CN102417882A - Expression method of pseudomonas stutzeri A1501 rpoN gene - Google Patents
Expression method of pseudomonas stutzeri A1501 rpoN geneDownload PDFInfo
- Publication number
- CN102417882A CN102417882ACN2011103267044ACN201110326704ACN102417882ACN 102417882 ACN102417882 ACN 102417882ACN 2011103267044 ACN2011103267044 ACN 2011103267044ACN 201110326704 ACN201110326704 ACN 201110326704ACN 102417882 ACN102417882 ACN 102417882A
- Authority
- CN
- China
- Prior art keywords
- chlamydomonas
- gene
- rpon
- chloroplast
- expression
- Prior art date
- Legal status (The legal status is an assumption and is not a legal conclusion. Google has not performed a legal analysis and makes no representation as to the accuracy of the status listed.)
- Pending
Links
- 101150011750rpoN geneProteins0.000titleclaimsabstractdescription61
- 230000014509gene expressionEffects0.000titleclaimsabstractdescription35
- 238000000034methodMethods0.000titleclaimsabstractdescription35
- 241000212210Pseudomonas stutzeri A1501Species0.000titleabstractdescription13
- 241000195585ChlamydomonasSpecies0.000claimsabstractdescription101
- 210000003763chloroplastAnatomy0.000claimsabstractdescription58
- 108090000623proteins and genesProteins0.000claimsabstractdescription21
- 230000009261transgenic effectEffects0.000claimsabstractdescription19
- 239000013604expression vectorSubstances0.000claimsabstractdescription17
- 239000012634fragmentSubstances0.000claimsabstractdescription12
- 241000589614Pseudomonas stutzeriSpecies0.000claimsabstractdescription10
- 102000004169proteins and genesHuman genes0.000claimsabstractdescription8
- 238000012216screeningMethods0.000claimsabstractdescription8
- 241000195597Chlamydomonas reinhardtiiSpecies0.000claimsdescription12
- 101150026213atpB geneProteins0.000claimsdescription8
- 101100301006Allochromatium vinosum (strain ATCC 17899 / DSM 180 / NBRC 103801 / NCIMB 10441 / D) cbbL2 geneProteins0.000claimsdescription7
- 101150004101cbbL geneProteins0.000claimsdescription7
- 239000002245particleSubstances0.000claimsdescription7
- 101150074945rbcL geneProteins0.000claimsdescription7
- 101150088806atpA geneProteins0.000claimsdescription6
- 230000033228biological regulationEffects0.000claimsdescription5
- 239000002773nucleotideSubstances0.000claimsdescription5
- 125000003729nucleotide groupChemical group0.000claimsdescription5
- 108700031407Chloroplast GenesProteins0.000claimsdescription3
- 230000012010growthEffects0.000claimsdescription3
- 101150035600atpD geneProteins0.000claimsdescription2
- 101150038923atpF geneProteins0.000claimsdescription2
- 101150075980psbA geneProteins0.000claimsdescription2
- 101150010007psbD geneProteins0.000claimsdescription2
- IJGRMHOSHXDMSA-UHFFFAOYSA-NAtomic nitrogenChemical compoundN#NIJGRMHOSHXDMSA-UHFFFAOYSA-N0.000abstractdescription45
- 229910052757nitrogenInorganic materials0.000abstractdescription23
- 101150067683rpo10 geneProteins0.000abstractdescription16
- 239000000463materialSubstances0.000abstractdescription7
- 238000000746purificationMethods0.000abstractdescription4
- 230000007246mechanismEffects0.000abstractdescription2
- 230000001131transforming effectEffects0.000abstractdescription2
- 238000003306harvestingMethods0.000abstract1
- 230000001939inductive effectEffects0.000abstract1
- 108020004414DNAProteins0.000description20
- 239000002609mediumSubstances0.000description17
- 239000012528membraneSubstances0.000description16
- 239000013612plasmidSubstances0.000description15
- 239000006228supernatantSubstances0.000description14
- LFQSCWFLJHTTHZ-UHFFFAOYSA-NEthanolChemical compoundCCOLFQSCWFLJHTTHZ-UHFFFAOYSA-N0.000description13
- XLYOFNOQVPJJNP-UHFFFAOYSA-NwaterChemical compoundOXLYOFNOQVPJJNP-UHFFFAOYSA-N0.000description13
- 239000000243solutionSubstances0.000description11
- 210000004027cellAnatomy0.000description10
- 239000000203mixtureSubstances0.000description10
- 239000000047productSubstances0.000description10
- 238000006243chemical reactionMethods0.000description9
- 238000012546transferMethods0.000description9
- QTBSBXVTEAMEQO-UHFFFAOYSA-MAcetateChemical compoundCC([O-])=OQTBSBXVTEAMEQO-UHFFFAOYSA-M0.000description7
- 241000894006BacteriaSpecies0.000description7
- 241000589516PseudomonasSpecies0.000description7
- PCHJSUWPFVWCPO-UHFFFAOYSA-NgoldChemical compound[Au]PCHJSUWPFVWCPO-UHFFFAOYSA-N0.000description7
- 230000009466transformationEffects0.000description7
- 239000013598vectorSubstances0.000description7
- HEDRZPFGACZZDS-UHFFFAOYSA-NChloroformChemical compoundClC(Cl)ClHEDRZPFGACZZDS-UHFFFAOYSA-N0.000description6
- 238000001514detection methodMethods0.000description6
- 239000002244precipitateSubstances0.000description6
- 108010020943NitrogenaseProteins0.000description5
- 238000004178biological nitrogen fixationMethods0.000description5
- 239000000499gelSubstances0.000description5
- 238000005406washingMethods0.000description5
- CSCPPACGZOOCGX-UHFFFAOYSA-NAcetoneChemical compoundCC(C)=OCSCPPACGZOOCGX-UHFFFAOYSA-N0.000description4
- 108090000790EnzymesProteins0.000description4
- 102000004190EnzymesHuman genes0.000description4
- FAPWRFPIFSIZLT-UHFFFAOYSA-MSodium chlorideChemical compound[Na+].[Cl-]FAPWRFPIFSIZLT-UHFFFAOYSA-M0.000description4
- 239000000872bufferSubstances0.000description4
- 238000010276constructionMethods0.000description4
- 230000007812deficiencyEffects0.000description4
- 230000000694effectsEffects0.000description4
- 238000001962electrophoresisMethods0.000description4
- 238000011160researchMethods0.000description4
- PEDCQBHIVMGVHV-UHFFFAOYSA-NGlycerineChemical compoundOCC(O)COPEDCQBHIVMGVHV-UHFFFAOYSA-N0.000description3
- 230000000903blocking effectEffects0.000description3
- 238000005119centrifugationMethods0.000description3
- 239000003153chemical reaction reagentSubstances0.000description3
- 230000029087digestionEffects0.000description3
- 230000006801homologous recombinationEffects0.000description3
- 238000002744homologous recombinationMethods0.000description3
- 239000007788liquidSubstances0.000description3
- 239000008188pelletSubstances0.000description3
- 238000002360preparation methodMethods0.000description3
- 238000012163sequencing techniqueMethods0.000description3
- 239000007787solidSubstances0.000description3
- QGZKDVFQNNGYKY-UHFFFAOYSA-NAmmoniaChemical compoundNQGZKDVFQNNGYKY-UHFFFAOYSA-N0.000description2
- 241000283707CapraSpecies0.000description2
- 108091026890Coding regionProteins0.000description2
- 241000195493CryptophytaSpecies0.000description2
- 241000196324EmbryophytaSpecies0.000description2
- 241000588724Escherichia coliSpecies0.000description2
- KFZMGEQAYNKOFK-UHFFFAOYSA-NIsopropanolChemical compoundCC(C)OKFZMGEQAYNKOFK-UHFFFAOYSA-N0.000description2
- 241001465754MetazoaSpecies0.000description2
- KWYHDKDOAIKMQN-UHFFFAOYSA-NN,N,N',N'-tetramethylethylenediamineChemical compoundCN(C)CCN(C)CKWYHDKDOAIKMQN-UHFFFAOYSA-N0.000description2
- 239000001888PeptoneSubstances0.000description2
- 108010080698PeptonesProteins0.000description2
- 239000007983Tris bufferSubstances0.000description2
- 238000004458analytical methodMethods0.000description2
- 229940041514candida albicans extractDrugs0.000description2
- 238000010367cloningMethods0.000description2
- 238000010586diagramMethods0.000description2
- 238000005516engineering processMethods0.000description2
- ZMMJGEGLRURXTF-UHFFFAOYSA-Nethidium bromideChemical compound[Br-].C12=CC(N)=CC=C2C2=CC=C(N)C=C2[N+](CC)=C1C1=CC=CC=C1ZMMJGEGLRURXTF-UHFFFAOYSA-N0.000description2
- 229960005542ethidium bromideDrugs0.000description2
- 239000000284extractSubstances0.000description2
- 238000000605extractionMethods0.000description2
- 239000011521glassSubstances0.000description2
- 235000021374legumesNutrition0.000description2
- 244000005700microbiomeSpecies0.000description2
- 235000019319peptoneNutrition0.000description2
- 239000012071phaseSubstances0.000description2
- SCVFZCLFOSHCOH-UHFFFAOYSA-Mpotassium acetateChemical compound[K+].CC([O-])=OSCVFZCLFOSHCOH-UHFFFAOYSA-M0.000description2
- 239000011780sodium chlorideSubstances0.000description2
- PFNFFQXMRSDOHW-UHFFFAOYSA-NspermineChemical compoundNCCCNCCCCNCCCNPFNFFQXMRSDOHW-UHFFFAOYSA-N0.000description2
- 239000008223sterile waterSubstances0.000description2
- 239000000126substanceSubstances0.000description2
- LENZDBCJOHFCAS-UHFFFAOYSA-NtrisChemical compoundOCC(N)(CO)COLENZDBCJOHFCAS-UHFFFAOYSA-N0.000description2
- 238000001262western blotMethods0.000description2
- 239000012138yeast extractSubstances0.000description2
- QKNYBSVHEMOAJP-UHFFFAOYSA-N2-amino-2-(hydroxymethyl)propane-1,3-diol;hydron;chlorideChemical compoundCl.OCC(N)(CO)COQKNYBSVHEMOAJP-UHFFFAOYSA-N0.000description1
- FWMNVWWHGCHHJJ-SKKKGAJSSA-N4-amino-1-[(2r)-6-amino-2-[[(2r)-2-[[(2r)-2-[[(2r)-2-amino-3-phenylpropanoyl]amino]-3-phenylpropanoyl]amino]-4-methylpentanoyl]amino]hexanoyl]piperidine-4-carboxylic acidChemical compoundC([C@H](C(=O)N[C@H](CC(C)C)C(=O)N[C@H](CCCCN)C(=O)N1CCC(N)(CC1)C(O)=O)NC(=O)[C@H](N)CC=1C=CC=CC=1)C1=CC=CC=C1FWMNVWWHGCHHJJ-SKKKGAJSSA-N0.000description1
- OPIFSICVWOWJMJ-AEOCFKNESA-N5-bromo-4-chloro-3-indolyl beta-D-galactosideChemical compoundO[C@@H]1[C@@H](O)[C@@H](O)[C@@H](CO)O[C@H]1OC1=CNC2=CC=C(Br)C(Cl)=C12OPIFSICVWOWJMJ-AEOCFKNESA-N0.000description1
- HRPVXLWXLXDGHG-UHFFFAOYSA-NAcrylamideChemical compoundNC(=O)C=CHRPVXLWXLXDGHG-UHFFFAOYSA-N0.000description1
- 229920001817AgarPolymers0.000description1
- 229920000936AgarosePolymers0.000description1
- 244000153158Ammi visnagaSpecies0.000description1
- 235000010585Ammi visnagaNutrition0.000description1
- QGZKDVFQNNGYKY-UHFFFAOYSA-OAmmoniumChemical compound[NH4+]QGZKDVFQNNGYKY-UHFFFAOYSA-O0.000description1
- 241000203069ArchaeaSpecies0.000description1
- 241000589152Azotobacter chroococcumSpecies0.000description1
- 101000623895Bos taurus Mucin-15Proteins0.000description1
- 241000193469Clostridium pasteurianumSpecies0.000description1
- 108020004705CodonProteins0.000description1
- 241000192700CyanobacteriaSpecies0.000description1
- 102000012410DNA LigasesHuman genes0.000description1
- 108010061982DNA LigasesProteins0.000description1
- 102000004163DNA-directed RNA polymerasesHuman genes0.000description1
- 108090000626DNA-directed RNA polymerasesProteins0.000description1
- KCXVZYZYPLLWCC-UHFFFAOYSA-NEDTAChemical compoundOC(=O)CN(CC(O)=O)CCN(CC(O)=O)CC(O)=OKCXVZYZYPLLWCC-UHFFFAOYSA-N0.000description1
- 108010067770Endopeptidase KProteins0.000description1
- 241000206602EukaryotaSpecies0.000description1
- 102000008857FerritinHuman genes0.000description1
- 108050000784FerritinProteins0.000description1
- 238000008416FerritinMethods0.000description1
- 241000588748KlebsiellaSpecies0.000description1
- 108010038629MolybdoferredoxinProteins0.000description1
- 241000108056MonasSpecies0.000description1
- 244000061176Nicotiana tabacumSpecies0.000description1
- 235000002637Nicotiana tabacumNutrition0.000description1
- 239000000020NitrocelluloseSubstances0.000description1
- 239000004677NylonSubstances0.000description1
- 238000012408PCR amplificationMethods0.000description1
- ISWSIDIOOBJBQZ-UHFFFAOYSA-NPhenolChemical compoundOC1=CC=CC=C1ISWSIDIOOBJBQZ-UHFFFAOYSA-N0.000description1
- 241000589194Rhizobium leguminosarumSpecies0.000description1
- 108010006785Taq PolymeraseProteins0.000description1
- 101150067314aadA geneProteins0.000description1
- 239000008272agarSubstances0.000description1
- 239000011543agarose gelSubstances0.000description1
- 238000000246agarose gel electrophoresisMethods0.000description1
- 229910021529ammoniaInorganic materials0.000description1
- 229960000723ampicillinDrugs0.000description1
- AVKUERGKIZMTKX-NJBDSQKTSA-NampicillinChemical compoundC1([C@@H](N)C(=O)N[C@H]2[C@H]3SC([C@@H](N3C2=O)C(O)=O)(C)C)=CC=CC=C1AVKUERGKIZMTKX-NJBDSQKTSA-N0.000description1
- 230000003321amplificationEffects0.000description1
- 230000003698anagen phaseEffects0.000description1
- 239000003242anti bacterial agentSubstances0.000description1
- 229940088710antibiotic agentDrugs0.000description1
- 239000008346aqueous phaseSubstances0.000description1
- QVGXLLKOCUKJST-UHFFFAOYSA-Natomic oxygenChemical compound[O]QVGXLLKOCUKJST-UHFFFAOYSA-N0.000description1
- 239000002585baseSubstances0.000description1
- 230000002457bidirectional effectEffects0.000description1
- 238000010170biological methodMethods0.000description1
- 238000006555catalytic reactionMethods0.000description1
- 210000002421cell wallAnatomy0.000description1
- 238000012512characterization methodMethods0.000description1
- 239000003638chemical reducing agentSubstances0.000description1
- 239000003795chemical substances by applicationSubstances0.000description1
- 229930002875chlorophyllNatural products0.000description1
- 235000019804chlorophyllNutrition0.000description1
- ATNHDLDRLWWWCB-AENOIHSZSA-Mchlorophyll aChemical compoundC1([C@@H](C(=O)OC)C(=O)C2=C3C)=C2N2C3=CC(C(CC)=C3C)=[N+]4C3=CC3=C(C=C)C(C)=C5N3[Mg-2]42[N+]2=C1[C@@H](CCC(=O)OC\C=C(/C)CCC[C@H](C)CCC[C@H](C)CCCC(C)C)[C@H](C)C2=C5ATNHDLDRLWWWCB-AENOIHSZSA-M0.000description1
- 239000012141concentrateSubstances0.000description1
- NKLPQNGYXWVELD-UHFFFAOYSA-Mcoomassie brilliant blueChemical compound[Na+].C1=CC(OCC)=CC=C1NC1=CC=C(C(=C2C=CC(C=C2)=[N+](CC)CC=2C=C(C=CC=2)S([O-])(=O)=O)C=2C=CC(=CC=2)N(CC)CC=2C=C(C=CC=2)S([O-])(=O)=O)C=C1NKLPQNGYXWVELD-UHFFFAOYSA-M0.000description1
- 238000012136culture methodMethods0.000description1
- 230000009089cytolysisEffects0.000description1
- 230000002950deficientEffects0.000description1
- 239000008367deionised waterSubstances0.000description1
- 229910021641deionized waterInorganic materials0.000description1
- 238000013461designMethods0.000description1
- 238000011161developmentMethods0.000description1
- 230000018109developmental processEffects0.000description1
- 238000001035dryingMethods0.000description1
- 238000006911enzymatic reactionMethods0.000description1
- 238000001976enzyme digestionMethods0.000description1
- 239000012458free baseSubstances0.000description1
- 108091008053gene clustersProteins0.000description1
- 230000030279gene silencingEffects0.000description1
- 238000012226gene silencing methodMethods0.000description1
- 230000002068genetic effectEffects0.000description1
- 239000001963growth mediumSubstances0.000description1
- 239000001307heliumSubstances0.000description1
- 229910052734heliumInorganic materials0.000description1
- SWQJXJOGLNCZEY-UHFFFAOYSA-Nhelium atomChemical compound[He]SWQJXJOGLNCZEY-UHFFFAOYSA-N0.000description1
- 238000000265homogenisationMethods0.000description1
- 238000009396hybridizationMethods0.000description1
- 230000006698inductionEffects0.000description1
- 238000003780insertionMethods0.000description1
- 230000037431insertionEffects0.000description1
- BPHPUYQFMNQIOC-NXRLNHOXSA-Nisopropyl beta-D-thiogalactopyranosideChemical compoundCC(C)S[C@@H]1O[C@H](CO)[C@H](O)[C@H](O)[C@H]1OBPHPUYQFMNQIOC-NXRLNHOXSA-N0.000description1
- 238000009630liquid cultureMethods0.000description1
- 239000012160loading bufferSubstances0.000description1
- 239000006166lysateSubstances0.000description1
- 230000010534mechanism of actionEffects0.000description1
- 239000008267milkSubstances0.000description1
- 210000004080milkAnatomy0.000description1
- 235000013336milkNutrition0.000description1
- 238000010369molecular cloningMethods0.000description1
- 230000009456molecular mechanismEffects0.000description1
- 238000002703mutagenesisMethods0.000description1
- 231100000350mutagenesisToxicity0.000description1
- 230000035772mutationEffects0.000description1
- ZIUHHBKFKCYYJD-UHFFFAOYSA-Nn,n'-methylenebisacrylamideChemical compoundC=CC(=O)NCNC(=O)C=CZIUHHBKFKCYYJD-UHFFFAOYSA-N0.000description1
- 229920001220nitrocellulosPolymers0.000description1
- QJGQUHMNIGDVPM-UHFFFAOYSA-Nnitrogen groupChemical group[N]QJGQUHMNIGDVPM-UHFFFAOYSA-N0.000description1
- 238000003199nucleic acid amplification methodMethods0.000description1
- 108020004707nucleic acidsProteins0.000description1
- 102000039446nucleic acidsHuman genes0.000description1
- 150000007523nucleic acidsChemical class0.000description1
- 229920001778nylonPolymers0.000description1
- 239000001301oxygenSubstances0.000description1
- 229910052760oxygenInorganic materials0.000description1
- 230000000243photosynthetic effectEffects0.000description1
- 230000008635plant growthEffects0.000description1
- 235000011056potassium acetateNutrition0.000description1
- 238000001556precipitationMethods0.000description1
- 230000008569processEffects0.000description1
- 239000011535reaction bufferSubstances0.000description1
- 230000006798recombinationEffects0.000description1
- 238000005215recombinationMethods0.000description1
- 238000011084recoveryMethods0.000description1
- 230000001105regulatory effectEffects0.000description1
- 230000008844regulatory mechanismEffects0.000description1
- 230000004044responseEffects0.000description1
- 108091008146restriction endonucleasesProteins0.000description1
- 230000035939shockEffects0.000description1
- 229940063675spermineDrugs0.000description1
- 230000010473stable expressionEffects0.000description1
- 239000000725suspensionSubstances0.000description1
- 238000012360testing methodMethods0.000description1
- 238000013518transcriptionMethods0.000description1
- 230000035897transcriptionEffects0.000description1
- 230000002103transcriptional effectEffects0.000description1
- 238000011426transformation methodMethods0.000description1
- 238000013519translationMethods0.000description1
- YNJBWRMUSHSURL-UHFFFAOYSA-Ntrichloroacetic acidChemical compoundOC(=O)C(Cl)(Cl)ClYNJBWRMUSHSURL-UHFFFAOYSA-N0.000description1
- 239000012137tryptoneSubstances0.000description1
- 229910021642ultra pure waterInorganic materials0.000description1
- 239000012498ultrapure waterSubstances0.000description1
- 238000012795verificationMethods0.000description1
- 238000005303weighingMethods0.000description1
- DGVVWUTYPXICAM-UHFFFAOYSA-Nβ‐MercaptoethanolChemical compoundOCCSDGVVWUTYPXICAM-UHFFFAOYSA-N0.000description1
Images
Landscapes
- Micro-Organisms Or Cultivation Processes Thereof (AREA)
Abstract
Translated fromChineseDescription
Translated fromChinese技术领域technical field
本发明涉及固氮斯氏假单胞菌(Pseudomonas stutzeri)A1501的rpoN基因的表达方法,尤其涉及利用衣藻叶绿体表达系统表达斯氏假单胞菌Pseudomonas stutzeriA1501的rpoN基因的方法,属于固氮斯氏假单胞菌A1501 rpoN基因的表达领域。The invention relates to an expression method of the rpoN gene of Pseudomonas stutzeri (Pseudomonas stutzeri) A1501, in particular to a method for expressing the rpoN gene of Pseudomonas stutzeri A1501 using the Chlamydomonas chloroplast expression system, which belongs to the pseudomonas stutzeri pseudomonas. Expression domain of the rpoN gene of Monas spp. A1501.
背景技术Background technique
氮是构成蛋白质和核酸的主要物质,是自然界中动物、植物、微生物不可缺少的生命元素。氮素在自然界有多种存在形式,数量最大的是大气中的氮气占大气体积的79%,总量约3.9×107亿吨,但是不能为大多数生物(包括所有植物和动物)直接利用,因此氨态氮的供应成为生物界繁荣发展的主要决定因素。目前,固氮主要有以下两种方式,即工业固氮和生物固氮。工业固氮指用高温、高压、化学催化的方法;生物固氮是指某些种类的原核生物利用体内的固氮酶将空气中的氮气还原为氨,为植物生长提供氮素。Nitrogen is the main substance that constitutes protein and nucleic acid, and is an indispensable life element for animals, plants, and microorganisms in nature. Nitrogen exists in many forms in nature. The largest amount is nitrogen in the atmosphere, which accounts for 79% of the volume of the atmosphere, with a total amount of about 3.9×107 billion tons, but it cannot be directly used by most organisms (including all plants and animals). Therefore, the supply of ammoniacal nitrogen has become the main determinant of the prosperity and development of the biological world. At present, nitrogen fixation mainly has the following two methods, namely industrial nitrogen fixation and biological nitrogen fixation. Industrial nitrogen fixation refers to the use of high temperature, high pressure, and chemical catalysis; biological nitrogen fixation refers to the use of nitrogenase in certain types of prokaryotes to reduce nitrogen in the air to ammonia, providing nitrogen for plant growth.
人类对于生物固氮的认识要追溯到十九世纪,距今已有100多年的历史。最早在1830年,Boussingault报道豆科植物可以固氮。Hellriegel & Wilfarth于1886年报道并于1888年发表关于豆科植物能够固氮的可靠依据,并且确定了豆科植物的根瘤是固氮的主要场所。随后不久,厌氧的Clostridium pasteurianum,好养的Azotobacter chroococcum,以及cyanobacteria,photosynthetic bacteria,Klebsiella spp.,Archaebacteria,Desulfovibriosp.,等都加入到固氮的行列。到目前为止,随着研究的不断深入,发现自然界中可进行生物固氮的微生物种属范围分布比较广,可以分成自生固氮、共生固氮和联合共生固氮3种。Human understanding of biological nitrogen fixation can be traced back to the nineteenth century, more than 100 years ago. As early as 1830, Boussingault reported that legumes could fix nitrogen. Hellriegel & Wilfarth reported in 1886 and published in 1888 a reliable basis for the nitrogen fixation of legumes, and determined that the root nodules of legumes are the main site of nitrogen fixation. Soon afterwards, the anaerobic Clostridium pasteurianum, the easy-to-raise Azotobacter chroococcum, and cyanobacteria, photosynthetic bacteria, Klebsiella spp., Archaebacteria, Desulfovibriosp., etc. all joined the ranks of nitrogen fixation. So far, with the deepening of research, it has been found that the microbial species that can carry out biological nitrogen fixation in nature are widely distributed, and can be divided into three types: authigenic nitrogen fixation, symbiotic nitrogen fixation and joint symbiotic nitrogen fixation.
生物固氮主要依靠多数原核生物,这些原核生物之所以能够固氮,是因为固氮酶的缘故。固氮酶是一种复杂的金属酶,由钼铁蛋白和铁蛋白组成(或称为组分I和组分II,也有称之为Dinitrogenase和Dinitrogenase reductant)。固氮作用是一个复杂的过程,其调节是在转录水平上实现的,主要是根据环境中氧气的含量和铵的水平进行调控的。Biological nitrogen fixation mainly relies on most prokaryotes, and the reason why these prokaryotes can fix nitrogen is because of nitrogenase. Nitrogenase is a complex metalloenzyme composed of molybdenum iron protein and ferritin (or called component I and component II, also known as Dinitrogenase and Dinitrogenase reductant). Nitrogen fixation is a complex process that is regulated at the transcriptional level, mainly in response to the amount of oxygen and ammonium in the environment.
固氮斯氏假单胞菌(Pseudomonas stutzeri)A1501的rpoN基因在斯氏假单胞菌A1501的固氮调节作用中起着非常重要的作用。RNA聚合酶需要依靠σ54(rpoN基因的产物)才能启动主要nif基因簇的转录,因此才能保证某些原核生物的固氮作用。The rpoN gene of Pseudomonas stutzeri A1501 plays a very important role in the nitrogen fixation regulation of Pseudomonas stutzeri A1501. RNA polymerase needs to rely on σ54 (the product of the rpoN gene) to initiate the transcription of the major nif gene cluster, thus ensuring nitrogen fixation in some prokaryotes.
为了深入的研究固氮斯氏假单胞菌的固氮作用机理以及rpoN蛋白的功能、特性及纯化方式等,需要有大量的rpoN蛋白作为物质基础。因此,提供一种稳定、高效的表达rpoN基因的方法能够为固氮斯氏假单胞菌的固氮调节作用机制以及rpoN蛋白的功能、特性及纯化方式等研究奠定物质基础。In order to deeply study the mechanism of nitrogen fixation of Pseudomonas azotii and the function, characteristics and purification methods of rpoN protein, a large amount of rpoN protein is needed as the material basis. Therefore, providing a stable and efficient method for expressing the rpoN gene can lay a material foundation for the research on the regulation mechanism of nitrogen fixation of Pseudomonas azostirii and the function, characteristics and purification methods of the rpoN protein.
发明内容Contents of the invention
本发明所要解决的技术问题是克服现有技术的不足,提供一种固氮斯氏假单胞菌A1501的rpoN基因的表达方法,该方法以衣藻叶绿体为表达系统,能够稳定、高效的在衣藻叶绿体中表达rpoN基因。The technical problem to be solved by the present invention is to overcome the deficiencies in the prior art, and to provide a method for expressing the rpoN gene of Pseudomonas azotii A1501, which uses the Chlamydomonas chloroplast as the expression system, and can stably and efficiently express The rpoN gene was expressed in algal chloroplasts.
本发明所要解决的技术问题是通过以下技术方案来实现的:The technical problem to be solved by the present invention is achieved through the following technical solutions:
固氮斯氏假单胞菌(Pseudomonas stutzeri)A1501的rpoN基因的表达方法,包括:构建rpoN基因的表达盒;将所构建的rpoN基因的表达盒插入到衣藻叶绿体表达载体上的衣藻叶绿体基因组的同源片段之间,构建得到重组衣藻叶绿体表达载体;将重组衣藻叶绿体表达载体转化衣藻;筛选转基因衣藻,诱导转基因衣藻中的rpoN基因表达,收获并纯化所表达的蛋白。The expression method of the rpoN gene of Pseudomonas stutzeri (Pseudomonas stutzeri) A1501, comprising: constructing the expression cassette of the rpoN gene; inserting the expression cassette of the constructed rpoN gene into the Chlamydomonas chloroplast genome on the Chlamydomonas chloroplast expression vector The recombinant Chlamydomonas chloroplast expression vector was constructed between the homologous fragments; the recombinant Chlamydomonas chloroplast expression vector was transformed into Chlamydomonas; the transgenic Chlamydomonas was screened, the rpoN gene expression in the transgenic Chlamydomonas was induced, and the expressed protein was harvested and purified.
其中,所述的斯氏假单胞菌Pseudomonas stutzeri A1501的rpoN基因的核苷酸序列为SEQ ID NO:1所示;Wherein, the nucleotide sequence of the rpoN gene of the Pseudomonas stutzeri A1501 is shown in SEQ ID NO: 1;
所述的rpoN基因的表达盒由衣藻叶绿体特异启动子、rpoN基因和衣藻叶绿体特异终止子所组成,其中,rpoN基因置于衣藻叶绿体基因特异启动子和终止子的调控之下;所述的衣藻叶绿体特异启动子包括但不限于atpA、atpB、psbA、psbD或rbcL,优选为atpA启动子;所述的衣藻叶绿体特异终止子优选为rbcL终止子。The expression cassette of the rpoN gene is composed of a Chlamydomonas chloroplast-specific promoter, an rpoN gene and a Chlamydomonas chloroplast-specific terminator, wherein the rpoN gene is placed under the regulation of the Chlamydomonas chloroplast gene-specific promoter and terminator; The Chlamydomonas chloroplast-specific promoter includes but not limited to atpA, atpB, psbA, psbD or rbcL, preferably the atpA promoter; the Chlamydomonas chloroplast-specific terminator is preferably the rbcL terminator.
所述的衣藻叶绿体基因组的同源片段为trnE2-psbH基因,其核苷酸序列为SEQID NO:2所示;The homologous fragment of the Chlamydomonas chloroplast genome is the trnE2-psbH gene, and its nucleotide sequence is shown in SEQ ID NO: 2;
本发明中所用到的衣藻(Chlamydomonas reinhardtii)可以为任何一种野生型莱茵衣藻,均能实现本发明的技术效果;为了达到更好的技术效果,本发明所述的衣藻(Chlamydomonas reinhardtii)更优选为品系cw15;Chlamydomonas reinhardtii used in the present invention can be any wild type Chlamydomonas reinhardtii, all can realize technical effect of the present invention; In order to achieve better technical effect, Chlamydomonas reinhardtii described in the present invention ) is more preferably strain cwl5;
其中,所述的重组衣藻叶绿体表达载体转化衣藻的方式优选采用包裹有重组衣藻叶绿体表达载体的基因枪轰击处于生长对数期后期的衣藻并通过无醋酸盐的培养基筛选得到转基因衣藻(衣藻转化子),实验结果发现,本发明所筛选得到的转基因衣藻aadA的抗性消失。Wherein, the method of transforming the recombinant Chlamydomonas chloroplast expression vector into Chlamydomonas is preferably obtained by bombarding the Chlamydomonas in the late logarithmic phase of growth with a particle gun wrapped with the recombinant Chlamydomonas chloroplast expression vector and screening with an acetate-free medium As for the transgenic Chlamydomonas (Chlamydomonas transformant), the experimental results show that the resistance of the transgenic Chlamydomonas aadA screened by the present invention disappears.
本发明采用基因同源重组技术,将来源于斯氏假单胞菌Pseudomonas stutzeriA1501的rpoN基因构建到由衣藻叶绿体特异启动子atpA和终止子rbcL组合所驱动的表达盒,利用衣藻叶绿体基因组中的trnE2-psbH作为同源片段,发生同源重组,通过基因枪转化法导入衣藻叶绿体中,通过PCR、Western blot等分子生物学手段检测最终获得转基因衣藻,证明斯氏假单胞菌Pseudomonas stutzeri A1501的rpoN基因在衣藻叶绿体中能够稳定、高效的表达。The present invention adopts gene homologous recombination technology to construct the rpoN gene derived from Pseudomonas stutzeriA1501 into an expression cassette driven by the combination of the Chlamydomonas chloroplast-specific promoter atpA and the terminator rbcL, and utilizes the rpoN gene in the Chlamydomonas chloroplast genome As a homologous fragment, trnE2-psbH undergoes homologous recombination and is introduced into the Chlamydomonas chloroplast by the gene gun transformation method. The transgenic Chlamydomonas is finally obtained by PCR, Western blot and other molecular biological methods, proving that Pseudomonas stutzeri The rpoN gene of A1501 can be expressed stably and efficiently in Chlamydomonas chloroplast.
衣藻(Chlamydomonas reinhardtii)作为较简单的单细胞真核生物,日益吸引了细胞和分子生物学等研究领域的注意力。衣藻(Chlamydomonas reinhardtii)是单细胞绿藻,呈椭圆形,细胞大小大约长10μm,宽3μm,具有单个杯状叶绿体,约占细胞总体积的40%-60%。本发明采用衣藻叶绿体作为rpoN基因的表达系统,不仅弥补了核转化的不足,而且还具有以下几方面的优点:第一,本发明通过在转化载体上设计叶绿体基因组的同源片段,通过同源重组作用实现rpoN基因定点插入,避免位置效应和基因沉默的产生,保证了rpoN基因的稳定表达。第二,衣藻叶绿体基因组遗传表达体系具有原核性。莱茵衣藻叶绿体基因组基因的排列方式、调控方式、GC碱基对含量及翻译所偏爱的密码子与原核生物相接近,这有利于直接表达rpoN基因。第三,rpoN基因在衣藻叶绿体中可稳定、高效的进行表达。Chlamydomonas reinhardtii, as a relatively simple unicellular eukaryote, has increasingly attracted the attention of research fields such as cell and molecular biology. Chlamydomonas reinhardtii is a single-celled green alga, oval in shape, with a cell size of about 10 μm in length and 3 μm in width, with a single cup-shaped chloroplast, accounting for about 40%-60% of the total cell volume. The present invention adopts Chlamydomonas chloroplast as the expression system of rpoN gene, which not only makes up for the deficiencies of nuclear transformation, but also has the following advantages: first, the present invention designs homologous fragments of chloroplast genome on the transformation vector, through simultaneous Source recombination realizes site-specific insertion of rpoN gene, avoids position effect and gene silencing, and ensures stable expression of rpoN gene. Second, the Chlamydomonas chloroplast genome genetic expression system is prokaryotic. The arrangement, regulation, GC base pair content and translation preference codons of Chlamydomonas reinhardtii chloroplast genome genes are close to those of prokaryotes, which is conducive to the direct expression of rpoN gene. Third, the rpoN gene can be stably and efficiently expressed in Chlamydomonas chloroplasts.
本发明方法成功的将固氮斯氏假单胞菌Pseudomonas stutzeri A1501的rpoN基因在衣藻叶绿体中稳定、高效的表达,采用本发明方法能够通过衣藻叶绿体表达系统获得大量的重组rpoN蛋白,为展开固氮斯氏假单胞菌的作用机制以及rpoN蛋白的纯化、功能及特性等研究奠定了物质基础。The method of the present invention successfully expresses the rpoN gene of Pseudomonas stutzeri A1501 in the Chlamydomonas chloroplast stably and efficiently, and adopts the method of the present invention to obtain a large amount of recombinant rpoN protein through the Chlamydomonas chloroplast expression system. The mechanism of action of Pseudomonas azotii and the purification, function and characteristics of rpoN protein have laid a material foundation.
附图说明Description of drawings
图1转目的基因rpoN的转基因衣藻PCR检测结果;1、野生型衣藻;2、转化子样品1;3、转化子样品2。Fig. 1 PCR detection results of transgenic Chlamydomonas transgenic with target gene rpoN; 1, wild-type Chlamydomonas; 2,
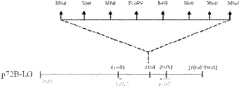
图2载体p72B-LG的结构简图。Fig. 2 Schematic diagram of the structure of the vector p72B-LG.
图3质粒paptY的酶切图谱。Fig. 3 Restriction map of plasmid paptY.
具体实施方式Detailed ways
下面结合具体实施例来进一步描述本发明,本发明的优点和特点将会随着描述而更为清楚。应了解本发明不限于本文中所述的特定方法、实验方案、细胞系、构建体和试剂且因此可加以变化。也应了解本文中所用的方法是仅用于描述特定实施例的目的,且并不意欲限制本发明的范畴,本发明的范畴将仅由随附权利要求书所限制。The present invention will be further described below in conjunction with specific embodiments, and the advantages and characteristics of the present invention will become clearer along with the description. It is to be understood that this invention is not limited to the particular methodology, protocols, cell lines, constructs and reagents described herein and as such may vary. It should also be understood that the methodologies used herein are for the purpose of describing particular embodiments only, and are not intended to limit the scope of the invention which will be limited only by the appended claims.
说明:在以下实施例中未作详细介绍的技术,均按照以下实验手册或文献中的相关章节或部分来进行,包括:Sambrook等人,Molecular Cloning,A Laboratory Manual(第3版.2001);Kriegler,Gene Transfer and Expression:A Laboratory Manual(1990);和Current Protocols in Molecular Biology(Ausubel等人编,1994)。Explanation: the technology that does not introduce in detail in the following examples is all carried out according to the relevant chapters or parts in the following experimental manuals or documents, including: Sambrook et al., Molecular Cloning, A Laboratory Manual (3rd edition. 2001); Kriegler, Gene Transfer and Expression: A Laboratory Manual (1990); and Current Protocols in Molecular Biology (eds. Ausubel et al., 1994).
除非另外定义,否则本发明所用的所有技术及科学术语都具有与本发明所属领域的普通技术人员通常所了解相同的含义。虽然在本发明的实践或测试中可使用与本文所述者类似或等效的任何方法、装置和材料,但现在描述优选方法、装置和材料。Unless otherwise defined, all technical and scientific terms used herein have the same meaning as commonly understood by one of ordinary skill in the art to which this invention belongs. Although any methods, devices and materials similar or equivalent to those described herein can be used in the practice or testing of the present invention, the preferred methods, devices and materials are now described.
实验材料Experimental Materials
1.E.coli TOP10购自Promega公司;固氮斯氏假单胞菌Pseudomonas stutzeriA1501的rpoN基因由中国农业科学院生物技术所保存;野生型莱茵衣藻(Chlamydomonas reinhardtti)CW-15(说明:本发明中所述的CW-15可以由任何一种野生型莱茵衣藻所代替,均能实现本发明的技术效果)由中国农业科学院生物技术研究所保存,具体培养方法和特性可以参考相关的文献(Hyams,J.and D.R.Davies,Theinduction and characterisation of cell wall mutants of Chlamydomonas reinhardtii.Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis,1972.14(4):p.381-389);paptY、p72B-LG由中国农业科学院生物技术研究所保存;1.E.coli TOP10 is purchased from Promega Company; The rpoN gene of Pseudomonas stutzeriA1501 is preserved by Biotechnology Institute of Chinese Academy of Agricultural Sciences; Wild type Chlamydomonas reinhardtti CW-15 (illustration: in the present invention Described CW-15 can be replaced by any kind of wild-type Chlamydomonas reinhardtii, all can realize the technical effect of the present invention) is preserved by Institute of Biotechnology, Chinese Academy of Agricultural Sciences, specific culture method and characteristic can refer to relevant literature (Hyams , J.and D.R.Davies, The induction and characterization of cell wall mutants of Chlamydomonas reinhardtii.Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis, 1972.14(4): p.381-389); paptY, p72B-LG by the Chinese Academy of Agricultural Sciences kept by the Institute of Technology;
载体p72B-LG的插入核苷酸序列为SEQ ID No.3所示,其结构简图见图1,其中,插入序列为2.2kb,pUC9载体为2.67kb,载体p72B-LG的全序列为4.87kb。The inserted nucleotide sequence of the vector p72B-LG is shown in SEQ ID No.3, and its structure diagram is shown in Figure 1. Among them, the inserted sequence is 2.2kb, the pUC9 vector is 2.67kb, and the full sequence of the vector p72B-LG is 4.87 kb.
质粒paptY(pBlutscript II-aptY)的载体图谱见图2,关于paptY的构建可按照有关文献构建得到该质粒paptY(Expression of human soluble TRAIL in Chlamydomonasreinhardtii chloroplast.Chinese Science Bulletin 2006 Vol.51 No.14 1703-1709)。The vector map of plasmid paptY (pBlutscript II-aptY) is shown in Figure 2. The construction of paptY can be constructed according to relevant literature to obtain the plasmid paptY (Expression of human soluble TRAIL in Chlamydomonasreinhardtii chloroplast. Chinese Science Bulletin 2006 Vol.51 No.14 1703- 1709).
酶与试剂:各种限制性内切酶、T4DNA连接酶、Klenow酶及其配套缓冲液购自Promega公司;Enzymes and reagents: various restriction endonucleases,T4 DNA ligase, Klenow enzyme and supporting buffers were purchased from Promega;
2.生化试剂:IPTG、X-Gal为Promega公司产品;琼脂糖为Sun biotech公司产品;Tris、dNTPs购自Sigma公司;蛋白胨、酵母抽提物、胰蛋白胨均购自OXOID公司;溴化乙锭(EB)购自Fluka公司;HRP标记的羊抗兔rpoN抗体;TEMED(N,N,N’,N’-四甲基乙二胺)购自瑞典LKB BROMMA公司;SDS为BDH公司产品;β-巯基乙醇,Tris购自Sigma公司;考马斯亮兰R-250、N,N′-甲叉双丙烯酰胺购自瑞士FlukaChemie AG公司;丙烯酰胺为Aldrich化学有限公司产品;硝酸纤维素膜Hybond-N和尼龙膜Hybond-C均为Amersham产品;2. Biochemical reagents: IPTG and X-Gal are products of Promega Company; agarose is a product of Sun Biotech Company; Tris and dNTPs are purchased from Sigma Company; peptone, yeast extract and tryptone are purchased from OXOID Company; ethidium bromide (EB) was purchased from Fluka; HRP-labeled goat anti-rabbit rpoN antibody; TEMED (N, N, N', N'-tetramethylethylenediamine) was purchased from LKB BROMMA, Sweden; SDS was the product of BDH; -Mercaptoethanol, Tris was purchased from Sigma; Coomassie Brilliant Blue R-250 and N,N′-methylenebisacrylamide were purchased from FlukaChemie AG, Switzerland; acrylamide was a product of Aldrich Chemical Co., Ltd.; nitrocellulose membrane Hybond-N And nylon membrane Hybond-C are Amersham products;
3.培养基:大肠杆菌培养基为LB培养基(1%蛋白胨、0.5%酵母提取物、1% NaCl,pH7.0);衣藻培养基为TAP培养基;3. Medium: Escherichia coli medium is LB medium (1% peptone, 0.5% yeast extract, 1% NaCl, pH7.0); Chlamydomonas medium is TAP medium;
实施例固氮斯氏假单胞菌Pseudomonas stutzeri A1501的rpoN基因的获得、重组衣藻叶绿体表达载体的构建和转化以及转基因衣藻的鉴定Example: Obtaining the rpoN gene of Pseudomonas stutzeri A1501, construction and transformation of recombinant Chlamydomonas chloroplast expression vector and identification of transgenic Chlamydomonas
1.固氮斯氏假单胞菌Pseudomonas stutzeri A1501的rpoN基因的克隆和表达载体的构建1. Cloning of the rpoN gene of Pseudomonas stutzeri A1501 and construction of an expression vector
1.1目的基因的获得1.1 Obtaining the target gene
根据已知的固氮斯氏假单胞菌Pseudomonas stutzeri A1501的rpoN的序列,分别设计引物,并且分别引入XbaI和HindIII位点,两对引物如下:According to the sequence of the known rpoN of Pseudomonas stutzeri A1501, primers were designed respectively, and XbaI and HindIII sites were introduced respectively, and two pairs of primers were as follows:
F rpoN:5′-ATCTAGAATGAAGCAAGGTTTGCAATTAAG-3′F rpoN: 5′-ATCTAGA ATGAAGCAAGGTTTGCAATTAAG-3′
XbaI酶切位点XbaI restriction site
R rpoN:5′-AAAGCTTTCAGACCAGTTGTTTACGCTG-3′R rpoN: 5′-AAAGCTT TCAGACCAGTTGTTTACGCTG-3′
HindIII酶切位点HindIII restriction site
在0.5mL Eppendorf管中加入Add to 0.5mL Eppendorf tube
反应程序:Reaction procedure:
1.2产物的回收1.2 Recovery of products
用普通凝胶进行电泳,切下所需DNA条带,称重后放入Eppendorf管中,加入3倍体积(v/w)的6mol/L NaI溶液,37℃下使凝胶充分溶解,再加入10μL玻璃奶吸附DNA,室温下放置5min,稍离心5s,去上清,沉淀用New Wash洗液洗涤一次,重复离心、洗涤三次,晾干后用30μL 0.1×TE Buffer溶解DNA,离心后取上清,将DNA保存于-20℃备用。Perform electrophoresis with ordinary gel, cut out the required DNA band, put it into an Eppendorf tube after weighing, add 3 times the volume (v/w) of 6mol/L NaI solution, and fully dissolve the gel at 37°C, then Add 10 μL of glass milk to absorb DNA, place it at room temperature for 5 minutes, centrifuge for 5 seconds, remove the supernatant, wash the precipitate once with New Wash washing solution, repeat centrifugation and washing three times, dissolve DNA with 30 μL 0.1×TE Buffer after drying, and take out after centrifugation. The supernatant and the DNA were stored at -20°C for later use.
1.3酶切和连接反应1.3 Digestion and ligation reaction
酶切和连接反应参照试剂盒产品说明中各种酶的最适反应条件进行。反应体系包含质粒DNA 1~2μg,1/10体积的10×酶反应缓冲液,酶量为2-4M/μg DNA,总体积为15~20μL。最适温度(一般为37℃)下反应1~2h,用琼脂糖凝胶电泳鉴定酶切是否完全。The digestion and ligation reactions were carried out referring to the optimal reaction conditions for various enzymes in the product instructions of the kit. The reaction system contains 1-2 μg of plasmid DNA, 1/10 volume of 10× enzyme reaction buffer, the amount of enzyme is 2-4M/μg DNA, and the total volume is 15-20 μL. React at the optimum temperature (generally 37°C) for 1 to 2 hours, and use agarose gel electrophoresis to identify whether the digestion is complete.
1.4质粒DNA或连接产物的转化1.4 Transformation of plasmid DNA or ligation product
取-70℃冻存的感受态细胞,用双手搓至大部分融化后(一定不能完全融化)迅速置冰上。感受态细胞完全融化后加入质粒DNA20~100ng或连接混合物5μL,轻轻混匀,冰浴30min。42℃热激90sec,迅速置冰上1~2min。加入1mL已温育至37℃的LB培养基,37℃振荡(≤150rpm)培养1h。4,000g离心5min,去部分上清后(留150~200μL)涂布于含适当抗生素的LB平板上。37℃倒置培养过夜。Take the competent cells frozen at -70°C, rub them with both hands until most of them melt (it must not be completely melted), and put them on ice quickly. After the competent cells were completely thawed, add 20-100ng of plasmid DNA or 5μL of the ligation mixture, mix gently, and ice-bath for 30min. Heat shock at 42°C for 90 sec, and quickly place on ice for 1-2 min. Add 1 mL of LB medium that has been incubated to 37°C, and incubate at 37°C with shaking (≤150rpm) for 1 hour. Centrifuge at 4,000 g for 5 min, remove part of the supernatant (retain 150-200 μL) and smear it on an LB plate containing appropriate antibiotics. Incubate overnight at 37°C.
1.5碱裂解法提取质粒DNA1.5 Alkaline lysis method to extract plasmid DNA
(1)用无菌牙签从LB平板上挑取单菌落,接种于4mL含100μg/mL氨卡青霉素的LB液体培养基中,37℃,230r/min,振荡培养过夜;(1) Use a sterile toothpick to pick a single colony from the LB plate, inoculate it in 4 mL of LB liquid medium containing 100 μg/mL ampicillin, culture at 37°C, 230 r/min, with shaking overnight;
(2)取1.5mL过夜培养物于Eppendorf管内,5,000rpm离心5min收集菌体,弃上清;(2) Take 1.5 mL of the overnight culture in an Eppendorf tube, centrifuge at 5,000 rpm for 5 min to collect the bacteria, and discard the supernatant;
(3)用150μL Sol I悬浮沉淀,冰浴10min;(3) Suspend the precipitate with 150 μL Sol I, and ice-bath for 10 minutes;
(4)加300μL Sol II和150μL氯仿,轻轻倒转混匀后冰浴5min;(4) Add 300 μL Sol II and 150 μL chloroform, gently invert and mix well, then ice bath for 5 minutes;
(5)加450μL SolIII,倒转混匀后冰浴10min;(5) Add 450 μL SolIII, invert and mix well, then ice bath for 10 min;
(6)12,000rpm离心10min,取上清,加0.6倍体积异丙醇,混匀后于4℃放置20min;(6) Centrifuge at 12,000rpm for 10min, take the supernatant, add 0.6 times the volume of isopropanol, mix well and place at 4°C for 20min;
(7)12,000rpm离心10min,弃上清,沉淀溶于250μL TER(含20μg/mL RNaseA的1×TE)中,37℃消化20min,加入300μL PPt沉淀Buffer,混匀后置4℃20min;(7) Centrifuge at 12,000rpm for 10min, discard the supernatant, dissolve the pellet in 250μL TER (1×TE containing 20μg/mL RNaseA), digest at 37℃ for 20min, add 300μL PPt precipitation buffer, mix well and place at 4℃ for 20min;
(8)12,000rpm离心10min,弃上清,70%乙醇洗一次,真空干燥沉淀,用80μL0.1×TE(pH8.0)溶解,-20℃保存备用。(8) Centrifuge at 12,000 rpm for 10 min, discard the supernatant, wash once with 70% ethanol, dry the precipitate in vacuo, dissolve in 80 μL 0.1×TE (pH 8.0), and store at -20°C for later use.
1.6重组子的鉴定1.6 Identification of recombinants
酶切鉴定:首先小量抽提质粒DNA,然后利用XbaI和HindIII进行酶切反应,电泳后出现约1.37kb的DNA条带的质粒为重组质粒pEASY-T3-rpoN。将重组质粒pEASY-T3-rpoN进行双向测序,测序结果与预期结果一致。Enzyme digestion identification: First, a small amount of plasmid DNA was extracted, and then digested with XbaI and HindIII. After electrophoresis, the plasmid with a DNA band of about 1.37kb was the recombinant plasmid pEASY-T3-rpoN. The recombinant plasmid pEASY-T3-rpoN was subjected to bidirectional sequencing, and the sequencing results were consistent with the expected results.
2 重组衣藻叶绿体表达载体的构建2 Construction of recombinant Chlamydomonas chloroplast expression vector
质粒patpY含有衣藻叶绿体基因组特异启动子5’atpA和终止子3’rbcL及由BamHI起始的一组多克隆位点。将测序正确的阳性重组质粒pEASY-T3-rpoN,用XbaI和HindIII双酶切,将回收的目的片段,连入patpY载体XbaI和HindIII之间,使rpoN片段置于衣藻叶绿体基因特异启动子5’atpA和终止子3’rbcL调控之下,得到质粒patpY-rpoN;再用EcoRV和Not I双酶切,并补平NotI位点,回收含有rpoN编码区表达盒的2.6kbDNA片段,将其克隆入同源重组片段trnE-psbH的p72B-LG质粒EcoRV位点,构建成编码衣藻叶绿体表达载体p72B-LG-rpoN。Plasmid patpY contains Chlamydomonas chloroplast genome-specific promoter 5'atpA, terminator 3'rbcL and a set of multiple cloning sites initiated by BamHI. The positive recombinant plasmid pEASY-T3-rpoN with correct sequencing was digested with XbaI and HindIII, and the recovered target fragment was connected between XbaI and HindIII of the patpY vector, so that the rpoN fragment was placed in the Chlamydomonas chloroplast gene-
3 基因枪转化3 Biolistic transformation
3.1衣藻的培养及衣藻受体细胞的准备3.1 Cultivation of Chlamydomonas and preparation of Chlamydomonas recipient cells
野生型衣藻生长在TAP液体培养基中,20~25℃,160rpm,光周期12hr/12hr,参阅文献Harris(1989)(The Chlamydomonas soursebooke,1989 Academic Press,San Diego)。Wild type Chlamydomonas grows in TAP liquid medium, 20~25 ℃, 160rpm, photoperiod 12hr/12hr, refer to literature Harris (1989) (The Chlamydomonas soursebooke, 1989 Academic Press, San Diego).
取1.5mL生长至对数后期(5~6×106cells/mL)的衣藻于离心管中浓缩(5,000rpm,20℃,5min),去上清,留200μL,涂于TAP固体平板上,3000Lux的光照条件下培养1~2天,用无菌铲子挖出均匀地长有一层衣藻的直径为2~3cm的培养基块,放入9cm无菌平皿中央,以备轰击。Take 1.5 mL of Chlamydomonas grown to late logarithmic phase (5-6×106 cells/mL) and concentrate in a centrifuge tube (5,000 rpm, 20°C, 5 min), remove the supernatant, leave 200 μL, and spread on the TAP solid plate , under the light condition of 3000Lux, cultivated for 1-2 days, dug out with a sterile shovel a medium block with a diameter of 2-3 cm that evenly grew a layer of Chlamydomonas, and put it into the center of a 9 cm sterile plate for bombardment.
3.2微粒子弹(DNA coated microprojectiles)的制备3.2 Preparation of DNA coated microprojectiles
称60mg 1.0μm的金粉(Bio-Rad)于1.5mL Eppendorf管中;加入1mL无水乙醇,充分涡旋(vortex),然后静置10min,10,000rpm离心10sec;小心去除上清液,加入1mL无菌水,充分涡旋,10,000rpm离心10sec,弃上清液;重复用无菌水洗涤2次;用1mL无菌的50%(v/v)甘油重悬金粉颗粒。处理后的金粉可在-20℃长期保存;Weigh 60mg of 1.0μm gold powder (Bio-Rad) in a 1.5mL Eppendorf tube; add 1mL of absolute ethanol, fully vortex (vortex), then let it stand for 10min, and centrifuge at 10,000rpm for 10sec; carefully remove the supernatant, add 1mL of Sterilized water, vortex fully, centrifuge at 10,000rpm for 10sec, discard supernatant; repeat washing with sterile water twice; resuspend gold powder particles with 1mL sterile 50% (v/v) glycerol. The treated gold powder can be stored at -20°C for a long time;
取50μl上述制备好并已涡旋的金粉颗粒,转入一无菌的1.5mL eppendorf管中,按顺序加入5μL DNA(1μg/μL)、50μl 2.5mol/L CaCl2和20μL 0.1mol/L亚精胺(Freebase,现配现用);将混合物涡旋1min,置于冰上1min;重复10次;置于冰上30min以上;15,000rpm离心5sec,去除上清液;用250μL无水乙醇洗涤包被好的金粉颗粒2次,10,000rpm离心10sec,去除上清液;60μL无水乙醇重悬金粉颗粒。Take 50 μl of the above-prepared and vortexed gold powder particles, transfer to a sterile 1.5mL eppendorf tube, add 5 μL DNA (1 μg/μL), 50 μl 2.5mol/L CaCl2 and 20 μL 0.1mol/L sub Spermine (Freebase, ready to use); vortex the mixture for 1 min, place on ice for 1 min; repeat 10 times; place on ice for more than 30 min; centrifuge at 15,000 rpm for 5 sec, remove the supernatant; wash with 250 μL absolute ethanol The coated gold powder particles were centrifuged twice at 10,000 rpm for 10 sec, and the supernatant was removed; the gold powder particles were resuspended in 60 μL of absolute ethanol.
3.3装备基因枪3.3 Equip gene gun
(1)基因枪的准备(1) Preparation of gene gun
将基因枪放置于一个较大型号的超净工作台上,打开超净台紫外灯灭菌30min以上;用70%酒精擦净基因枪真空室及各种元件;用浸泡消毒法将可裂膜(Ruptμre disk1,100psi)、载体膜(Macrocarrier)、阻挡网(Stopping screen)及Macrocarrier holders于无水乙醇中消毒15min,然后将Rupture disk和Macrocarrier置于无菌滤纸上,在超净台中吹干;用镊子夹住Macrocarrier holder,在酒精灯火焰上稍事灼烧后置于无菌滤纸上;待Macrocarrier holder冷却后,用镊子将Macrocarrier置于其中并展平。取10μL包被DNA的金粉颗粒无水乙醇悬浮液,点于Macrocarrier中心位置,于超净工作台中吹干;打开电源开关、真空泵及氦气瓶阀门;取出Macrocarrier launch assembly,拧开盖子,将Stopping screen在酒精灯火焰上灼烧后置于凹槽内,把Macrocarrier holder倒扣于凹槽上,旋紧盖子;将Rupture disk装于固定盖中并旋紧;将组装好的Macrocarrier launchassembly置于基因枪真空室的上数第二个格子中;将装有烟草培养基块的培养皿置于Target shelf中心,并将Target shelf置于真空室的上数第四个格子中,使轰击距离为9cm;关紧真空室小门。Place the gene gun on a large ultra-clean workbench, turn on the ultra-clean bench ultraviolet lamp to sterilize for more than 30 minutes; clean the vacuum chamber and various components of the gene gun with 70% alcohol; (Ruptμre disk1, 100psi), carrier film (Macrocarrier), blocking screen (Stopping screen) and Macrocarrier holders were sterilized in absolute ethanol for 15 minutes, then placed Rupture disk and Macrocarrier on sterile filter paper, and dried in an ultra-clean bench; Clamp the Macrocarrier holder with tweezers, burn it on the flame of an alcohol lamp and place it on sterile filter paper; after the Macrocarrier holder cools down, place the Macrocarrier in it with tweezers and flatten it. Take 10 μL of DNA-coated gold powder particle suspension in absolute ethanol, point it at the center of the Macrocarrier, and blow it dry on the ultra-clean workbench; turn on the power switch, vacuum pump and helium cylinder valve; take out the Macrocarrier launch assembly, unscrew the cover, and put the Stopping After burning the screen on the flame of an alcohol lamp, place it in the groove, buckle the Macrocarrier holder upside down on the groove, and screw the cover tightly; put the Rupture disk in the fixed cover and screw it tightly; put the assembled Macrocarrier launch assembly in the gene In the second grid from the top of the gun vacuum chamber; place the petri dish with the tobacco culture block in the center of the Target shelf, and place the Target shelf in the fourth grid from the top of the vacuum chamber, so that the bombardment distance is 9cm ; Close the small door of the vacuum chamber.
(2)轰击(2) Bombardment
抽真空:按VAC键,当真空度达到25~28inches Hg时,将VAC键直接锁定在HOLD位置;Vacuuming: Press the VAC button, when the vacuum reaches 25-28 inches Hg, directly lock the VAC button in the HOLD position;
轰击:按住FIRE键,直至Rupture disk爆裂;再按VENT键,使真空表指针回零。打开真空室小门,取出样品。通常每个样品轰击2次。Bombardment: Press and hold the FIRE key until the Rupture disk bursts; then press the VENT key to return the pointer of the vacuum gauge to zero. Open the small door of the vacuum chamber and take out the sample. Typically each sample is bombarded twice.
4 转基因衣藻的筛选4 Screening of transgenic Chlamydomonas
4.1转化后衣藻的过度培养4.1 Overculture of Chlamydomonas after transformation
轰击后的衣藻置于3000Lux的光照条件下培养8h(20~25℃),再用1mL无菌水把琼脂块上的衣藻洗下,涂于5皿不含醋酸盐的抗性选择培养基(TAP)上培养。After bombardment, Chlamydomonas was cultured under 3000 Lux light conditions for 8 hours (20-25°C), and then washed with 1 mL of sterile water to wash Chlamydomonas on the agar block, and applied to 5 dishes for resistance selection without acetate. culture medium (TAP).
4.2转化衣藻的筛选培养4.2 Screening and cultivation of transformed Chlamydomonas
将涂于醋酸盐缺失的选择培养基上的衣藻在3000Lux的连续光照条件下培养约10天左右,挑取单藻落于醋酸盐缺失的体选择培养基(TAP)中培养。Cultivate Chlamydomonas smeared on acetate-depleted selection medium for about 10 days under continuous light conditions of 3000 Lux, pick single algae and culture them in acetate-depleted selection medium (TAP).
4.3转化衣藻的验证反应4.3 Verification reaction for transformation of Chlamydomonas
取部分于醋酸盐缺失的选择培养基(TAP)中正常生长的衣藻,涂布于固体抗性选择培养基(TAP+Spc 100μg/mL)上培养,大概7-10天衣藻死亡。Take part of Chlamydomonas that normally grows in acetate-deficient selection medium (TAP), and spread it on solid resistance selection medium (TAP+Spc 100μg/mL) for culture, and Chlamydomonas dies in about 7-10 days.
4.4衣藻转化子的同质化培养4.4 Homogeneous culture of Chlamydomonas transformants
尽管衣藻细胞内只有一个叶绿体,但是叶绿体内却含有80~100个叶绿体基因组拷贝,最初转化的衣藻叶绿体内既含有转基因的基因组,也含有野生型基因组拷贝,因此它的叶绿体基因组是异质的,需要进一步的同质化筛选。Although there is only one chloroplast in the Chlamydomonas cell, the chloroplast contains 80 to 100 copies of the chloroplast genome. The chloroplast of the initially transformed Chlamydomonas contains both the transgenic genome and the wild-type genome copy, so its chloroplast genome is heterogeneous. Yes, further homogenization screening is required.
将TAP液体选择培养基(醋酸盐缺失)中培养7天左右的转化子,重新涂布于固体选择培养基TAP(醋酸盐缺失)上培养,而后再挑取单藻落转到液体培养基(醋酸盐缺失)中培养,如此重复多轮进行继代培养,以达到提高外源基因在叶绿体基因组中的同质化程度。Transformants cultured in TAP liquid selection medium (acetate deficiency) for about 7 days were re-coated on solid selection medium TAP (acetate deficiency) for culture, and then single algae colonies were picked and transferred to liquid culture Cultured in medium (without acetate), repeated several rounds of subculture in order to improve the degree of homogeneity of exogenous genes in the chloroplast genome.
5 转基因衣藻的检测5 Detection of transgenic Chlamydomonas
5.1衣藻DNA的提取5.1 Extraction of Chlamydomonas DNA
(1)取10mL处于对数生长后期的衣藻培养液,5,000rpm,4℃离心5min;(1) Take 10 mL of Chlamydomonas culture solution in late logarithmic growth, centrifuge at 5,000 rpm, 4°C for 5 min;
(2)衣藻细胞沉淀用350μL NET溶液(0.1mol/LNaCl,50mM EDTA,20mM Tris-HCl,pH 8.0)悬浮后,加入25μL Proteinase K(10mg/mL),25μL 20% SDS,混匀,55℃水浴2h。(2) Suspend Chlamydomonas cell pellet with 350 μL NET solution (0.1mol/L NaCl, 50mM EDTA, 20mM Tris-HCl, pH 8.0), add 25μL Proteinase K (10mg/mL), 25μL 20% SDS, mix well, 55 ℃ water bath for 2h.
(3)置于冰上冷却,加入200μL 5M乙酸钾(KAc),冰上静置30min。(3) Cool on ice, add 200 μL of 5M potassium acetate (KAc), and let stand on ice for 30 minutes.
(4)12,000rpm,4℃离心5min,上清液加入等体积的酚/氯仿(1∶1)抽提两次,再用等体积的氯仿抽提一次。水相加入2倍体积的无水乙醇,混匀后置于-20℃静置20min。(4) Centrifuge at 12,000 rpm at 4°C for 5 min, add an equal volume of phenol/chloroform (1:1) to the supernatant to extract twice, and then extract once with an equal volume of chloroform. Add 2 times the volume of absolute ethanol to the aqueous phase, mix well, and then place it at -20°C for 20 minutes.
(5)12,000rpm,4℃离心5min,沉淀用70%乙醇洗涤,真空干燥后溶于30μL超纯水中。(5) Centrifuge at 12,000 rpm at 4°C for 5 min, wash the precipitate with 70% ethanol, dry in vacuum and dissolve in 30 μL of ultrapure water.
5.2转基因衣藻PCR检测5.2 PCR detection of transgenic Chlamydomonas
(1)衣藻转化子rpoN编码区DNA的PCR检测(1) PCR detection of rpoN coding region DNA of Chlamydomonas transformants
PCR体系:PCR system:
反应程序为:The reaction procedure is:
取4μL PCR产物上样于1%琼脂糖凝胶上电泳,在衣藻转化子中分别扩增获得与预期大小完全一致的片段,而在野生型衣藻中则没有相应的条带出现(图1)。Take 4 μL of the PCR product and load it on a 1% agarose gel for electrophoresis. In the Chlamydomonas transformants, the fragments with the expected size were amplified respectively, but no corresponding bands appeared in the wild-type Chlamydomonas (Fig. 1).
(2)叶绿体转基因衣藻同质化程度的PCR检测(2) PCR detection of homogeneity degree of chloroplast transgenic Chlamydomonas
P1:GCCTCGCCTATCGGCTAACAAGP1: GCCTCGCCTATCGGCTAACAAG
P2:GTAAATTCAGACTTCCAAGAACP2: GTAAATTCAGACTTCCAAGAAC
以转基因衣藻DNA为模板,用LA Taq DNA聚合酶进行PCR扩增。Using the transgenic Chlamydomonas DNA as a template, PCR amplification was performed with LA Taq DNA polymerase.
PCR体系:PCR system:
反应程序为:The reaction procedure is:
扩增结果,经过4轮抗性筛选后的分别转rpoN基因的衣藻叶绿体转化子,都扩出了一条0.86kb的条带和一条3.5kb的条带,大小与预期结果相符,说明斯氏假单胞菌Pseudomonas stutzeri A1501 rpoN基因定点插入到衣藻叶绿体基因组中。As a result of the amplification, after four rounds of resistance screening, a 0.86 kb band and a 3.5 kb band were expanded in the Chlamydomonas chloroplast transformants that were respectively transfected with the rpoN gene, and the size was in line with the expected result, indicating that Stellaris The rpoN gene of Pseudomonas stutzeri A1501 was site-directedly inserted into the Chlamydomonas chloroplast genome.
5.3衣藻总蛋白的提取5.3 Extraction of Chlamydomonas total protein
(1)将生长期为对数后期的10mL衣藻培养液,离心收集;(1) Collect 10 mL of the Chlamydomonas culture solution whose growth phase is in the late logarithmic period, and collect it by centrifugation;
(2)沉淀用200μL裂解液(2% SDS,50mM Tris-Cl pH 7.5,5%β-巯基乙醇)重悬后煮沸5min;(2) The precipitate was resuspended in 200 μL lysate (2% SDS, 50mM Tris-Cl pH 7.5, 5% β-mercaptoethanol) and boiled for 5 minutes;
(3)12000rpm,离心5min,上清加入三氯乙酸至终浓度为10%,混匀;(3) Centrifuge at 12000 rpm for 5 minutes, add trichloroacetic acid to the supernatant to a final concentration of 10%, and mix well;
(4)12000rpm,离心10min,沉淀用90%丙酮洗涤,将大部分叶绿素抽提掉;(4) Centrifuge at 12000rpm for 10min, wash the precipitate with 90% acetone, and extract most of the chlorophyll;
(5)待丙酮挥发掉后,沉淀重悬于加样缓冲液,以备蛋白分析。(5) After the acetone evaporates, the pellet is resuspended in loading buffer for protein analysis.
5.4Western blot检测5.4 Western blot detection
(1)取总蛋白进行电泳;(1) Take the total protein for electrophoresis;
(2)剪下所需大小的NC膜,然后将膜转入转移缓冲液中至少5min;同时也将凝胶浸入转移缓冲液中;(2) Cut off the NC membrane of the required size, and then transfer the membrane to the transfer buffer for at least 5 minutes; at the same time, immerse the gel in the transfer buffer;
(3)在电转仪上依次铺上滤纸、NC膜、凝胶、滤纸,用玻璃棒赶去气泡,以1.5mA/cm2凝胶面积的恒定电流转印1.5h;(3) Lay filter paper, NC membrane, gel, and filter paper on the electroporator in sequence, drive out air bubbles with a glass rod, and transfer with a constant current of 1.5mA/cm2 gel area for 1.5h;
(4)取出转印好的膜,放入PBS中轻轻摇动洗膜10min;(4) Take out the transferred membrane, put it into PBS and shake gently to wash the membrane for 10min;
(5)倾去PBS,加封闭液室温轻摇1h;(5) Pour off PBS, add blocking solution and shake at room temperature for 1 hour;
(6)将膜浸入含第一抗体(1∶1500)的封闭液中,4℃轻摇过夜;(6) Immerse the membrane in the blocking solution containing the primary antibody (1:1500), shake gently at 4°C overnight;
(7)将膜放入洗涤液中轻轻摇动洗膜3次,每次10min;(7) Put the membrane into the washing solution and shake gently to wash the
(8)将膜转入含HRP标记的羊抗兔二抗(1∶1000)的PBS中,室温轻摇1h;(8) Transfer the membrane into PBS containing HRP-labeled goat anti-rabbit secondary antibody (1:1000), shake gently at room temperature for 1 h;
(9)将膜放入洗涤液中轻轻摇动洗膜3次,每次10min;(9) Put the membrane into the washing solution and shake gently to wash the
(10)将膜转入PBS中,轻摇洗膜2次,每次5min;(10) Transfer the membrane into PBS, gently shake and wash the membrane twice, 5 min each time;
(11)将膜转入新配的DAB显色液中,室温下暗处显色5-10min,待有明显的显色条带时,用去离子水漂洗终止反应。(11) Transfer the membrane into the newly prepared DAB color developing solution, and develop the color in the dark at room temperature for 5-10 minutes. When there are obvious color bands, rinse with deionized water to terminate the reaction.
结果在52kDa位置处都出现特异杂交信号条带,而未转基因的野生型衣藻则没有出现可见的信号条带。这说明rpoN蛋白在三株衣藻转化子的叶绿体中得到了表达。The results showed specific hybridization signal bands at the position of 52kDa, but no visible signal bands appeared in the non-transgenic wild-type Chlamydomonas. This indicated that the rpoN protein was expressed in the chloroplasts of the three strains of Chlamydomonas transformants.
结果分析表明,本发明利用衣藻的叶绿体成功表达了斯氏假单胞菌Pseudomonasstutzeri A1501 rpoN的蛋白。The result analysis shows that the present invention successfully expresses the protein of Pseudomonasstutzeri A1501 rpoN by using the Chlamydomonas chloroplast.
<110> 中国农业科学院生物技术研究所<110> Institute of Biotechnology, Chinese Academy of Agricultural Sciences
the
<120> 固氮斯氏假单胞菌A1501 rpoN基因的表达方法<120> Expression method of Pseudomonas azotii A1501 rpoN gene
the
<130> dqxl0026<130> dqxl0026
the
<160> 3 <160> 3
the
<170> PatentIn version 3.5<170> PatentIn version 3.5
the
<210> 1<210> 1
<211> 1374<211> 1374
<212> DNA<212> DNA
<213> Pseudomonas stutzeri<213> Pseudomonas stutzeri
the
<400> 1<400> 1
atgctcgagc gtgaagagga cggtgacgac ttcgacggct ccgaccccat ggccgaggcc atgctcgagc gtgaagagga cggtgacgac ttcgacggct ccgaccccat ggccgaggcc
6060
the
gtcgatcgtt cgagcgacgc tggcagcgaa aagatcagta ctcaggaaag cacctatgaa gtcgatcgtt cgagcgacgc tggcagcgaa aagatcagta ctcaggaaag cacctatgaa
120120
the
gagccttcgc acagcggcga aggcctcgac gaggccgact ggggtgagcg cattcccagc gagccttcgc acagcggcga aggcctcgac gaggccgact ggggtgagcg cattcccagc
180180
the
gagctaccgg tcgacaccgc ctgggaagac atctaccaga ccagcgccag cagcctgccg gagctaccgg tcgacaccgc ctgggaagac atctaccaga ccagcgccag cagcctgccg
240240
the
agcaacgacg acgacgaatg ggacttcacc agccgtacgt ccagcggtgt cagcctgcag agcaacgacg acgacgaatg ggacttcacc agccgtacgt ccagcggtgt cagcctgcag
300300
the
agccatctgc tgtggcagct caacctcgcc cccatgagcg acaccgaccg cctgatcgcc agccatctgc tgtggcagct caacctcgcc cccatgagcg acaccgaccg cctgatcgcc
360360
the
accacactga tcgactgcat caacgatcag ggctatctcg aggagtctct gcaggaaatc accacactga tcgactgcat caacgatcag ggctatctcg aggagtctct gcaggaaatc
420420
the
gtggacagct tcgacccgga gcttgaaatc gagctcgacg aggtcgaagt cgtgctgcgt gtggacagct tcgacccgga gcttgaaatc gagctcgacg aggtcgaagt cgtgctgcgt
480480
the
cgcgtacagc agttcgaacc tgccggtatc ggcgcacgag atttgcgcga atgcctgctc cgcgtacagc agttcgaacc tgccggtatc ggcgcacgag atttgcgcga atgcctgctc
540540
the
ctgcagctgc gccagctccc tgaccgcacc ccttggctca gcgaagccca gcggctggtg ctgcagctgc gccagctccc tgaccgcacc ccttggctca gcgaagccca gcggctggtg
600600
the
agtgaccatc tggacctgtt gggtagccgc gactacagcc tgctgatgcg ccgcatgaag agtgaccatc tggacctgtt gggtagccgc gactacagcc tgctgatgcg ccgcatgaag
660660
the
ctcaaggaag acgagctgcg ccaggtcatc gagctcatcc agagcctcaa ccctcgcccg ctcaaggaag acgagctgcg ccaggtcatc gagctcatcc agagcctcaa ccctcgcccg
720720
the
ggctcgcaga tcgaggcgag cgagccggag tacgtggtgc cggacgtcat cgtacgcaag ggctcgcaga tcgaggcgag cgagccggag tacgtggtgc cggacgtcat cgtacgcaag
780780
the
cacaacgatc gctggttggt ggagctcaac caggacgcca tgccccgact tcgcgtcaat cacaacgatc gctggttggt ggagctcaac caggacgcca tgccccgact tcgcgtcaat
840840
the
gcccagtacg ccagcttcgt aaagcgcgcc gattcgagtg ccgacaatac cttcatgcgc gcccagtacg ccagcttcgt aaagcgcgcc gattcgagtg ccgacaatac cttcatgcgc
900900
the
aaccagctgc aggaagcgcg ctggttcatc aagagcctgc tcagtcgcaa cgaaacactg aaccagctgc aggaagcgcg ctggttcatc aagagcctgc tcagtcgcaa cgaaacactg
960960
the
atgaaggtcg ccacccagat cgtcgagcac cagcgtgcct tcctcgaaca cggcgacgag atgaaggtcg ccaccgat cgtcgagcac cagcgtgcct tcctcgaaca cggcgacgag
10201020
the
gcgatgaagc cgctggtgct gcacgacatc gccgaagccg tggggatgca tgagtcgacg gcgatgaagc cgctggtgct gcacgacatc gccgaagccg tggggatgca tgagtcgacg
10801080
the
atttcccgcg tcaccacgca gaaatacatg catacgccgc gtggcatcta cgaactcaaa atttcccgcg tcaccacgca gaaatacatg catacgccgc gtggcatcta cgaactcaaa
11401140
the
tacttcttct ccagccacgt cagcacggcg gaaggcggcg aatgttcgtc caccgcgatc tacttcttct ccagccacgt cagcacggcg gaaggcggcg aatgttcgtc caccgcgatc
12001200
the
cgcgcgatca tcaagaagct ggtcgccgcg gaaaacccga aaaagccgtt gagcgacagc cgcgcgatca tcaagaagct ggtcgccgcg gaaaacccga aaaagccgtt gagcgacagc
12601260
the
aagatcgctg gtttactgga ggcgcagggc atccaggtgg cccgccgcac cgttgcgaaa aagatcgctg gtttactgga ggcgcagggc atccaggtgg cccgccgcac cgttgcgaaa
13201320
the
taccgcgagt cgctcggtat cgcaccgtcc agcgagcgta aacgattgct gtga taccgcgagt cgctcggtat cgcaccgtcc agcgagcgta aacgattgct gtga
13741374
the
the
<210> 2<210> 2
<211> 1214<211> 1214
<212> DNA<212> DNA
<213> Chlamydomonas reinhardtii<213> Chlamydomonas reinhardtii
the
<400> 2<400> 2
gaatccgcgt tttctccgtg aaagggaggt gtcctaggcc tctagacgat gggggctttt gaatccgcgt tttctccgtg aaagggaggt gtcctaggcc tctagacgat gggggctttt
6060
the
tgttatattt tactaaatat atattataat taaaaaaaat tgaattgtca atttttaatg tgttatattt tactaaatat atattataat taaaaaaaat tgaattgtca atttttaatg
120120
the
tacacttagt tgaaagtgcc cctgtcccct tggccatatt taacagaagt tatttataac tacacttagt tgaaagtgcc cctgtcccct tggccatatt taacagaagt tattataac
180180
the
gcagctgttt tttggagtct ataaatttat aacatcagtt actatggatt tccctttagt gcagctgttt tttggagtct ataaatttat aacatcagtt actatggatt tccctttagt
240240
the
tttatggcct aggacgtccc cttccccttc gatgctggag gcatcctttt acgggacaat tttatggcct aggacgtccc cttccccttc gatgctggag gcatcctttt acgggacaat
300300
the
aaataaattt gttgcctcgc ctatcggcta acaagttcct tcggagtata taaatatagg aaataaattt gttgcctcgc ctatcggcta acaagttcct tcggagtata taaatatagg
360360
the
atgttaatac tgctataaac tttagttgcc caatatttat attaggacgc cagtggcagt atgttaatac tgctataaac tttagttgcc caatatttat attaggacgc cagtggcagt
420420
the
ggtaccgcca ctgcctgctt cgcagtatat aaatataggc agttggcagg caactgccac ggtaccgcca ctgcctgctt cgcagtatat aaatataggc agttggcagg caactgccac
480480
the
tgacgtccta ttttaatact cccaagttta cttgcctagg cagttggcag gcaacaaatt tgacgtccta ttttaatact cccaagttta cttgcctagg cagttggcag gcaacaaatt
540540
the
tatttattgt ccactaaaat ttatttgccc gaaggggacg tccactaaaa tttatttacc tatttattgt ccactaaaat ttatttgccc gaaggggacg tccactaaaa tttatttacc
600600
the
cgaaggggac gtcctaatat aaatatgggg atgtcaatgc tccgttagga agtaactaac cgaaggggac gtcctaatat aaatatgggg atgtcaatgc tccgttagga agtaactaac
660660
the
gtttttcaaa taaattttat cccggaggga agtaggcagt agcccgccac tgtcatcctt gtttttcaaa taaattttat cccggaggga agtaggcagt agcccgccac tgtcatcctt
720720
the
taagtggatc tctcgtcagg caatttgctt acacctttaa attaaaaatt aaatttaaag taagtggatc tctcgtcagg caatttgctt acacctttaa attaaaaatt aaatttaaag
780780
the
aaaagtgagc tattaacgcg tttatcttaa cggaaggcca gtggcagttg gcggtgccac aaaagtgagc tattaacgcg tttatcttaa cggaaggcca gtggcagttg gcggtgccac
840840
the
tgccgaatat aaatatggtt gagttgctta gtttacctta gcgaaaagaa gacttagcag tgccgaatat aaatatggtt gagttgctta gtttacctta gcgaaaagaa gacttagcag
900900
the
ctagccttaa caaacagttt tatattttat gtttgtgtta aataaaatta agaaacttta ctagccttaa caaacagttt tatattttat gtttgtgtta aataaaatta agaaacttta
960960
the
gctaaagttt cccaactcat agaaacgtca tctaaaatta aagaactgtt gtaaatttct gctaaagttt cccaactcat agaaacgtca tctaaaatta aagaactgtt gtaaatttct
10201020
the
aaaatgatta ataagaatgc tgcaaataaa aggataaata cagccattaa aacagttgta aaaatgatta ataagaatgc tgcaaataaa aggataaata cagccattaa aacagttgta
10801080
the
ccccagcctg gtaatacttt acctgcttct gagttaagtg gacgtaataa agtacctaat ccccagcctg gtaatacttt acctgcttct gagttaagtg gacgtaataa agtacctaat
11401140
the
ggtgtaacta aaccaggttc ttggaagtct gaatttactt ttgatggttt agctttagaa ggtgtaacta aaccaggttc ttggaagtct gaatttactt ttgatggttt agctttagaa
12001200
the
gttcctgttg ccat gttcctgttg ccat
12141214
the
the
<210> 3<210> 3
<211> 2201<211> 2201
<212> DNA<212> DNA
<213> Artifical sequence<213> Artificial sequence
the
<400> 3<400> 3
gaattcgaat ccgcgttttc tccgtgaaag ggaggtgtcc taggcctcta gacgatgggg gaattcgaat ccgcgttttc tccgtgaaag ggaggtgtcc taggcctcta gacgatgggg
6060
the
gctttttgtt atattttact aaatatatat tataattaaa aaaaattgaa ttgtcaattt gctttttgtt atattttact aaatatatat tataattaaa aaaaattgaa ttgtcaattt
120120
the
ttaatgtaca cttagttgaa agtgcccctg tccccttggc catatttaac agaagttatt ttaatgtaca cttagttgaa agtgcccctg tccccttggc catatttaac agaagttat
180180
the
tataacgcag ctgttttttg gagtctataa atttataaca tcagttacta tggatttccc tataacgcag ctgttttttg gagtctataa atttataaca tcagttacta tggatttccc
240240
the
tttagtttta tggcctagga cgtccccttc cccttcgatg ctggaggcat ccttttacgg tttagtttta tggcctagga cgtccccttc cccttcgatg ctggaggcat ccttttacgg
300300
the
gacaataaat aaatttgttg cctcgcctat cggctaacaa gttccttcgg agtatataaa gacaataaat aaatttgttg cctcgcctat cggctaacaa gttccttcgg agtatataaa
360360
the
tataggatgt taatactgct ataaacttta gttgcccaat atttatatta ggacgccagt tataggatgt taatactgct ataaacttta gttgcccaat atttatatta ggacgccagt
420420
the
ggcagtggta ccgccactgc ctgcttcgca gtatataaat ataggcagtt ggcaggcaac ggcagtggta ccgccactgc ctgcttcgca gtatataaat ataggcagtt ggcaggcaac
480480
the
tgccactgac gtcctatttt aatactccca agtttacttg cctaggcagt tggcaggcaa tgccactgac gtcctatttt aatactccca agtttacttg cctaggcagt tggcaggcaa
540540
the
caaatttatt tattgtccac taaaatttat ttgcccgaag gggacgtcca ctaaaattta caaatttatt tattgtccac taaaatttat ttgcccgaag gggacgtcca ctaaaattta
600600
the
tttacccgaa ggggacgtcc taatataaat atggggatgt caatgctccg ttaggaagta tttacccgaa ggggacgtcc taatataaat atggggatgt caatgctccg ttaggaagta
660660
the
actaacgttt ttcaaataaa ttttatcccg gagggaagta ggcagtagcc cgccactgtc actaacgttt ttcaaataaa ttttatcccg gagggaagta ggcagtagcc cgccactgtc
720720
the
atcctttaag tggatctctc gtcaggcaat ttgcttacac ctttaaatta aaaattaaat atcctttaag tggatctctc gtcaggcaat ttgcttacac ctttaaatta aaaattaaat
780780
the
ttaaagaaaa gtgagctatt aacgcgtact agtcaattgg atatcagatc tgcggccgcc ttaaagaaaa gtgagctatt aacgcgtact agtcaattgg atatcagatc tgcggccgcc
840840
the
tcgagacgcg tttatcttaa cggaaggcca gtggcagttg gcggtgccac tgccgaatat tcgagacgcg tttatcttaa cggaaggcca gtggcagttg gcggtgccac tgccgaatat
900900
the
aaatatggtt gagttgctta gtttacctta gcgaaaagaa gacttagcag ctagccttaa aaatatggtt gagttgctta gtttacctta gcgaaaagaa gacttagcag ctagccttaa
960960
the
caaacagttt tatattttat gtttgtgtta aataaaatta agaaacttta gctaaagttt caaacagttt tatattttat gtttgtgtta aataaaatta agaaacttta gctaaagttt
10201020
the
cccaactcat agaaacgtca tctaaaatta aagaactgtt gtaaatttct aaaatgatta cccaactcat agaaacgtca tctaaaatta aagaactgtt gtaaatttct aaaatgatta
10801080
the
ataagaatgc tgcaaataaa aggataaata cagccattaa aacagttgta ccccagcctg ataagaatgc tgcaaataaa aggataaata cagccattaa aacagttgta ccccagcctg
11401140
the
gtaatacttt acctgcttct gagttaagtg gacgtaataa agtacctaat ggtgtaacta gtaatacttt acctgcttct gagttaagtg gacgtaataa agtacctaat ggtgtaacta
12001200
the
aaccaggttc ttggaagtct gaatttactt ttgatggttt agctttagaa gttcctgttg aaccaggttc ttggaagtct gaatttactt ttgatggttt agctttagaa gttcctgttg
12601260
the
ccataattga ttaaatgaat taagcgttat tagcgctatt ttatttactt tctgtaaaaa ccataattga ttaaatgaat taagcgttat tagcgctatt ttatttactt tctgtaaaaa
13201320
the
ataaggaaaa tattcttcag tgcattccct ctcaggatta taaatactct gaggataacg ataaggaaaa tattcttcag tgcattccct ctcaggatta taaatactct gaggataacg
13801380
the
ttctctcgtc aaggggttgc ttcttgtgag tatagaaacc tactagcaca agaaataaat ttctctcgtc aaggggttgc ttcttgtgag tatagaaacc tactagcaca agaaataaat
14401440
the
tgcataaaaa tgtatttacc taggaccgca gtaggcagtc ccttttcccc ttcagaactg tgcataaaaa tgtatttacc taggaccgca gtaggcagtc ccttttcccc ttcagaactg
15001500
the
cctgctttaa aagaatgaaa aaactgcctt gtctggtaag taaaactctt taattactca cctgctttaa aagaatgaaa aaactgcctt gtctggtaag taaaactctt taattactca
15601560
the
ctaaagacga tcttagaagt tctttgttca ttttttattt aatataatat ttgttatata ctaaagacga tcttagaagt tctttgttca ttttttatt aatataatat ttgttatata
16201620
the
aaaattaaat aatttttaat taatgtttaa ctttgtaagg acagtttcaa agtgacatga aaaattaaat aatttttaat taatgtttaa ctttgtaagg acagtttcaa agtgacatga
16801680
the
atggctactg caaaaacgaa gtaagttatt ctttctcagg gcaaaatttt gagtagatta atggctactg caaaaacgaa gtaagttat ctttctcagg gcaaaatttt gagtagatta
17401740
the
attttgttta aaaatgtggg acacagtcgt caagtctttt gaactatcta agagatatgt attttgttta aaaatgtggg acagtcgt caagtctttt gaactatcta agagatatgt
18001800
the
tgaaaagaga ataattttat tattaaatga gctatggaaa gtccagcttt tttctttacc tgaaaagaga ataattttat tattaaatga gctatggaaa gtccagcttt tttctttacc
18601860
the
ttttttttat ggtttcttct gttaagtgta actggctatt cagtttatgt tagttttggt ttttttttat ggtttcttct gttaagtgta actggctatt cagtttatgt tagttttggt
19201920
the
ccaccttcaa aaaaattacg tgatcctttt gaagaacacg aagattaaac aagttaaaaa ccaccttcaa aaaaattacg tgatcctttt gaagaacacg aagattaaac aagttaaaaa
19801980
the
gtactatttt tacaagtgac ttcggtgcct ctgagaaccc tagttatagt gatataaaat gtactatttt tacaagtgac ttcggtgcct ctgagaaccc tagttatagt gatataaaat
20402040
the
aactagctaa ctactttata tttttatgaa agtcattttg tcgagcatat aaacaaaaac aactagctaa ctactttata tttttatgaa agtcattttg tcgagcatat aaacaaaaac
21002100
the
aaaattgcta tactaggcag tcacagtgca actgtctccg tctccttaac cgagaaaggg aaaattgcta tactaggcag tcacagtgca actgtctccg tctccttaac cgagaaaggg
21602160
the
taaacgtctt cggtaaagta acaaacttta gttatgttcc c taaacgtctt cggtaaagta acaaacttta gttatgttcc c
22012201
Claims (9)
Translated fromChinesePriority Applications (1)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| CN2011103267044ACN102417882A (en) | 2011-10-25 | 2011-10-25 | Expression method of pseudomonas stutzeri A1501 rpoN gene |
Applications Claiming Priority (1)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| CN2011103267044ACN102417882A (en) | 2011-10-25 | 2011-10-25 | Expression method of pseudomonas stutzeri A1501 rpoN gene |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| CN102417882Atrue CN102417882A (en) | 2012-04-18 |
Family
ID=45942526
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| CN2011103267044APendingCN102417882A (en) | 2011-10-25 | 2011-10-25 | Expression method of pseudomonas stutzeri A1501 rpoN gene |
Country Status (1)
| Country | Link |
|---|---|
| CN (1) | CN102417882A (en) |
Cited By (10)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN103614410A (en)* | 2013-11-20 | 2014-03-05 | 中国农业科学院生物技术研究所 | Application of rsmA gene in colonization and growth promotion of root surface of plant |
| CN111527057A (en)* | 2017-10-25 | 2020-08-11 | 皮沃特生物股份有限公司 | Gene targets for nitrogen fixation targeting improved plant traits |
| US11479516B2 (en) | 2015-10-05 | 2022-10-25 | Massachusetts Institute Of Technology | Nitrogen fixation using refactored NIF clusters |
| US11565979B2 (en) | 2017-01-12 | 2023-01-31 | Pivot Bio, Inc. | Methods and compositions for improving plant traits |
| US11739032B2 (en) | 2015-07-13 | 2023-08-29 | Pivot Bio, Inc. | Methods and compositions for improving plant traits |
| US11946162B2 (en) | 2012-11-01 | 2024-04-02 | Massachusetts Institute Of Technology | Directed evolution of synthetic gene cluster |
| US11993778B2 (en) | 2017-10-25 | 2024-05-28 | Pivot Bio, Inc. | Methods and compositions for improving engineered microbes that fix nitrogen |
| US12209245B2 (en) | 2011-06-16 | 2025-01-28 | The Regents Of The University Of California | Synthetic gene clusters |
| US12281299B2 (en) | 2019-03-19 | 2025-04-22 | Massachusetts Institute Of Technology | Control of nitrogen fixation in rhizobia that associate with cereals |
| US12281980B2 (en) | 2020-05-01 | 2025-04-22 | Pivot Bio, Inc. | Measurement of nitrogen fixation and incorporation |
Citations (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO2010061404A1 (en)* | 2008-11-27 | 2010-06-03 | International Centre For Genetic Engineering And Biotechnology | A method of expressing foreign protein in plastids |
| CN101812468A (en)* | 2009-11-20 | 2010-08-25 | 中国农业科学院生物技术研究所 | Method for preparing rabies virus antigen by chloroplast |
- 2011
- 2011-10-25CNCN2011103267044Apatent/CN102417882A/enactivePending
Patent Citations (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO2010061404A1 (en)* | 2008-11-27 | 2010-06-03 | International Centre For Genetic Engineering And Biotechnology | A method of expressing foreign protein in plastids |
| CN101812468A (en)* | 2009-11-20 | 2010-08-25 | 中国农业科学院生物技术研究所 | Method for preparing rabies virus antigen by chloroplast |
Non-Patent Citations (3)
| Title |
|---|
| YAN Y. 等: "登录号:CP000304.1", 《GENBANK》, 28 May 2008 (2008-05-28)* |
| 刘国宪 等: "衣藻叶绿体表达系统的研究", 《生物技术通报》, no. 9, 31 December 2009 (2009-12-31)* |
| 韩四海 等: "外源基因在莱茵衣藻叶绿体中的表达", 《生物技术通报》, no. 1, 31 December 2007 (2007-12-31)* |
Cited By (13)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US12209245B2 (en) | 2011-06-16 | 2025-01-28 | The Regents Of The University Of California | Synthetic gene clusters |
| US11946162B2 (en) | 2012-11-01 | 2024-04-02 | Massachusetts Institute Of Technology | Directed evolution of synthetic gene cluster |
| CN103614410B (en)* | 2013-11-20 | 2016-02-24 | 中国农业科学院生物技术研究所 | RsmA gene plant root table surely grow with growth-promoting in application |
| CN103614410A (en)* | 2013-11-20 | 2014-03-05 | 中国农业科学院生物技术研究所 | Application of rsmA gene in colonization and growth promotion of root surface of plant |
| US11739032B2 (en) | 2015-07-13 | 2023-08-29 | Pivot Bio, Inc. | Methods and compositions for improving plant traits |
| US11479516B2 (en) | 2015-10-05 | 2022-10-25 | Massachusetts Institute Of Technology | Nitrogen fixation using refactored NIF clusters |
| US11565979B2 (en) | 2017-01-12 | 2023-01-31 | Pivot Bio, Inc. | Methods and compositions for improving plant traits |
| JP2023100703A (en)* | 2017-10-25 | 2023-07-19 | ピボット バイオ, インコーポレイテッド | Gene targets for nitrogen fixation to improve plant traits |
| US11993778B2 (en) | 2017-10-25 | 2024-05-28 | Pivot Bio, Inc. | Methods and compositions for improving engineered microbes that fix nitrogen |
| US12151988B2 (en) | 2017-10-25 | 2024-11-26 | Pivot Bio, Inc. | Gene targets for nitrogen fixation targeting for improving plant traits |
| CN111527057A (en)* | 2017-10-25 | 2020-08-11 | 皮沃特生物股份有限公司 | Gene targets for nitrogen fixation targeting improved plant traits |
| US12281299B2 (en) | 2019-03-19 | 2025-04-22 | Massachusetts Institute Of Technology | Control of nitrogen fixation in rhizobia that associate with cereals |
| US12281980B2 (en) | 2020-05-01 | 2025-04-22 | Pivot Bio, Inc. | Measurement of nitrogen fixation and incorporation |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| CN102417882A (en) | Expression method of pseudomonas stutzeri A1501 rpoN gene | |
| CN107400677B (en) | Bacillus licheniformis genome editing vector based on CRISPR-Cas9 system and preparation method thereof | |
| CN113604490B (en) | Kiwi canker susceptibility gene AcBXL1 and its application | |
| Ianiri et al. | Approaches for genetic discoveries in the skin commensal and pathogenic Malassezia yeasts | |
| CN109971773A (en) | A kind of gene dsmI encoding gentisic acid dioxygenase DsmI that can degrade 3-chlorogentisic acid and its application | |
| CN113403209A (en) | Application of aspartic protease gene in improving beauveria bassiana variety | |
| CN105505950A (en) | Novel application of five tobacco metallothionein genes | |
| KR20160111947A (en) | Method of producing a recombinant microorganism | |
| CN101696414A (en) | Gene capable of improving radiation resistance of organisms and application thereof | |
| CN116121288B (en) | Vector for cloning pseudomonas putida large fragment DNA and application thereof | |
| CN107164256A (en) | A kind of method of Sphingol single-cell genetic transformation | |
| CN110591993A (en) | A method based on hydrochloric acid-stimulated homologous recombination gene knockout of Vibrio harvey | |
| CN111808832A (en) | A cation transporting ATPase gene of Rhizoctonia oryzae and its fragment Rscta and its application | |
| US20250084424A1 (en) | Chlamydomonas reinhardtii chloroplast-saccharomyces cerevisiae-escherichia coli shuttle vector and construction method and use thereof | |
| CN112175981B (en) | Method for site-directed gene knockout of vibrio harveyi based on stimulation of absolute ethyl alcohol or sodium dodecyl sulfonate | |
| CN105980564A (en) | A modified microorganism having enhanced biomass synthesis capacity and a method thereof | |
| CN112592926A (en) | CRISPR system and application thereof in mortierella alpina | |
| JP6893820B2 (en) | Methods to improve resistance of nitric acid to substrate analogs in microalgae | |
| CN109837280A (en) | A kind of endogenous temperature inducible promoter of seaweed and its application | |
| CN105177049B (en) | A kind of method of transformed bacillus Bacillus strain | |
| CN114875031B (en) | A nodule-specific expression promoter LjNAD1 and its application in nodule-specific expression | |
| CN110592081A (en) | Xylose Inducible Promoter and Its Plasmid Vector and Application | |
| CN111549039B (en) | A LbKN1 gene derived from "Lubao No. 1" and its application | |
| CN101948856B (en) | Recombinant lactobacillus rhamnosus engineering strain and preparation method thereof | |
| CN102311972B (en) | Method and product for preparing porcine circovirus type Ⅱ antigen by using chloroplast |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| C06 | Publication | ||
| PB01 | Publication | ||
| C10 | Entry into substantive examination | ||
| SE01 | Entry into force of request for substantive examination | ||
| C12 | Rejection of a patent application after its publication | ||
| RJ01 | Rejection of invention patent application after publication | Application publication date:20120418 |