21)Forward Inward Primer FIP-AACCTAAATGTTAAAATAAGTCAGGGAGACGACCGT (Seq ID No.21)
反向内引物BIP-AGTTAATAAAGTAGGAGTACTTTATTTTGTTCTAGTATGGAATCGG(Seq IDNo.22)Reverse inner primer BIP-AGTTAATAAAGTAGGAGTACTTTATTTTGTTCTAGTATGGAATCGG(Seq IDNo.22)
正向环引物LF-CGATGACATAATAAATAACCG(Seq ID No.23)Forward loop primer LF-CGATGACATAATAAATAACCG (Seq ID No.23)
反向环引物LB-TATATATACCACTACCA(Seq ID No.24)Reverse loop primer LB-TATATATACCACTACCA (Seq ID No.24)
用于扩增肺炎克雷伯菌特异性序列;Used to amplify Klebsiella pneumonia-specific sequences;
一种铜绿假单胞菌检测用引物组,由下列引物组成:A primer set for detection of Pseudomonas aeruginosa, consisting of the following primers:
正向外引物F3-TGTTATGGAAATGTCCACCTT(Seq ID No.25)Forward primer F3-TGTTATGGAAATGTCCACCTT (Seq ID No.25)
反向外引物B3-TCTTATGATATTTCTGAG(Seq ID No.26)Reverse outer primer B3-TCTTATGATATTTCTGAG (Seq ID No.26)
正向内引物FIP-GTACAGAACATAATACAGAGGAACACGATGAACAACGT(Seq ID No.27)Forward Inward Primer FIP-GTACAGACATAATACAGAGGAACACGATGAACAACGT (Seq ID No.27)
反向内引物BIP-ATATCGTCTGACCTATACCCTTCGTCATACTTTAGAT(Seq ID No.28)Reverse inner primer BIP-ATATCGTCTGACCTATACCCTTCGTCATACTTTAGAT (Seq ID No.28)
正向环引物LF-CCAGATAAGAGAATTTCAGA(Seq ID No.29)Forward loop primer LF-CCAGATAAGAGAATTTCAGA (Seq ID No.29)
反向环引物LB-ATATTATCGTTATCAGG(Seq ID No.30)Reverse loop primer LB-ATATTATCGTTATCAGG (Seq ID No.30)
用于扩增铜绿假单胞菌特异性序列;Used to amplify Pseudomonas aeruginosa specific sequences;
一种鲍曼不动杆菌检测用引物组,由下列引物组成:A primer set for detection of Acinetobacter baumannii consists of the following primers:
正向外引物F3-TGTTACAATTAACTTCTTATA(Seq ID No.31)Forward primer F3-TGTACAATTAACTTCTTATA (Seq ID No.31)
反向外引物B3-CTTGTAATATCGTTATAGTAGTT(Seq ID No.32)Reverse outer primer B3-CTTGTAATATCGTTATAGTAGTT (Seq ID No.32)
正向内引物FIP-Forward Inward Primer FIP-
AGTGAATTCGGTTATCGTAATATCGAATAATAAAAAAAAATTAGTCAC(Seq ID No.33)AGTGAATTCGGTTATCGTAATATCGAATAATAAAAAAAAAATTAGTCAC(Seq ID No.33)
反向内引物BIPreverse inner primer BIP
AATGATATGTATCAAAATATAGAGTATTAACTCATGTTAGATGG(Seq ID No.34)AATGATATGTATCAAAATATAGAGTATTAACTCATGTTAGATGG(Seq ID No.34)
正向环引物LF-TATAAGAGAGGTCTAC(Seq ID No.35)Forward loop primer LF-TATAAGAGAGGTCTAC (Seq ID No.35)
反向环引物LB-CCGTTAAAAATTATAAGG(Seq ID No.36)Reverse loop primer LB-CCGTTAAAAATTATAAGG (Seq ID No.36)
用于扩增鲍曼不动杆菌特异性序列;Used to amplify Acinetobacter baumannii specific sequences;
一种白色念珠菌检测用引物组,由下列引物组成:A primer set for detecting Candida albicans, consisting of the following primers:
正向外引物F3-TCAACTTGTCACACCAGAT(Seq ID No.37)Forward primer F3-TCAACTTGTCACACCAGAT (Seq ID No.37)
反向外引物B3-CTCAACACCAAACCCAGC(Seq ID No.38)Reverse outer primer B3-CTCAACACCAAACCCAGC (Seq ID No.38)
正向内引物FIP-TCGCTGCGTTCTTCATCGATAATAGTCAAAACTTTCAACAACG(Seq IDNo.39)Forward inner primer FIP-TCGCTGCGTTCTTCATCGATAATAGTCAAAACTTTCAACAACG(Seq IDNo.39)
反向内引物BIP-TGCAGATATTCGTGAATCATCGAATAAACGACGCTCAAACAGG(Seq IDNo.40)Reverse inner primer BIP-TGCAGATATTCGTGAATCATCGAATAAACGACGCTCAAACAGG (Seq IDNo.40)
正向环引物LF-GCGAGAACCAAGAGATC(Seq ID No.41)Forward loop primer LF-GCGAGAACCAAGAGATC (Seq ID No.41)
反向环引物LB-CTTTGAACGCACATTGC(Seq ID No.42)Reverse loop primer LB-CTTTGAACGCACATTGC (Seq ID No.42)
可以扩增白色念珠菌特异性序列;Can amplify Candida albicans specific sequences;
一种光滑念珠菌检测用引物组,由下列引物组成:A primer set for detecting Candida glabrata, consisting of the following primers:
正向外引物F3-CCATTAAAAAGTGCAATGGTG(Seq ID No.43)Forward primer F3-CCATTAAAAAGTGCAATGGTG (Seq ID No.43)
反向外引物B3-CTGTCTTTAGAATCCAACTGC(Seq ID No.44)Reverse outer primer B3-CTGTCTTTAGAATCCAACTGC (Seq ID No.44)
正向内引物FIP-TTCTTAGCACTGTTGTCAGAAACACAATGCCTTCCTGTACCA(Seq IDNo.45)Forward inner primer FIP-TTCTTAGCACTGTTGTCAGAAACACAATGCCTTCCTGTACCA(Seq IDNo.45)
反向内引物BIP-GTCGGCGAGGGTAAATTCAACGTTTGATTCCTTAAGCCCGATA(Seq IDNo.46)Reverse inner primer BIP-GTCGGCGAGGGTAAATTCAACGTTTGATTCCTTAAGCCCGATA (Seq IDNo.46)
正向环引物LF-CGGTGGTGGTATCTGAGACAT(Seq ID No.47)Forward loop primer LF-CGGTGGTGGTATCTGAGACAT (Seq ID No.47)
反向环引物LB-TGATACACTAACTGCATCTCCCT(Seq ID No.48)Reverse loop primer LB-TGATACACTAACTGCATCTCCCT (Seq ID No.48)
可以扩增光滑念珠菌特异性序列;Can amplify Candida glabrata-specific sequences;
一种毛霉菌检测用引物组,由下列引物组成:A primer set for detection of Mucor fungi, consisting of the following primers:
正向外引物F3-CTTCATCTTATTTGGTCTTTTGC(Seq ID No.49)Forward primer F3-CTTCATCTTATTTGGTCTTTTGC (Seq ID No.49)
反向外引物B3-TTTATGCACGTGGTTGGT(Seq ID No.50)Reverse outer primer B3-TTTATGCACGTGGTTGGT (Seq ID No.50)
正向内引物FIP-CAGTTTCTTCGTATGCATTGTCACTACGAACACAAGATGTCACT(Seq IDNo.51)Forward inner primer FIP-CAGTTTCTCGTATGCATTGTCACTACGAACACAAGATGTCACT (Seq IDNo.51)
反向内引物BIP-CAGATCCAAGGATCCTATGGTCTTGGCCCTTGTTGGTATTTTGAG(Seq IDNo.52)Reverse inner primer BIP-CAGATCCAAGGATCCTATGGTCTTGGCCCTTGTTGGTATTTTGAG(Seq IDNo.52)
正向环引物LF-GTTGCACTTGCAAGTTGATG(Seq ID No.53)Forward loop primer LF-GTTGCACTTGCAAGTTGATG (Seq ID No.53)
反向环引物LB-CACATGGGCTTTAGAAGATACA(Seq ID No.54)Reverse loop primer LB-CACATGGGCTTTAGAAGATACA (Seq ID No.54)
可以扩增毛霉菌特异性序列。Mucor specific sequences can be amplified.
一种阳性质控用引物组,由下列引物组成:A primer set for positive quality control, consisting of the following primers:
正向外引物F3-TTACTGGTCGCCCATCTC(Seq ID No.55)Forward primer F3-TTACTGGTCGCCCATCTC (Seq ID No.55)
反向外引物B3-CTATAGGCACTTCTCCTGTAG(Seq ID No.56)Reverse outer primer B3-CTATAGGCACTTCTCCTGTAG (Seq ID No.56)
正向内引物FIP-ACAACCCGTCCCCATAGTAATAACACTCATGCCTTCCACA(Seq ID No.57)Forward inner primer FIP-ACAACCCGTCCCCATAGTAATAACACTCATGCCTTCCACA (Seq ID No.57)
反向内引物BIP-CGATGCTATACTTCGTATAGGAAGCCATTCTTGTGCTATGGAACA(Seq IDNo.58)Reverse inner primer BIP-CGATGCTATACTTCGTATAGGAAGCCATTCTTGTGCTATGGAACA (Seq IDNo.58)
正向环引物LF-GACTTAGTCGCTAAGTGGAA(Seq ID No.59)Forward loop primer LF-GACTTAGTCGCTAAGTGGAA (Seq ID No.59)
反向环引物LB-CGTTTTTTTATGCCCCATCC(Seq ID No.60)Reverse loop primer LB-CGTTTTTTATGCCCCATCC (Seq ID No.60)
用于扩增阳性质控特异性序列。Used to amplify positive control-specific sequences.
本发明所述每组引物都经过多次筛选,均为扩增效率高,特异性好的引物序列。Each set of primers described in the present invention has been screened for many times, and all are primer sequences with high amplification efficiency and good specificity.
本发明的另一目的是提供一种创面分泌物快速检测芯片,是针对耐甲氧西林金黄色葡萄球菌、金黄色葡萄球菌、大肠埃希菌、肺炎克雷伯菌、铜绿假单胞菌、鲍曼不动杆菌、白色念珠菌、光滑念珠菌、毛霉菌特异性基因片段进行检测的微流控芯片。Another object of the present invention is to provide a rapid detection chip for wound secretions, which is suitable for methicillin-resistant Staphylococcus aureus, Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Microfluidic chip for detection of specific gene fragments of Acinetobacter baumannii, Candida albicans, Candida glabrata, and Mucor.
所述微流控芯片快速检测芯片,固定有上述9种菌的引物组及阳性质控和阴性质控阵列。具体构建样式详见附图及说明。所述检测芯片在恒温核酸扩增分析仪上工作运行。The microfluidic chip rapid detection chip is immobilized with the above-mentioned 9 kinds of bacteria primer sets and positive quality control and negative quality control arrays. For specific construction styles, please refer to the attached drawings and descriptions. The detection chip works on a constant temperature nucleic acid amplification analyzer.
所述芯片包含阴性质控通道和阳性质控通道,阴性质控通道中不固定任何引物,阳性质控通道中固定酵母DNA片段及可以扩增该片段的引物。所述阴性质控使用TE作为模板扩增。The chip includes a negative quality control channel and a positive quality control channel. No primer is fixed in the negative quality control channel, and a yeast DNA fragment and a primer capable of amplifying the fragment are fixed in the positive quality control channel. The negative control was amplified using TE as template.
所述检测方法是基于核酸等温扩增法检测耐甲氧西林金黄色葡萄球菌、金黄色葡萄球菌、大肠埃希菌、肺炎克雷伯菌、铜绿假单胞菌、鲍曼不动杆菌、白色念珠菌、光滑念珠菌、毛霉菌的检测方法。The detection method is based on nucleic acid isothermal amplification method to detect methicillin-resistant Staphylococcus aureus, Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, white Methods for the detection of Candida, Candida glabrata, and Mucor.
上述目的是通过如下技术实现:The above objectives are achieved through the following technologies:
通过针对上述9种病原菌特定基因片段设计引物,并利用核酸等温扩增技术,以微流控芯片为载体,在恒温核酸扩增分析仪上进行扩增,并根据显色反应进行结构判读。By designing primers for the specific gene fragments of the above 9 pathogenic bacteria, and using nucleic acid isothermal amplification technology, the microfluidic chip is used as the carrier, and the amplification is carried out on a constant temperature nucleic acid amplification analyzer, and the structure is interpreted according to the color reaction.
本发明的另一个目的在于,提供基于核酸等温扩增技术和微流控芯片的试剂盒,可实现耐甲氧西林金黄色葡萄球菌、金黄色葡萄球菌、大肠埃希菌、肺炎克雷伯菌、铜绿假单胞菌、鲍曼不动杆菌、白色念珠菌、光滑念珠菌、毛霉菌的快速检测。特征在于,所述试剂盒包含有扩增反应液及固定有上述病原菌引物的基因芯片。Another object of the present invention is to provide a kit based on nucleic acid isothermal amplification technology and microfluidic chip, which can realize methicillin-resistant Staphylococcus aureus, Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae , Rapid detection of Pseudomonas aeruginosa, Acinetobacter baumannii, Candida albicans, Candida glabrata, and Mucor. It is characterized in that the kit comprises an amplification reaction solution and a gene chip immobilized with the above-mentioned pathogenic bacteria primers.
扩增反应液具体组分及浓度如下:The specific components and concentrations of the amplification reaction solution are as follows:
所述酶为每微升含8-16个活性单位的Bst DNA聚合酶The enzyme is Bst DNA polymerase containing 8-16 active units per microliter
本发明另一个目的是提供一种前述试剂盒的使用方法,具体步骤如下:Another object of the present invention is to provide a method of using the aforementioned kit, the specific steps are as follows:
(1)样品处理和模板提取:样品范围为于创面分泌物样品标本,取1mL待检样品加入1.5mL离心管中,12000rpm离心2min后,弃上清,加入600μL超纯水,12000rpm离心2min后,弃上清对菌液进行清洗,在离心管中加入200ul超纯水充分混匀后加至装有玻璃珠的离心管中,震荡5min后,恒温金属浴100℃加热5min,12000rpm离心2min,取上清做待检模板;(1) Sample processing and template extraction: The sample range is the wound secretion sample. Take 1 mL of the sample to be tested and add it to a 1.5 mL centrifuge tube. After centrifugation at 12,000 rpm for 2 minutes, discard the supernatant, add 600 μL of ultrapure water, and centrifuge at 12,000 rpm for 2 minutes. , discard the supernatant to wash the bacterial liquid, add 200ul ultrapure water to the centrifuge tube and mix well, add it to the centrifuge tube with glass beads, shake for 5min, heat the thermostatic metal bath at 100 ℃ for 5min, centrifuge at 12000rpm for 2min, Take the supernatant as a template to be tested;
(2)取试剂盒中的分别的至少两种菌的24μL内引物FIP/BIP,3μL外引物F3/B,10μl环引物LF/LB三对引物混合,取0.296μL混合引物和超纯水0.159μL及0.545μL 0.1%琼脂糖混合,点在相应位置;(2) Mix three pairs of primers of 24 μL inner primer FIP/BIP, 3 μL outer primer F3/B, 10 μL loop primer LF/LB of at least two bacteria in the kit, and take 0.296 μL mixed primer and 0.159 μL of ultrapure water. Mix μL and 0.545 μL 0.1% agarose, and place them on the corresponding positions;
(3)取0.159μL 1×105酵母DNA,0.545μL l0.1%琼脂糖和0.296μL引物混合之后,点在相应位置;(3) Mix 0.159 μL of 1×105 yeast DNA, 0.545 μL of 10.1% agarose and 0.296 μL of primers, and place them on the corresponding positions;
(4)取扩增反应液,加入待检模板,形成如下的反应体系,振荡混匀后离心,将离心后的液体注入芯片,将芯片放入芯片恒温扩增核酸分析仪中,温度65℃进行反应,时间50min~60min,反应体系组分及浓度如下,反应总体积70μL;(4) Take the amplification reaction solution, add the template to be tested to form the following reaction system, vibrate and mix well, centrifuge, inject the centrifuged liquid into the chip, put the chip into the chip constant temperature amplification nucleic acid analyzer, and the temperature is 65°C Carry out the reaction, the time is 50min~60min, the components and concentrations of the reaction system are as follows, and the total reaction volume is 70μL;
(5)检测:检测有S形曲线判定为阳性,未见S形曲线为阴性。(5) Detection: if there is a sigmoid curve in the detection, it is judged to be positive, and if no sigmoid curve is found, it is negative.
其中,反应体系中具体组分及浓度如下表Among them, the specific components and concentrations in the reaction system are as follows
本发明的有益效果是:本方案提供的9种创面常见病原菌微流控芯片快速检测技术及试剂盒具有灵敏度高、特异性强、方便快捷、适用范围广等优点,可解决创面病原菌的现场快速检测和基层普及应用难题;特别是现场检测及战时野外应用。同时,还拥有试剂消耗量小、污染小、成本低、且便携等诸多优点。The beneficial effects of the present invention are as follows: the microfluidic chip rapid detection technology and kit for 9 kinds of common pathogenic bacteria on wound surface provided by this scheme have the advantages of high sensitivity, strong specificity, convenience and rapidity, wide application range, etc., and can solve the on-site rapid detection of pathogenic bacteria on wound surface. Detection and grassroots popularization and application problems; especially on-site detection and wartime field applications. At the same time, it also has many advantages such as low reagent consumption, low pollution, low cost, and portability.
附图说明Description of drawings
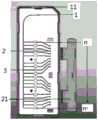
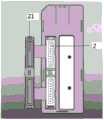
图1A为本发明微流控芯片一实施例的剖面示意图;图1B为本发明微流控芯片一实施例的外观示意图,其中,1为进样孔,12为进样孔卡扣,2为出气孔,21为出气孔卡扣,3为反应孔,n,n’为反应孔序号。1A is a schematic cross-sectional view of an embodiment of the microfluidic chip of the present invention; FIG. 1B is a schematic appearance diagram of an embodiment of the microfluidic chip of the present invention, wherein 1 is the injection hole, 12 is the injection hole buckle, and 2 is the injection hole buckle. For the air outlet, 21 is the air outlet buckle, 3 is the reaction hole, and n, n' are the reaction hole serial numbers.
图2为用图1所示的基因芯片对耐甲氧西林金黄色葡萄球菌进行实际检测的结果;Fig. 2 is the result of carrying out actual detection to methicillin-resistant Staphylococcus aureus with the gene chip shown in Fig. 1;
图3为用图1所示的基因芯片对金黄色葡萄球菌进行实际检测的结果;Fig. 3 is the result that Staphylococcus aureus is actually detected with the gene chip shown in Fig. 1;
图4为用图1所示的基因芯片对大肠埃希菌进行实际检测的结果;Fig. 4 is the result of carrying out actual detection to Escherichia coli with the gene chip shown in Fig. 1;
图5为用图1所示的基因芯片对肺炎克雷伯菌进行实际检测的结果;Fig. 5 is the result that Klebsiella pneumoniae is actually detected with the gene chip shown in Fig. 1;
图6为用图1所示的基因芯片对铜绿假单胞菌进行实际检测的结果;Fig. 6 is the result of actually detecting Pseudomonas aeruginosa with the gene chip shown in Fig. 1;
图7为用图1所示的基因芯片对鲍曼不动杆菌进行实际检测的结果;Fig. 7 is the result of actually detecting Acinetobacter baumannii with the gene chip shown in Fig. 1;
图8为用图1所示的基因芯片对白色念珠菌进行实际检测的结果;Fig. 8 is the result that candida albicans is actually detected with the gene chip shown in Fig. 1;
图9为用图1所示的基因芯片对光滑念珠菌进行实际检测的结果;Fig. 9 is the result that candida glabrata is actually detected with the gene chip shown in Fig. 1;
图10为用图1所示的基因芯片对毛霉菌进行实际检测的结果;Fig. 10 is the result that the gene chip shown in Fig. 1 is actually detected to Mucor;
图中横坐标单位是分钟(min),纵坐标没有单位;1-12通道分别为:1为阳性质控,2为阴性质控,3为耐甲氧西林金黄色葡萄球菌,4为金黄色葡萄球菌,5为大肠埃希氏菌,6为肺炎克雷伯菌,7为铜绿假单胞菌,8为鲍曼不动杆菌,9为白色念珠菌,10为光滑念珠菌,11为毛霉菌,12为阴性质控;图中有S形曲线判定为阳性,未见S形曲线为阴性。The unit of abscissa in the figure is minutes (min), and the ordinate has no unit; channels 1-12 are: 1 is positive quality control, 2 is negative quality control, 3 is methicillin-resistant Staphylococcus aureus, and 4 is golden yellow Staphylococcus, 5 is Escherichia coli, 6 is Klebsiella pneumoniae, 7 is Pseudomonas aeruginosa, 8 is Acinetobacter baumannii, 9 is Candida albicans, 10 is Candida glabrata, 11 is Hairy Mold, 12 is a negative control; if there is an S-shaped curve in the figure, it is judged to be positive, and no S-shaped curve is found to be negative.
具体实施方式Detailed ways
为进一步理解本发明,下面结合具体实施例对本发明优选方案进行描述,但是应当理解,这些描述只是为进一步说明本发明的特征和优点,而不是对本发明权利要求的限制。In order to further understand the present invention, the preferred embodiments of the present invention are described below with reference to specific embodiments, but it should be understood that these descriptions are only for further illustrating the features and advantages of the present invention, rather than limiting the claims of the present invention.
以下结合具体实施例对上述方案做进一步说明。应理解,这些实施例是用于说明本发明而不限于限制本发明的范围。实施例中采用的实施条件可以根据具体厂家的条件做进一步调整,未注明的实施条件通常为常规实验中的条件。The above scheme will be further described below in conjunction with specific embodiments. It should be understood that these examples are intended to illustrate the invention and not to limit the scope of the invention. The implementation conditions used in the examples can be further adjusted according to the conditions of specific manufacturers, and the implementation conditions not specified are usually the conditions in routine experiments.
下面通过具体的实施例和附图对本发明进行说明,以耐甲氧西林金黄色葡萄球菌、金黄色葡萄球菌、大肠埃希菌、肺炎克雷伯菌、铜绿假单胞菌、鲍曼不动杆菌、白色念珠菌、光滑念珠菌、毛霉菌检测过程通过基因芯片来对本发明进行说明,如无特别说明,均为常规方法。The present invention will be described below through specific examples and accompanying drawings, with methicillin-resistant Staphylococcus aureus, Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Baumannii The detection process of Bacillus, Candida albicans, Candida glabrata, and Mucor spp. is used to illustrate the present invention through a gene chip, and all are conventional methods unless otherwise specified.
1.引物制备,以上述的核酸序列合成,得到引物组,由上海生工合成。1. Primer preparation, synthesized with the above-mentioned nucleic acid sequence, to obtain a primer set, which was synthesized by Shanghai Shenggong.
2.基因芯片制备,芯片是由上述Seq ID No.1~Seq ID No.54引物、阳性质控品Seq ID No.55-Seq ID No.60引物、阴性质控品组成的阵列;具体方法为:2. Gene chip preparation, the chip is an array composed of the above-mentioned Seq ID No.1~Seq ID No.54 primers, positive quality control substance Seq ID No.55-Seq ID No.60 primer, and negative quality control substance; specific method for:
取24μl耐甲氧西林金黄色葡萄球菌的内引物FIP/BIP,3μL外引物F3/B,10μL环引物LF/LB三对引物混合,取0.296μL混合引物和超纯水0.159μL及0.545μL 0.1%琼脂糖混合,得到反应混和物,然后点在相应位置,自然晾干;Take 24 μl of the inner primer FIP/BIP of methicillin-resistant Staphylococcus aureus, 3 μL of the outer primer F3/B, 10 μL of the loop primer LF/LB three pairs of primers and mix, take 0.296 μL of mixed primers and ultrapure water 0.159 μL and 0.545 μL 0.1 % agarose to get the reaction mixture, then point it in the corresponding position and let it dry naturally;
取24μl金黄色葡萄球菌的内引物FIP/BIP,3μL外引物F3/B,10μl环引物LF/LB三对引物混合,取0.296μL混合引物和超纯水0.159μL及0.545μL0.1%琼脂糖混合,得到反应混和物,然后点在相应位置,自然晾干;Take 24 μl of Staphylococcus aureus inner primer FIP/BIP, 3 μL outer primer F3/B, 10 μl loop primer LF/LB three pairs of primers and mix, take 0.296 μL of mixed primers, 0.159 μL of ultrapure water and 0.545 μL of 0.1% agarose Mix to get the reaction mixture, then point it in the corresponding position and let it dry naturally;
取24μL大肠埃希菌的内引物FIP/BIP,3μL外引物F3/B,10μL环引物LF/LB三对引物混合,取0.296μL混合引物和超纯水0.159μL及0.545μL0.1%琼脂糖混合,得到反应混和物,然后点在相应位置,自然晾干;Take 24μL of the inner primer FIP/BIP of Escherichia coli, 3μL of the outer primer F3/B, and 10μL of the loop primer LF/LB to mix three pairs of primers, take 0.296μL of mixed primers, 0.159μL of ultrapure water and 0.545μL of 0.1% agarose Mix to get the reaction mixture, then point it in the corresponding position and let it dry naturally;
取24μL肺炎克雷伯菌的内引物FIP/BIP,3μL外引物F3/B,10μL环引物LF/LB三对引物混合,取0.296μl混合引物和超纯水0.159μL及0.545μl 0.1%琼脂糖混合,得到反应混和物,然后点在相应位置,自然晾干;Take 24μL of Klebsiella pneumoniae inner primer FIP/BIP, 3μL outer primer F3/B, 10μL loop primer LF/LB three pairs of primers and mix, take 0.296μl mixed primers and ultrapure water 0.159μL and 0.545μl 0.1% agarose Mix to get the reaction mixture, then point it in the corresponding position and let it dry naturally;
取24μL铜绿假单胞菌的内引物FIP/BIP,3μL外引物F3/B,10μL环引物LF/LB三对引物混合,取0.296μL混合引物和超纯水0.159μL及0.545μL 0.1%琼脂糖混合,得到反应混和物,然后点在相应位置,自然晾干;Take 24μL of the inner primer FIP/BIP of Pseudomonas aeruginosa, 3μL of the outer primer F3/B, and 10μL of the loop primer LF/LB to mix three pairs of primers, take 0.296μL of mixed primers and 0.159μL of ultrapure water and 0.545μL of 0.1% agarose Mix to get the reaction mixture, then point it in the corresponding position and let it dry naturally;
取24μl鲍曼不动杆菌的内引物FIP/BIP,3μL外引物F3/B,10μL环引物LF/LB三对引物混合,取0.296μL混合引物和超纯水0.159μL及0.545μL 0.1%琼脂糖混合,得到反应混和物,然后点在相应位置,自然晾干;Take 24 μl of Acinetobacter baumannii inner primer FIP/BIP, 3 μL outer primer F3/B, 10 μL loop primer LF/LB three pairs of primers and mix, take 0.296 μL mixed primers and 0.159 μL ultrapure water and 0.545 μL 0.1% agarose Mix to get the reaction mixture, then point it in the corresponding position and let it dry naturally;
取24μl白色念珠菌的内引物FIP/BIP,3μL外引物F3/B,10μL环引物LF/LB三对引物混合,取0.296μL混合引物和超纯水0.159μL及0.545μL 0.1%琼脂糖混合,得到反应混和物,然后点在相应位置,自然晾干;Take 24 μL of Candida albicans inner primer FIP/BIP, 3 μL outer primer F3/B, 10 μL loop primer LF/LB three primer pairs and mix, take 0.296 μL mixed primer and mix with 0.159 μL ultrapure water and 0.545 μL 0.1% agarose, Get the reaction mixture, then point it in the corresponding position and let it dry naturally;
取24μl光滑念珠菌的内引物FIP/BIP,3μL外引物F3/B,10μL环引物LF/LB三对引物混合,取0.296μL混合引物和超纯水0.159μL及0.545μL 0.1%琼脂糖混合,得到反应混和物,然后点在相应位置,自然晾干;Take 24 μl of the inner primer FIP/BIP of Candida glabrata, 3 μL of the outer primer F3/B, and 10 μL of the loop primer LF/LB and mix three pairs of primers. Get the reaction mixture, then point it in the corresponding position and let it dry naturally;
取24μl毛霉菌菌株的内引物FIP/BIP,3μL外引物F3/B,10μL环引物LF/LB三对引物混合,取0.296μL混合引物和超纯水0.159μL及0.545μL 0.1%琼脂糖混合,得到反应混和物,然后点在相应位置,自然晾干;Take 24 μl of the inner primer FIP/BIP of the Mucor strain, 3 μL of the outer primer F3/B, and 10 μL of the loop primer LF/LB, and mix three pairs of primers. Take 0.296 μL of the mixed primer and mix it with 0.159 μL of ultrapure water and 0.545 μL of 0.1% agarose. Get the reaction mixture, then point it in the corresponding position and let it dry naturally;
取0.159μL 1×105酵母DNA,0.545μL 0.1%琼脂糖和0.296μL引物混合之后,点在相应位置,自然晾干。After mixing 0.159 μL of 1×105 yeast DNA, 0.545 μL of 0.1% agarose and 0.296 μL of primers, place them on the corresponding positions and let them dry naturally.
3.分别提取耐甲氧西林金黄色葡萄球菌、金黄色葡萄球菌、大肠埃希菌、肺炎克雷伯菌、铜绿假单胞菌、鲍曼不动杆菌、白色念珠菌、光滑念珠菌、毛霉菌菌株基因DNA:3. Respectively extract methicillin-resistant Staphylococcus aureus, Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, Candida albicans, Candida glabrata, trichomes Mold strain gene DNA:
取1mL菌液加入1.5mL离心管中,12000rpm,离心2min后弃上清,加入600μl超纯水,再次12000rpm,离心2min后弃上清对菌液进行清洗,在离心管中加入200μl超纯水充分混匀后加至装有玻璃珠的离心管中,震荡5min后,恒温金属浴加热5min,12000rpm离心2min取上清。Take 1 mL of bacterial liquid into a 1.5 mL centrifuge tube, centrifuge at 12,000 rpm for 2 minutes, discard the supernatant, add 600 μl of ultrapure water, centrifuge again at 12,000 rpm for 2 minutes, discard the supernatant to wash the bacterial liquid, and add 200 μl of ultrapure water to the centrifuge tube. After fully mixing, it was added to a centrifuge tube containing glass beads, shaken for 5 min, heated in a constant temperature metal bath for 5 min, and centrifuged at 12,000 rpm for 2 min to take the supernatant.
4.基于核酸扩增法的基因扩增。4. Gene amplification based on nucleic acid amplification method.
取扩增反应液28μl,加入35μl待检模板及7μl水,形成如下表总体积为70μl的反应体系,反应体系如下:(反应总体积为70μl)。在温度为65℃的iChip-400上反应50分钟,反应体系中各组分及浓度如下表。Take 28 μl of the amplification reaction solution, add 35 μl of template to be tested and 7 μl of water to form a reaction system with a total volume of 70 μl in the following table. The reaction system is as follows: (The total reaction volume is 70 μl). The reaction was carried out on iChip-400 at a temperature of 65° C. for 50 minutes. The components and concentrations in the reaction system were as follows.
扩增产物检测:与阴性对照孔比较,检测孔在iChip-400设备上有S形曲线,判定LAMP测试结果为阳性。在iChip-400设备上未见S形曲线,判定LAMP测试结果为阴性。Amplification product detection: Compared with the negative control well, the detection well has an S-shaped curve on the iChip-400 device, and the LAMP test result is judged to be positive. No S-shaped curve was seen on the iChip-400 device, and the LAMP test result was judged to be negative.
以上所述的实施例只是本发明的一种较佳的方案,并非对本发明作任何形式上的限制,在不超过权利要求所记载的技术方案前提下,还有其他变体或改型。The above-mentioned embodiment is only a preferred solution of the present invention, and does not limit the present invention in any form, and there are other variations or modifications under the premise of not exceeding the technical solution recorded in the claims.
本发明的技术内容及技术特征已揭示如上,然而熟悉本领域的技术人员仍可能基于本发明的教示及揭示而作种种不背离本发明精神的替换及修饰,因此,本发明保护范围应不限于实施例所揭示的内容,而应包括各种不背离本发明的替换及修饰,并为本专利申请权利要求所涵盖。The technical content and technical features of the present invention have been disclosed as above. However, those skilled in the art may still make various replacements and modifications based on the teaching and disclosure of the present invention without departing from the spirit of the present invention. Therefore, the protection scope of the present invention should not be limited to The contents disclosed in the embodiments should include various substitutions and modifications without departing from the present invention, and are covered by the claims of this patent application.
序列表sequence listing
<110> 上海蕴箬生物科技有限公司<110> Shanghai Yunluo Biotechnology Co., Ltd.
<120> 一种快速检测创面病原菌的微流控芯片试剂盒<120> A Microfluidic Chip Kit for Rapid Detection of Wound Pathogens
<160> 60<160> 60
<170> SIPOSequenceListing 1.0<170> SIPOSequenceListing 1.0
<210> 1<210> 1
<211> 19<211> 19
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 1<400> 1
ttatgtatca ggtacttat 19ttatgtatca ggtacttat 19
<210> 2<210> 2
<211> 25<211> 25
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 2<400> 2
ttttgttatt taacccaatc attta 25ttttgttatt taacccaatc attta 25
<210> 3<210> 3
<211> 49<211> 49
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 3<400> 3
attcttcgtt actcattaca tacatgtgaa ttattataac ttgtataac 49attcttcgtt actcattaca tacatgtgaa ttattataac ttgtataac 49
<210> 4<210> 4
<211> 50<211> 50
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 4<400> 4
aaccgaagat aaaaaagaac ctctgaatat tttttgagtt gaacctggtg 50aaccgaagat aaaaaagaac ctctgaatat tttttgagtt gaacctggtg 50
<210> 5<210> 5
<211> 24<211> 24
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 5<400> 5
aatggataga cgtcatatga aggt 24aatggataga cgtcatatga aggt 24
<210> 6<210> 6
<211> 25<211> 25
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 6<400> 6
ctcaacaagt tccagattac aactt 25ctcaacaagt tccagattac aactt 25
<210> 7<210> 7
<211> 20<211> 20
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 7<400> 7
gttactttac agatataatg 20
<210> 8<210> 8
<211> 22<211> 22
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 8<400> 8
gaaaaagtgt acgagttctt ga 22gaaaaagtgt acgagttctt ga 22
<210> 9<210> 9
<211> 44<211> 44
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 9<400> 9
gtttcataac cttcataaat atttccatac agtcatttca ctaa 44gtttcataac cttcataaat atttccatac agtcatttca ctaa 44
<210> 10<210> 10
<211> 40<211> 40
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 10<400> 10
gaggtcattt aatatttatt acttcgatca ctggacctag 40gaggtcattt aatatttatt acttcgatca ctggacctag 40
<210> 11<210> 11
<211> 18<211> 18
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 11<400> 11
aactcatagt gtacaaca 18aactcatagt gtacaaca 18
<210> 12<210> 12
<211> 22<211> 22
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 12<400> 12
gtacctgtta tgaaagtgtt ca 22gtacctgtta tgaaagtgtt ca 22
<210> 13<210> 13
<211> 20<211> 20
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 13<400> 13
gtaatcgtgg tgattgatga 20
<210> 14<210> 14
<211> 20<211> 20
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 14<400> 14
ggttcgttgt aaatactcca 20
<210> 15<210> 15
<211> 40<211> 40
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 15<400> 15
tctttcgtat tgtttaccta cttatgtcgt atttaacctc 40tctttcgtat tgtttaccta cttatgtcgt atttaacctc 40
<210> 16<210> 16
<211> 46<211> 46
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 16<400> 16
tacatagaag agtaagtcaa cgttttggtt tttgtcacta tatatc 46tacatagaag agtaagtcaa cgttttggtt tttgtcacta tatatc 46
<210> 17<210> 17
<211> 21<211> 21
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 17<400> 17
ttcgaaacca attactaaag a 21ttcgaaacca attactaaag a 21
<210> 18<210> 18
<211> 18<211> 18
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 18<400> 18
tataacttac agtagatt 18tataacttac agtagatt 18
<210> 19<210> 19
<211> 17<211> 17
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 19<400> 19
gtctctacaa taggaat 17gtctctacaa taggaat 17
<210> 20<210> 20
<211> 18<211> 18
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 20<400> 20
atgagaccta ctgggaca 18atgagaccta ctgggaca 18
<210> 21<210> 21
<211> 36<211> 36
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 21<400> 21
aacctaaatg ttaaaataag tcagggagac gaccgt 36aacctaaatg ttaaaataag tcagggagac gaccgt 36
<210> 22<210> 22
<211> 46<211> 46
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 22<400> 22
agttaataaa gtaggagtac tttattttgt tctagtatgg aatcgg 46agttaataaa gtaggagtac ttttattttgt tctagtatgg aatcgg 46
<210> 23<210> 23
<211> 21<211> 21
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 23<400> 23
cgatgacata ataaataacc g 21cgatgacata ataaataacc
<210> 24<210> 24
<211> 17<211> 17
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 24<400> 24
tatatatacc actacca 17tatatatacc actacca 17
<210> 25<210> 25
<211> 21<211> 21
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 25<400> 25
tgttatggaa atgtccacct t 21tgttatggaa atgtccacct
<210> 26<210> 26
<211> 18<211> 18
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 26<400> 26
tcttatgata tttctgag 18tcttatgata tttctgag 18
<210> 27<210> 27
<211> 38<211> 38
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 27<400> 27
gtacagaaca taatacagag gaacacgatg aacaacgt 38gtacagaaca taatacagag gaacacgatg aacaacgt 38
<210> 28<210> 28
<211> 37<211> 37
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 28<400> 28
atatcgtctg acctataccc ttcgtcatac tttagat 37atatcgtctg acctataccc ttcgtcatac tttagat 37
<210> 29<210> 29
<211> 20<211> 20
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 29<400> 29
ccagataaga gaatttcaga 20
<210> 30<210> 30
<211> 17<211> 17
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 30<400> 30
atattatcgt tatcagg 17atattatcgt tatcagg 17
<210> 31<210> 31
<211> 21<211> 21
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 31<400> 31
tgttacaatt aacttcttat a 21tgttacaatt aacttcttat a 21
<210> 32<210> 32
<211> 23<211> 23
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 32<400> 32
cttgtaatat cgttatagta gtt 23cttgtaatat cgttatagta gtt 23
<210> 33<210> 33
<211> 48<211> 48
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 33<400> 33
agtgaattcg gttatcgtaa tatcgaataa taaaaaaaaa ttagtcac 48agtgaattcg gttatcgtaa tatcgaataa taaaaaaaaa ttagtcac 48
<210> 34<210> 34
<211> 44<211> 44
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 34<400> 34
aatgatatgt atcaaaatat agagtattaa ctcatgttag atgg 44aatgatatgt atcaaaatat agagtattaa ctcatgttag atgg 44
<210> 35<210> 35
<211> 16<211> 16
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 35<400> 35
tataagagag gtctac 16tataagagag gtctac 16
<210> 36<210> 36
<211> 18<211> 18
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 36<400> 36
ccgttaaaaa ttataagg 18ccgttaaaaa ttataagg 18
<210> 37<210> 37
<211> 19<211> 19
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 37<400> 37
tcaacttgtc acaccagat 19tcaacttgtc acaccagat 19
<210> 38<210> 38
<211> 18<211> 18
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 38<400> 38
ctcaacacca aacccagc 18ctcaacacca aacccagc 18
<210> 39<210> 39
<211> 43<211> 43
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 39<400> 39
tcgctgcgtt cttcatcgat aatagtcaaa actttcaaca acg 43tcgctgcgtt cttcatcgat aatagtcaaa actttcaaca acg 43
<210> 40<210> 40
<211> 43<211> 43
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 40<400> 40
tgcagatatt cgtgaatcat cgaataaacg acgctcaaac agg 43tgcagatatt cgtgaatcat cgaataaacg acgctcaaac agg 43
<210> 41<210> 41
<211> 17<211> 17
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 41<400> 41
gcgagaacca agagatc 17gcgagaacca agagatc 17
<210> 42<210> 42
<211> 17<211> 17
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 42<400> 42
ctttgaacgc acattgc 17ctttgaacgc acattgc 17
<210> 43<210> 43
<211> 21<211> 21
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 43<400> 43
ccattaaaaa gtgcaatggt g 21ccattaaaaa gtgcaatggt
<210> 44<210> 44
<211> 21<211> 21
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 44<400> 44
ctgtctttag aatccaactg c 21ctgtctttag aatccaactg
<210> 45<210> 45
<211> 42<211> 42
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 45<400> 45
ttcttagcac tgttgtcaga aacacaatgc cttcctgtac ca 42ttcttagcac tgttgtcaga aacacaatgc cttcctgtac ca 42
<210> 46<210> 46
<211> 43<211> 43
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 46<400> 46
gtcggcgagg gtaaattcaa cgtttgattc cttaagcccg ata 43gtcggcgagg gtaaattcaa cgtttgattc cttaagcccg ata 43
<210> 47<210> 47
<211> 21<211> 21
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 47<400> 47
cggtggtggt atctgagaca t 21cggtggtggt atctgagaca
<210> 48<210> 48
<211> 23<211> 23
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 48<400> 48
tgatacacta actgcatctc cct 23tgatacacta actgcatctc cct 23
<210> 49<210> 49
<211> 23<211> 23
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 49<400> 49
cttcatctta tttggtcttt tgc 23cttcatctta tttggtcttt tgc 23
<210> 50<210> 50
<211> 18<211> 18
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 50<400> 50
tttatgcacg tggttggt 18tttatgcacg tggttggt 18
<210> 51<210> 51
<211> 44<211> 44
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 51<400> 51
cagtttcttc gtatgcattg tcactacgaa cacaagatgt cact 44cagtttcttc gtatgcattg tcactacgaa cacaagatgt cact 44
<210> 52<210> 52
<211> 45<211> 45
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 52<400> 52
cagatccaag gatcctatgg tcttggccct tgttggtatt ttgag 45cagatccaag gatcctatgg tcttggccct tgttggtatt ttgag 45
<210> 53<210> 53
<211> 20<211> 20
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 53<400> 53
gttgcacttg caagttgatg 20
<210> 54<210> 54
<211> 22<211> 22
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 54<400> 54
cacatgggct ttagaagata ca 22cacatgggct ttagaagata ca 22
<210> 55<210> 55
<211> 18<211> 18
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 55<400> 55
ttactggtcg cccatctc 18ttactggtcg cccatctc 18
<210> 56<210> 56
<211> 21<211> 21
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 56<400> 56
ctataggcac ttctcctgta g 21ctataggcac ttctcctgta
<210> 57<210> 57
<211> 40<211> 40
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 57<400> 57
acaacccgtc cccatagtaa taacactcat gccttccaca 40acaacccgtc cccatagtaa taacactcat gccttccaca 40
<210> 58<210> 58
<211> 45<211> 45
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 58<400> 58
cgatgctata cttcgtatag gaagccattc ttgtgctatg gaaca 45cgatgctata cttcgtatag gaagccattc ttgtgctatg gaaca 45
<210> 59<210> 59
<211> 20<211> 20
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 59<400> 59
gacttagtcg ctaagtggaa 20
<210> 60<210> 60
<211> 20<211> 20
<212> DNA<212> DNA
<213> 人工序列(Artificial Sequence)<213> Artificial Sequence
<400> 60<400> 60
cgttttttta tgccccatcc 20
Claims (7)
Priority Applications (1)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| CN202010169329.6ACN111235288A (en) | 2020-03-12 | 2020-03-12 | A microfluidic chip kit for rapid detection of pathogenic bacteria on wound surfaces |
Applications Claiming Priority (1)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| CN202010169329.6ACN111235288A (en) | 2020-03-12 | 2020-03-12 | A microfluidic chip kit for rapid detection of pathogenic bacteria on wound surfaces |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| CN111235288Atrue CN111235288A (en) | 2020-06-05 |
Family
ID=70867350
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| CN202010169329.6AWithdrawnCN111235288A (en) | 2020-03-12 | 2020-03-12 | A microfluidic chip kit for rapid detection of pathogenic bacteria on wound surfaces |
Country Status (1)
| Country | Link |
|---|---|
| CN (1) | CN111235288A (en) |
Cited By (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN112143821A (en)* | 2020-09-30 | 2020-12-29 | 北京师范大学 | Reagent for detecting candida albicans, cryptococcus neoformans and klebsiella pneumoniae by FRET and application thereof |
| CN119177304A (en)* | 2024-11-26 | 2024-12-24 | 北京大学第三医院(北京大学第三临床医学院) | Kit for rapid combined detection of multiple pathogen nucleic acids related to bad pregnancy ending and application thereof |
Citations (10)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US20090305274A1 (en)* | 2008-03-12 | 2009-12-10 | Medical Diagnostic Laboratories, Llc | Method for determining azole resistance in candida glabrata |
| CN104195030A (en)* | 2014-08-14 | 2014-12-10 | 国家纳米科学中心 | Micro-fluidic chip and method for evaluating wound dressing by using micro-fluidic chip |
| WO2016023397A1 (en)* | 2014-08-15 | 2016-02-18 | 广州医科大学附属第三医院 | Primer group for gonococci detection, kit comprising primer group and uses thereof |
| CN107119140A (en)* | 2017-06-16 | 2017-09-01 | 北京百康芯生物科技有限公司 | Respiratory tract micro-fluidic chip Fast Detection Technique and kit |
| CN107513582A (en)* | 2017-10-23 | 2017-12-26 | 蔡慧娜 | Authentication chip, kit and the authentication method of positive blood culture |
| US20180135108A1 (en)* | 2014-01-20 | 2018-05-17 | Board Of Trustees Of Michigan State University | Method for detecting bacterial and fungal pathogens |
| US20190071707A1 (en)* | 2016-01-21 | 2019-03-07 | T2 Biosystems, Inc. | Nmr methods and systems for the rapid detection of bacteria |
| CN109487000A (en)* | 2018-12-29 | 2019-03-19 | 博奥生物集团有限公司 | Primer combination and its application |
| CN110129316A (en)* | 2018-02-05 | 2019-08-16 | 北京智德医学检验所有限公司 | A kind of LAMP primer combination and application for detecting 4 kinds of Gram-negative bacteria in intraocular fluid |
| CN110551840A (en)* | 2019-08-20 | 2019-12-10 | 北京卓诚惠生生物科技股份有限公司 | Nucleic acid reagent, kit, system and method for detecting invasive fungi |
- 2020
- 2020-03-12CNCN202010169329.6Apatent/CN111235288A/ennot_activeWithdrawn
Patent Citations (10)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US20090305274A1 (en)* | 2008-03-12 | 2009-12-10 | Medical Diagnostic Laboratories, Llc | Method for determining azole resistance in candida glabrata |
| US20180135108A1 (en)* | 2014-01-20 | 2018-05-17 | Board Of Trustees Of Michigan State University | Method for detecting bacterial and fungal pathogens |
| CN104195030A (en)* | 2014-08-14 | 2014-12-10 | 国家纳米科学中心 | Micro-fluidic chip and method for evaluating wound dressing by using micro-fluidic chip |
| WO2016023397A1 (en)* | 2014-08-15 | 2016-02-18 | 广州医科大学附属第三医院 | Primer group for gonococci detection, kit comprising primer group and uses thereof |
| US20190071707A1 (en)* | 2016-01-21 | 2019-03-07 | T2 Biosystems, Inc. | Nmr methods and systems for the rapid detection of bacteria |
| CN107119140A (en)* | 2017-06-16 | 2017-09-01 | 北京百康芯生物科技有限公司 | Respiratory tract micro-fluidic chip Fast Detection Technique and kit |
| CN107513582A (en)* | 2017-10-23 | 2017-12-26 | 蔡慧娜 | Authentication chip, kit and the authentication method of positive blood culture |
| CN110129316A (en)* | 2018-02-05 | 2019-08-16 | 北京智德医学检验所有限公司 | A kind of LAMP primer combination and application for detecting 4 kinds of Gram-negative bacteria in intraocular fluid |
| CN109487000A (en)* | 2018-12-29 | 2019-03-19 | 博奥生物集团有限公司 | Primer combination and its application |
| CN110551840A (en)* | 2019-08-20 | 2019-12-10 | 北京卓诚惠生生物科技股份有限公司 | Nucleic acid reagent, kit, system and method for detecting invasive fungi |
Non-Patent Citations (5)
| Title |
|---|
| 李国锋;王晓娜;秦阳华;: "烧伤创面感染病原菌分布及耐药性分析"* |
| 芮勇宇;王前;裘宇容;: "伤口分泌物中1 640株病原菌主要种类及药敏结果分析"* |
| 芮勇宇;蔡贞;: "1659株伤口分泌物中分离病原菌种类及耐药性分析"* |
| 郑光辉;马瑞敏;李方强;张艳;唐明忠;张岩;吕虹;张国军;: "微流控芯片技术在神经外科术后细菌感染快速诊断中的应用"* |
| 钱进先;于文;吴健;杨爱祥;陶唯益;赵华;王耀东;: "昆山"8・2"特大烧伤合并吸入性损伤患者病原菌构成及耐药性分析"* |
Cited By (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN112143821A (en)* | 2020-09-30 | 2020-12-29 | 北京师范大学 | Reagent for detecting candida albicans, cryptococcus neoformans and klebsiella pneumoniae by FRET and application thereof |
| CN119177304A (en)* | 2024-11-26 | 2024-12-24 | 北京大学第三医院(北京大学第三临床医学院) | Kit for rapid combined detection of multiple pathogen nucleic acids related to bad pregnancy ending and application thereof |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| CN108192816A (en) | A kind of sample automatically processes and nucleic acid amplifier and application method | |
| CN107190079B (en) | Quick detection technology and kit for five food-borne pathogenic bacteria micro-fluidic chip | |
| CN107119140A (en) | Respiratory tract micro-fluidic chip Fast Detection Technique and kit | |
| US20120028819A1 (en) | Method for the detection and characterization of a toxinogenic clostridium difficile strain | |
| US10589276B2 (en) | Multi-functional microfluidics device for biological sample screening | |
| CN111235288A (en) | A microfluidic chip kit for rapid detection of pathogenic bacteria on wound surfaces | |
| CN113637781A (en) | LAMP primer group for detecting swine susceptibility related pathogenic bacteria, kit and LAMP chip based on LAMP primer group and application | |
| CN112126587B (en) | Nucleic acid detection chip device, nucleic acid detection chip and preparation method thereof | |
| CN110184386B (en) | RT-LAMP detection primer for detecting ANRSV and its application, detection reagent and detection method | |
| CN112538537A (en) | High-flux microfluidic LAMP chip for detecting livestock respiratory pathogens and detection method | |
| CN104911275B (en) | A kind of test kit for bacterial vaginosis | |
| CN109355407B (en) | Primer, kit and method for detecting pseudomonas aeruginosa through PSR isothermal amplification reaction | |
| CN115851990A (en) | A kind of amplification primer, kit and application thereof for rapid detection of Salmonella | |
| CN110079622A (en) | Kit based on LAMP method detection Klebsiella Pneumoniae | |
| CN112301158B (en) | RDA method and kit for rapidly detecting Classical Swine Fever Virus (CSFV) | |
| CN110656037B (en) | Micro-fluidic chip for pathogen nucleic acid detection and detection method | |
| CN109355408B (en) | Primer, kit and method for PSR (phosphosilicate receptor) detection of Escherichia coli type I Shiga toxin | |
| CN109355403B (en) | A kind of primer, kit and method for PSR detection of methicillin-resistant Staphylococcus aureus | |
| CN109355404B (en) | Primer, kit and detection method for isothermal detection of listeria monocytogenes based on polymerase helix reaction | |
| CN107475401B (en) | Method and primers for detection of foodborne Bacillus cereus by loop-mediated isothermal amplification | |
| CN105838825A (en) | Kit, primer and method for detecting Escherichia coli | |
| CN110578014A (en) | A kit and detection method for nucleic acid detection of Candida parapsilosis | |
| CN208200972U (en) | A kind of sample automatically processes and nucleic acid amplifier | |
| CN112852975A (en) | Primer probe set, kit and detection method for detecting schistosoma japonicum nucleic acid based on fluorescence RMA method | |
| CN113667766A (en) | CPA detection primer and detection kit and method for enterotoxin B-producing Staphylococcus aureus |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| PB01 | Publication | ||
| PB01 | Publication | ||
| SE01 | Entry into force of request for substantive examination | ||
| SE01 | Entry into force of request for substantive examination | ||
| WW01 | Invention patent application withdrawn after publication | ||
| WW01 | Invention patent application withdrawn after publication | Application publication date:20200605 |