CN101831429B - Identification of promoters and expression patterns of endosperm-specific genes in rice - Google Patents
Identification of promoters and expression patterns of endosperm-specific genes in riceDownload PDFInfo
- Publication number
- CN101831429B CN101831429BCN2010101510338ACN201010151033ACN101831429BCN 101831429 BCN101831429 BCN 101831429BCN 2010101510338 ACN2010101510338 ACN 2010101510338ACN 201010151033 ACN201010151033 ACN 201010151033ACN 101831429 BCN101831429 BCN 101831429B
- Authority
- CN
- China
- Prior art keywords
- enp3
- expression
- endosperm
- promoter
- promoters
- Prior art date
- Legal status (The legal status is an assumption and is not a legal conclusion. Google has not performed a legal analysis and makes no representation as to the accuracy of the status listed.)
- Expired - Fee Related
Links
- 230000014509gene expressionEffects0.000titleclaimsabstractdescription127
- 235000007164Oryza sativaNutrition0.000titleclaimsabstractdescription64
- 235000009566riceNutrition0.000titleclaimsabstractdescription56
- 108090000623proteins and genesProteins0.000titleabstractdescription38
- 240000007594Oryza sativaSpecies0.000titledescription9
- 241000209094OryzaSpecies0.000claimsabstractdescription55
- 239000002773nucleotideSubstances0.000claimsdescription18
- 125000003729nucleotide groupChemical group0.000claimsdescription18
- 230000002068genetic effectEffects0.000claimsdescription13
- 230000006872improvementEffects0.000claimsdescription6
- 241000196324EmbryophytaSpecies0.000abstractdescription39
- 238000000034methodMethods0.000abstractdescription23
- 241000589158AgrobacteriumSpecies0.000abstractdescription18
- 230000009261transgenic effectEffects0.000abstractdescription14
- 238000002360preparation methodMethods0.000abstractdescription10
- 238000010353genetic engineeringMethods0.000abstractdescription8
- 230000001404mediated effectEffects0.000abstractdescription8
- 210000001161mammalian embryoAnatomy0.000abstractdescription6
- 238000011161developmentMethods0.000abstractdescription3
- 239000013604expression vectorSubstances0.000abstractdescription3
- 238000011144upstream manufacturingMethods0.000abstractdescription3
- 108091028043Nucleic acid sequenceProteins0.000abstract1
- 239000011550stock solutionSubstances0.000description44
- KWYUFKZDYYNOTN-UHFFFAOYSA-MPotassium hydroxideChemical compound[OH-].[K+]KWYUFKZDYYNOTN-UHFFFAOYSA-M0.000description39
- 239000012634fragmentSubstances0.000description30
- XLYOFNOQVPJJNP-UHFFFAOYSA-NwaterSubstancesOXLYOFNOQVPJJNP-UHFFFAOYSA-N0.000description28
- 239000002609mediumSubstances0.000description25
- 239000012153distilled waterSubstances0.000description23
- 239000010413mother solutionSubstances0.000description20
- 239000000243solutionSubstances0.000description20
- 239000013598vectorSubstances0.000description18
- 108020004414DNAProteins0.000description16
- 238000010367cloningMethods0.000description16
- 238000012217deletionMethods0.000description16
- 230000037430deletionEffects0.000description16
- SEOVTRFCIGRIMH-UHFFFAOYSA-Nindole-3-acetic acidChemical compoundC1=CC=C2C(CC(=O)O)=CNC2=C1SEOVTRFCIGRIMH-UHFFFAOYSA-N0.000description16
- 206010020649HyperkeratosisDiseases0.000description14
- XEEYBQQBJWHFJM-UHFFFAOYSA-NIronChemical compound[Fe]XEEYBQQBJWHFJM-UHFFFAOYSA-N0.000description14
- 239000000047productSubstances0.000description13
- 101150054900gus geneProteins0.000description11
- 238000012546transferMethods0.000description11
- 2390000056312,4-Dichlorophenoxyacetic acidSubstances0.000description10
- IAZDPXIOMUYVGZ-UHFFFAOYSA-NDimethylsulphoxideChemical compoundCS(C)=OIAZDPXIOMUYVGZ-UHFFFAOYSA-N0.000description10
- KCXVZYZYPLLWCC-UHFFFAOYSA-NEDTAChemical compoundOC(=O)CN(CC(O)=O)CCN(CC(O)=O)CC(O)=OKCXVZYZYPLLWCC-UHFFFAOYSA-N0.000description10
- LFQSCWFLJHTTHZ-UHFFFAOYSA-NEthanolChemical compoundCCOLFQSCWFLJHTTHZ-UHFFFAOYSA-N0.000description10
- 230000000694effectsEffects0.000description10
- 230000021759endosperm developmentEffects0.000description10
- 229940088594vitaminDrugs0.000description10
- 239000011782vitaminSubstances0.000description10
- 235000013343vitaminNutrition0.000description10
- 229930003231vitaminNatural products0.000description10
- 150000003722vitamin derivativesChemical class0.000description10
- 229930006000SucroseNatural products0.000description9
- CZMRCDWAGMRECN-UGDNZRGBSA-NSucroseChemical compoundO[C@H]1[C@H](O)[C@@H](CO)O[C@@]1(CO)O[C@@H]1[C@H](O)[C@@H](O)[C@H](O)[C@@H](CO)O1CZMRCDWAGMRECN-UGDNZRGBSA-N0.000description9
- 229960003669carbenicillinDrugs0.000description9
- FPPNZSSZRUTDAP-UWFZAAFLSA-NcarbenicillinChemical compoundN([C@H]1[C@H]2SC([C@@H](N2C1=O)C(O)=O)(C)C)C(=O)C(C(O)=O)C1=CC=CC=C1FPPNZSSZRUTDAP-UWFZAAFLSA-N0.000description9
- 239000000203mixtureSubstances0.000description9
- 239000005720sucroseSubstances0.000description9
- 230000009466transformationEffects0.000description9
- 108700008625Reporter GenesProteins0.000description8
- 239000003617indole-3-acetic acidSubstances0.000description8
- NLKNQRATVPKPDG-UHFFFAOYSA-Mpotassium iodideChemical compound[K+].[I-]NLKNQRATVPKPDG-UHFFFAOYSA-M0.000description8
- 238000011160researchMethods0.000description8
- PRPINYUDVPFIRX-UHFFFAOYSA-N1-naphthaleneacetic acidChemical compoundC1=CC=C2C(CC(=O)O)=CC=CC2=C1PRPINYUDVPFIRX-UHFFFAOYSA-N0.000description7
- NWBJYWHLCVSVIJ-UHFFFAOYSA-NN-benzyladenineChemical compoundN=1C=NC=2NC=NC=2C=1NCC1=CC=CC=C1NWBJYWHLCVSVIJ-UHFFFAOYSA-N0.000description7
- 239000003153chemical reaction reagentSubstances0.000description7
- 230000004069differentiationEffects0.000description7
- 239000013642negative controlSubstances0.000description7
- 238000001514detection methodMethods0.000description6
- 235000013399edible fruitsNutrition0.000description6
- 210000000056organAnatomy0.000description6
- 238000003757reverse transcription PCRMethods0.000description6
- 238000010186stainingMethods0.000description6
- WQZGKKKJIJFFOK-GASJEMHNSA-NGlucoseNatural productsOC[C@H]1OC(O)[C@H](O)[C@@H](O)[C@@H]1OWQZGKKKJIJFFOK-GASJEMHNSA-N0.000description5
- 238000006243chemical reactionMethods0.000description5
- 238000001962electrophoresisMethods0.000description5
- 210000002257embryonic structureAnatomy0.000description5
- 239000008103glucoseSubstances0.000description5
- 230000001744histochemical effectEffects0.000description5
- 239000013612plasmidSubstances0.000description5
- 235000018102proteinsNutrition0.000description5
- 102000004169proteins and genesHuman genes0.000description5
- 230000017260vegetative to reproductive phase transition of meristemEffects0.000description5
- 108091032973(ribonucleotides)n+mProteins0.000description4
- 229920001817AgarPolymers0.000description4
- LZZYPRNAOMGNLH-UHFFFAOYSA-MCetrimonium bromideChemical compound[Br-].CCCCCCCCCCCCCCCC[N+](C)(C)CLZZYPRNAOMGNLH-UHFFFAOYSA-M0.000description4
- 108091062157Cis-regulatory elementProteins0.000description4
- 229920002148Gellan gumPolymers0.000description4
- CSNNHWWHGAXBCP-UHFFFAOYSA-LMagnesium sulfateChemical compound[Mg+2].[O-][S+2]([O-])([O-])[O-]CSNNHWWHGAXBCP-UHFFFAOYSA-L0.000description4
- 239000008272agarSubstances0.000description4
- 230000008901benefitEffects0.000description4
- 235000021186dishesNutrition0.000description4
- 108010048367enhanced green fluorescent proteinProteins0.000description4
- 238000001976enzyme digestionMethods0.000description4
- 230000004720fertilizationEffects0.000description4
- 239000001963growth mediumSubstances0.000description4
- 239000000843powderSubstances0.000description4
- 238000003860storageMethods0.000description4
- 239000000126substanceSubstances0.000description4
- 239000000725suspensionSubstances0.000description4
- 239000011573trace mineralSubstances0.000description4
- 235000013619trace mineralNutrition0.000description4
- 238000011426transformation methodMethods0.000description4
- QTBSBXVTEAMEQO-UHFFFAOYSA-NAcetic acidChemical compoundCC(O)=OQTBSBXVTEAMEQO-UHFFFAOYSA-N0.000description3
- 229920000936AgarosePolymers0.000description3
- 101100126948Arabidopsis thaliana FAE1 geneProteins0.000description3
- YQYJSBFKSSDGFO-UHFFFAOYSA-NEpihygromycinNatural productsOC1C(O)C(C(=O)C)OC1OC(C(=C1)O)=CC=C1C=C(C)C(=O)NC1C(O)C(O)C2OCOC2C1OYQYJSBFKSSDGFO-UHFFFAOYSA-N0.000description3
- 108050000784FerritinProteins0.000description3
- ONIBWKKTOPOVIA-BYPYZUCNSA-NL-ProlineChemical compoundOC(=O)[C@@H]1CCCN1ONIBWKKTOPOVIA-BYPYZUCNSA-N0.000description3
- 241000699670Mus sp.Species0.000description3
- ONIBWKKTOPOVIA-UHFFFAOYSA-NProlineNatural productsOC(=O)C1CCCN1ONIBWKKTOPOVIA-UHFFFAOYSA-N0.000description3
- 238000000246agarose gel electrophoresisMethods0.000description3
- 150000001413amino acidsChemical group0.000description3
- 238000009395breedingMethods0.000description3
- 230000001488breeding effectEffects0.000description3
- 229940027138cambiaDrugs0.000description3
- KRKNYBCHXYNGOX-UHFFFAOYSA-Ncitric acidChemical compoundOC(=O)CC(O)(C(O)=O)CC(O)=OKRKNYBCHXYNGOX-UHFFFAOYSA-N0.000description3
- 230000001276controlling effectEffects0.000description3
- 108010004073cysteinylcysteineProteins0.000description3
- KXZOIWWTXOCYKR-UHFFFAOYSA-Mdiclofenac potassiumChemical compound[K+].[O-]C(=O)CC1=CC=CC=C1NC1=C(Cl)C=CC=C1ClKXZOIWWTXOCYKR-UHFFFAOYSA-M0.000description3
- 230000000408embryogenic effectEffects0.000description3
- 238000002474experimental methodMethods0.000description3
- 238000000605extractionMethods0.000description3
- 230000006698inductionEffects0.000description3
- 230000001939inductive effectEffects0.000description3
- 238000002955isolationMethods0.000description3
- 238000004519manufacturing processMethods0.000description3
- 230000001737promoting effectEffects0.000description3
- LXNHXLLTXMVWPM-UHFFFAOYSA-NpyridoxineChemical compoundCC1=NC=C(CO)C(CO)=C1OLXNHXLLTXMVWPM-UHFFFAOYSA-N0.000description3
- 238000012163sequencing techniqueMethods0.000description3
- 230000001954sterilising effectEffects0.000description3
- 238000004659sterilization and disinfectionMethods0.000description3
- 238000012360testing methodMethods0.000description3
- FWMNVWWHGCHHJJ-SKKKGAJSSA-N4-amino-1-[(2r)-6-amino-2-[[(2r)-2-[[(2r)-2-[[(2r)-2-amino-3-phenylpropanoyl]amino]-3-phenylpropanoyl]amino]-4-methylpentanoyl]amino]hexanoyl]piperidine-4-carboxylic acidChemical compoundC([C@H](C(=O)N[C@H](CC(C)C)C(=O)N[C@H](CCCCN)C(=O)N1CCC(N)(CC1)C(O)=O)NC(=O)[C@H](N)CC=1C=CC=CC=1)C1=CC=CC=C1FWMNVWWHGCHHJJ-SKKKGAJSSA-N0.000description2
- 101710197633Actin-1Proteins0.000description2
- 101100107610Arabidopsis thaliana ABCF4 geneProteins0.000description2
- 241000894006BacteriaSpecies0.000description2
- UXVMQQNJUSDDNG-UHFFFAOYSA-LCalcium chlorideChemical compound[Cl-].[Cl-].[Ca+2]UXVMQQNJUSDDNG-UHFFFAOYSA-L0.000description2
- 241000701489Cauliflower mosaic virusSpecies0.000description2
- HEDRZPFGACZZDS-UHFFFAOYSA-NChloroformChemical compoundClC(Cl)ClHEDRZPFGACZZDS-UHFFFAOYSA-N0.000description2
- PGBLJHDDKCVSTC-CIUDSAMLSA-NCys-Met-GlnChemical compoundCSCC[C@H](NC(=O)[C@@H](N)CS)C(=O)N[C@@H](CCC(N)=O)C(O)=OPGBLJHDDKCVSTC-CIUDSAMLSA-N0.000description2
- 241000588724Escherichia coliSpecies0.000description2
- 241000620209Escherichia coli DH5[alpha]Species0.000description2
- 108700039691Genetic Promoter RegionsProteins0.000description2
- RRBLZNIIMHSHQF-FXQIFTODSA-NGln-Gln-CysChemical compoundC(CC(=O)N)[C@@H](C(=O)N[C@@H](CCC(=O)N)C(=O)N[C@@H](CS)C(=O)O)NRRBLZNIIMHSHQF-FXQIFTODSA-N0.000description2
- DHMQDGOQFOQNFH-UHFFFAOYSA-NGlycineChemical compoundNCC(O)=ODHMQDGOQFOQNFH-UHFFFAOYSA-N0.000description2
- FAIXYKHYOGVFKA-UHFFFAOYSA-NKinetinNatural productsN=1C=NC=2N=CNC=2C=1N(C)C1=CC=CO1FAIXYKHYOGVFKA-UHFFFAOYSA-N0.000description2
- KAFOIVJDVSZUMD-UHFFFAOYSA-NLeu-Gln-GlnNatural productsCC(C)CC(N)C(=O)NC(CCC(N)=O)C(=O)NC(CCC(N)=O)C(O)=OKAFOIVJDVSZUMD-UHFFFAOYSA-N0.000description2
- DPWGZWUMUUJQDT-IUCAKERBSA-NLeu-Gln-GlyChemical compoundCC(C)C[C@H](N)C(=O)N[C@@H](CCC(N)=O)C(=O)NCC(O)=ODPWGZWUMUUJQDT-IUCAKERBSA-N0.000description2
- JNDYEOUZBLOVOF-AVGNSLFASA-NLeu-Leu-GlnChemical compound[H]N[C@@H](CC(C)C)C(=O)N[C@@H](CC(C)C)C(=O)N[C@@H](CCC(N)=O)C(O)=OJNDYEOUZBLOVOF-AVGNSLFASA-N0.000description2
- MYKLINMAGAIRPJ-CIUDSAMLSA-NMet-Gln-AsnChemical compound[H]N[C@@H](CCSC)C(=O)N[C@@H](CCC(N)=O)C(=O)N[C@@H](CC(N)=O)C(O)=OMYKLINMAGAIRPJ-CIUDSAMLSA-N0.000description2
- HZLSUXCMSIBCRV-RVMXOQNASA-NMet-Ile-ProChemical compoundCC[C@H](C)[C@@H](C(=O)N1CCC[C@@H]1C(=O)O)NC(=O)[C@H](CCSC)NHZLSUXCMSIBCRV-RVMXOQNASA-N0.000description2
- 240000002582Oryza sativa Indica GroupSpecies0.000description2
- 101100068078Saccharomyces cerevisiae (strain ATCC 204508 / S288c) GCN4 geneProteins0.000description2
- FAPWRFPIFSIZLT-UHFFFAOYSA-MSodium chlorideChemical compound[Na+].[Cl-]FAPWRFPIFSIZLT-UHFFFAOYSA-M0.000description2
- 240000003768Solanum lycopersicumSpecies0.000description2
- HCHKCACWOHOZIP-UHFFFAOYSA-NZincChemical compound[Zn]HCHKCACWOHOZIP-UHFFFAOYSA-N0.000description2
- 108091007433antigensProteins0.000description2
- 230000033228biological regulationEffects0.000description2
- KGBXLFKZBHKPEV-UHFFFAOYSA-Nboric acidChemical compoundOB(O)OKGBXLFKZBHKPEV-UHFFFAOYSA-N0.000description2
- 239000001110calcium chlorideSubstances0.000description2
- 229910001628calcium chlorideInorganic materials0.000description2
- 239000012881co-culture mediumSubstances0.000description2
- 238000010276constructionMethods0.000description2
- 230000007423decreaseEffects0.000description2
- 210000005069earsAnatomy0.000description2
- 235000013305foodNutrition0.000description2
- 230000004927fusionEffects0.000description2
- 239000000499gelSubstances0.000description2
- 108010026364glycyl-glycyl-leucineProteins0.000description2
- 208000015181infectious diseaseDiseases0.000description2
- 229910052742ironInorganic materials0.000description2
- PHTQWCKDNZKARW-UHFFFAOYSA-NisoamylolChemical compoundCC(C)CCOPHTQWCKDNZKARW-UHFFFAOYSA-N0.000description2
- BPHPUYQFMNQIOC-NXRLNHOXSA-Nisopropyl beta-D-thiogalactopyranosideChemical compoundCC(C)S[C@@H]1O[C@H](CO)[C@H](O)[C@H](O)[C@H]1OBPHPUYQFMNQIOC-NXRLNHOXSA-N0.000description2
- QANMHLXAZMSUEX-UHFFFAOYSA-NkinetinChemical compoundN=1C=NC=2N=CNC=2C=1NCC1=CC=CO1QANMHLXAZMSUEX-UHFFFAOYSA-N0.000description2
- 229960001669kinetinDrugs0.000description2
- 229910052943magnesium sulfateInorganic materials0.000description2
- 235000019341magnesium sulphateNutrition0.000description2
- 229940099596manganese sulfateDrugs0.000description2
- 239000011702manganese sulphateSubstances0.000description2
- 235000007079manganese sulphateNutrition0.000description2
- SQQMAOCOWKFBNP-UHFFFAOYSA-Lmanganese(II) sulfateChemical compound[Mn+2].[O-]S([O-])(=O)=OSQQMAOCOWKFBNP-UHFFFAOYSA-L0.000description2
- 239000000463materialSubstances0.000description2
- 108020004999messenger RNAProteins0.000description2
- 108010085203methionylmethionineProteins0.000description2
- 238000010369molecular cloningMethods0.000description2
- 229910000402monopotassium phosphateInorganic materials0.000description2
- 235000019796monopotassium phosphateNutrition0.000description2
- 239000012452mother liquorSubstances0.000description2
- 239000006870ms-mediumSubstances0.000description2
- 235000015097nutrientsNutrition0.000description2
- PJNZPQUBCPKICU-UHFFFAOYSA-Nphosphoric acid;potassiumChemical compound[K].OP(O)(O)=OPJNZPQUBCPKICU-UHFFFAOYSA-N0.000description2
- 239000013641positive controlSubstances0.000description2
- FGIUAXJPYTZDNR-UHFFFAOYSA-Npotassium nitrateChemical compound[K+].[O-][N+]([O-])=OFGIUAXJPYTZDNR-UHFFFAOYSA-N0.000description2
- 230000008569processEffects0.000description2
- 108060006613prolaminProteins0.000description2
- 230000001681protective effectEffects0.000description2
- 238000011084recoveryMethods0.000description2
- 239000012882rooting mediumSubstances0.000description2
- 239000000523sampleSubstances0.000description2
- 238000012216screeningMethods0.000description2
- 239000011734sodiumSubstances0.000description2
- 239000012192staining solutionSubstances0.000description2
- 239000012879subculture mediumSubstances0.000description2
- 108010036387trimethionineProteins0.000description2
- 230000003612virological effectEffects0.000description2
- 229940011671vitamin b6Drugs0.000description2
- 239000011701zincSubstances0.000description2
- 229910052725zincInorganic materials0.000description2
- NWONKYPBYAMBJT-UHFFFAOYSA-Lzinc sulfateChemical compound[Zn+2].[O-]S([O-])(=O)=ONWONKYPBYAMBJT-UHFFFAOYSA-L0.000description2
- 229910000368zinc sulfateInorganic materials0.000description2
- 229960001763zinc sulfateDrugs0.000description2
- HXKWSTRRCHTUEC-UHFFFAOYSA-N2,4-DichlorophenoxyaceticacidChemical compoundOC(=O)C(Cl)OC1=CC=C(Cl)C=C1HXKWSTRRCHTUEC-UHFFFAOYSA-N0.000description1
- OVSKIKFHRZPJSS-DOMIDYPGSA-N2-(2,4-dichlorophenoxy)acetic acidChemical compoundOC(=O)[14CH2]OC1=CC=C(Cl)C=C1ClOVSKIKFHRZPJSS-DOMIDYPGSA-N0.000description1
- PAWQVTBBRAZDMG-UHFFFAOYSA-N2-(3-bromo-2-fluorophenyl)acetic acidChemical compoundOC(=O)CC1=CC=CC(Br)=C1FPAWQVTBBRAZDMG-UHFFFAOYSA-N0.000description1
- LMSDCGXQALIMLM-UHFFFAOYSA-N2-[2-[bis(carboxymethyl)amino]ethyl-(carboxymethyl)amino]acetic acid;ironChemical compound[Fe].OC(=O)CN(CC(O)=O)CCN(CC(O)=O)CC(O)=OLMSDCGXQALIMLM-UHFFFAOYSA-N0.000description1
- QKNYBSVHEMOAJP-UHFFFAOYSA-N2-amino-2-(hydroxymethyl)propane-1,3-diol;hydron;chlorideChemical compoundCl.OCC(N)(CO)COQKNYBSVHEMOAJP-UHFFFAOYSA-N0.000description1
- OPIFSICVWOWJMJ-AEOCFKNESA-N5-bromo-4-chloro-3-indolyl beta-D-galactosideChemical compoundO[C@@H]1[C@@H](O)[C@@H](O)[C@@H](CO)O[C@H]1OC1=CNC2=CC=C(Br)C(Cl)=C12OPIFSICVWOWJMJ-AEOCFKNESA-N0.000description1
- 2390000059726-BenzyladenineSubstances0.000description1
- 241000589155Agrobacterium tumefaciensSpecies0.000description1
- FOHXUHGZZKETFI-JBDRJPRFSA-NAla-Ile-CysChemical compoundCC[C@H](C)[C@@H](C(=O)N[C@@H](CS)C(=O)O)NC(=O)[C@H](C)NFOHXUHGZZKETFI-JBDRJPRFSA-N0.000description1
- OMDNCNKNEGFOMM-BQBZGAKWSA-NAla-Met-GlyChemical compound[H]N[C@@H](C)C(=O)N[C@@H](CCSC)C(=O)NCC(O)=OOMDNCNKNEGFOMM-BQBZGAKWSA-N0.000description1
- IPZQNYYAYVRKKK-FXQIFTODSA-NAla-Pro-AlaChemical compoundC[C@H](N)C(=O)N1CCC[C@H]1C(=O)N[C@@H](C)C(O)=OIPZQNYYAYVRKKK-FXQIFTODSA-N0.000description1
- DCVYRWFAMZFSDA-ZLUOBGJFSA-NAla-Ser-AlaChemical compound[H]N[C@@H](C)C(=O)N[C@@H](CO)C(=O)N[C@@H](C)C(O)=ODCVYRWFAMZFSDA-ZLUOBGJFSA-N0.000description1
- OEVCHROQUIVQFZ-YTLHQDLWSA-NAla-Thr-AlaChemical compoundC[C@H](N)C(=O)N[C@@H]([C@H](O)C)C(=O)N[C@@H](C)C(O)=OOEVCHROQUIVQFZ-YTLHQDLWSA-N0.000description1
- ISCYZXFOCXWUJU-KZVJFYERSA-NAla-Thr-MetChemical compound[H]N[C@@H](C)C(=O)N[C@@H]([C@@H](C)O)C(=O)N[C@@H](CCSC)C(O)=OISCYZXFOCXWUJU-KZVJFYERSA-N0.000description1
- 241000219195Arabidopsis thalianaSpecies0.000description1
- QDXQWFBLUVTOFL-FXQIFTODSA-NAsn-Met-AlaChemical compoundC[C@@H](C(=O)O)NC(=O)[C@H](CCSC)NC(=O)[C@H](CC(=O)N)NQDXQWFBLUVTOFL-FXQIFTODSA-N0.000description1
- UYRPHDGXHKBZHJ-CIUDSAMLSA-NAsn-Met-GlnChemical compoundCSCC[C@@H](C(=O)N[C@@H](CCC(=O)N)C(=O)O)NC(=O)[C@H](CC(=O)N)NUYRPHDGXHKBZHJ-CIUDSAMLSA-N0.000description1
- ZVUMKOMKQCANOM-AVGNSLFASA-NAsn-Phe-GlnChemical compound[H]N[C@@H](CC(N)=O)C(=O)N[C@@H](CC1=CC=CC=C1)C(=O)N[C@@H](CCC(N)=O)C(O)=OZVUMKOMKQCANOM-AVGNSLFASA-N0.000description1
- 108090000565Capsid ProteinsProteins0.000description1
- 102100023321CeruloplasminHuman genes0.000description1
- 108091026890Coding regionProteins0.000description1
- SQJSYLDKQBZQTG-FXQIFTODSA-NCys-Asn-MetChemical compoundCSCC[C@@H](C(=O)O)NC(=O)[C@H](CC(=O)N)NC(=O)[C@H](CS)NSQJSYLDKQBZQTG-FXQIFTODSA-N0.000description1
- LMXOUGMSGHFLRX-CIUDSAMLSA-NCys-Gln-MetChemical compoundCSCC[C@@H](C(=O)O)NC(=O)[C@H](CCC(=O)N)NC(=O)[C@H](CS)NLMXOUGMSGHFLRX-CIUDSAMLSA-N0.000description1
- CVLIHKBUPSFRQP-WHFBIAKZSA-NCys-Gly-AlaChemical compound[H]N[C@@H](CS)C(=O)NCC(=O)N[C@@H](C)C(O)=OCVLIHKBUPSFRQP-WHFBIAKZSA-N0.000description1
- XELISBQUZZAPQK-CIUDSAMLSA-NCys-His-CysChemical compoundC1=C(NC=N1)C[C@@H](C(=O)N[C@@H](CS)C(=O)O)NC(=O)[C@H](CS)NXELISBQUZZAPQK-CIUDSAMLSA-N0.000description1
- IDFVDSBJNMPBSX-SRVKXCTJSA-NCys-Lys-LeuChemical compound[H]N[C@@H](CS)C(=O)N[C@@H](CCCCN)C(=O)N[C@@H](CC(C)C)C(O)=OIDFVDSBJNMPBSX-SRVKXCTJSA-N0.000description1
- 239000003109Disodium ethylene diamine tetraacetateSubstances0.000description1
- ZGTMUACCHSMWAC-UHFFFAOYSA-LEDTA disodium salt (anhydrous)Chemical compound[Na+].[Na+].OC(=O)CN(CC([O-])=O)CCN(CC(O)=O)CC([O-])=OZGTMUACCHSMWAC-UHFFFAOYSA-L0.000description1
- VGGSQFUCUMXWEO-UHFFFAOYSA-NEtheneChemical compoundC=CVGGSQFUCUMXWEO-UHFFFAOYSA-N0.000description1
- 239000005977EthyleneSubstances0.000description1
- 241000282326Felis catusSpecies0.000description1
- 102000008857FerritinHuman genes0.000description1
- 238000008416FerritinMethods0.000description1
- ZRXBYKAOFHLTDN-GUBZILKMSA-NGln-Cys-HisChemical compoundC1=C(NC=N1)C[C@@H](C(=O)O)NC(=O)[C@H](CS)NC(=O)[C@H](CCC(=O)N)NZRXBYKAOFHLTDN-GUBZILKMSA-N0.000description1
- PKVWNYGXMNWJSI-CIUDSAMLSA-NGln-Gln-GlnChemical compound[H]N[C@@H](CCC(N)=O)C(=O)N[C@@H](CCC(N)=O)C(=O)N[C@@H](CCC(N)=O)C(O)=OPKVWNYGXMNWJSI-CIUDSAMLSA-N0.000description1
- ZBKUIQNCRIYVGH-SDDRHHMPSA-NGln-Leu-ProChemical compoundCC(C)C[C@@H](C(=O)N1CCC[C@@H]1C(=O)O)NC(=O)[C@H](CCC(=O)N)NZBKUIQNCRIYVGH-SDDRHHMPSA-N0.000description1
- YGNPTRVNRUKVLA-DCAQKATOSA-NGln-Met-MetChemical compoundCSCC[C@@H](C(=O)N[C@@H](CCSC)C(=O)O)NC(=O)[C@H](CCC(=O)N)NYGNPTRVNRUKVLA-DCAQKATOSA-N0.000description1
- OREPWMPAUWIIAM-ZPFDUUQYSA-NGln-Pro-IleChemical compoundCC[C@H](C)[C@@H](C(=O)O)NC(=O)[C@@H]1CCCN1C(=O)[C@H](CCC(=O)N)NOREPWMPAUWIIAM-ZPFDUUQYSA-N0.000description1
- 108010044091GlobulinsProteins0.000description1
- 102000006395GlobulinsHuman genes0.000description1
- 102000053187GlucuronidaseHuman genes0.000description1
- 108010060309GlucuronidaseProteins0.000description1
- LJPIRKICOISLKN-WHFBIAKZSA-NGly-Ala-SerChemical compoundNCC(=O)N[C@@H](C)C(=O)N[C@@H](CO)C(O)=OLJPIRKICOISLKN-WHFBIAKZSA-N0.000description1
- NMROINAYXCACKF-WHFBIAKZSA-NGly-Cys-CysChemical compoundNCC(=O)N[C@@H](CS)C(=O)N[C@@H](CS)C(O)=ONMROINAYXCACKF-WHFBIAKZSA-N0.000description1
- XLFHCWHXKSFVIB-BQBZGAKWSA-NGly-Gln-GlnChemical compoundNCC(=O)N[C@@H](CCC(N)=O)C(=O)N[C@@H](CCC(N)=O)C(O)=OXLFHCWHXKSFVIB-BQBZGAKWSA-N0.000description1
- XPJBQTCXPJNIFE-ZETCQYMHSA-NGly-Gly-LeuChemical compoundCC(C)C[C@@H](C(O)=O)NC(=O)CNC(=O)CNXPJBQTCXPJNIFE-ZETCQYMHSA-N0.000description1
- CCBIBMKQNXHNIN-ZETCQYMHSA-NGly-Leu-GlyChemical compoundNCC(=O)N[C@@H](CC(C)C)C(=O)NCC(O)=OCCBIBMKQNXHNIN-ZETCQYMHSA-N0.000description1
- FKYQEVBRZSFAMJ-QWRGUYRKSA-NGly-Ser-TyrChemical compoundNCC(=O)N[C@@H](CO)C(=O)N[C@H](C(O)=O)CC1=CC=C(O)C=C1FKYQEVBRZSFAMJ-QWRGUYRKSA-N0.000description1
- 239000004471GlycineSubstances0.000description1
- 244000068988Glycine maxSpecies0.000description1
- 235000010469Glycine maxNutrition0.000description1
- SQUHHTBVTRBESD-UHFFFAOYSA-NHexa-Ac-myo-InositolNatural productsCC(=O)OC1C(OC(C)=O)C(OC(C)=O)C(OC(C)=O)C(OC(C)=O)C1OC(C)=OSQUHHTBVTRBESD-UHFFFAOYSA-N0.000description1
- GRRNUXAQVGOGFE-UHFFFAOYSA-NHygromycin-BNatural productsOC1C(NC)CC(N)C(O)C1OC1C2OC3(C(C(O)C(O)C(C(N)CO)O3)O)OC2C(O)C(CO)O1GRRNUXAQVGOGFE-UHFFFAOYSA-N0.000description1
- PFTFEWHJSAXGED-ZKWXMUAHSA-NIle-Cys-GlyChemical compoundCC[C@H](C)[C@@H](C(=O)N[C@@H](CS)C(=O)NCC(=O)O)NPFTFEWHJSAXGED-ZKWXMUAHSA-N0.000description1
- XOZOSAUOGRPCES-STECZYCISA-NIle-Pro-TyrChemical compoundCC[C@H](C)[C@H](N)C(=O)N1CCC[C@H]1C(=O)N[C@H](C(O)=O)CC1=CC=C(O)C=C1XOZOSAUOGRPCES-STECZYCISA-N0.000description1
- NURNJECQNNCRBK-FLBSBUHZSA-NIle-Thr-ThrChemical compoundCC[C@H](C)[C@H](N)C(=O)N[C@@H]([C@@H](C)O)C(=O)N[C@@H]([C@@H](C)O)C(O)=ONURNJECQNNCRBK-FLBSBUHZSA-N0.000description1
- HGCNKOLVKRAVHD-UHFFFAOYSA-NL-Met-L-PheNatural productsCSCCC(N)C(=O)NC(C(O)=O)CC1=CC=CC=C1HGCNKOLVKRAVHD-UHFFFAOYSA-N0.000description1
- LHSGPCFBGJHPCY-UHFFFAOYSA-NL-leucine-L-tyrosineNatural productsCC(C)CC(N)C(=O)NC(C(O)=O)CC1=CC=C(O)C=C1LHSGPCFBGJHPCY-UHFFFAOYSA-N0.000description1
- QPXBPQUGXHURGP-UWVGGRQHSA-NLeu-Gly-MetChemical compoundCC(C)C[C@@H](C(=O)NCC(=O)N[C@@H](CCSC)C(=O)O)NQPXBPQUGXHURGP-UWVGGRQHSA-N0.000description1
- UFPLDOKWDNTTRP-ULQDDVLXSA-NLeu-Tyr-MetChemical compoundCSCC[C@@H](C(O)=O)NC(=O)[C@@H](NC(=O)[C@@H](N)CC(C)C)CC1=CC=C(O)C=C1UFPLDOKWDNTTRP-ULQDDVLXSA-N0.000description1
- 102000003960LigasesHuman genes0.000description1
- 108090000364LigasesProteins0.000description1
- 241000209510LiliopsidaSpecies0.000description1
- 235000007688Lycopersicon esculentumNutrition0.000description1
- KFSALEZVQJYHCE-AVGNSLFASA-NLys-Met-ValChemical compoundCC(C)[C@@H](C(=O)O)NC(=O)[C@H](CCSC)NC(=O)[C@H](CCCCN)NKFSALEZVQJYHCE-AVGNSLFASA-N0.000description1
- IHITVQKJXQQGLJ-LPEHRKFASA-NMet-Asn-ProChemical compoundCSCC[C@@H](C(=O)N[C@@H](CC(=O)N)C(=O)N1CCC[C@@H]1C(=O)O)NIHITVQKJXQQGLJ-LPEHRKFASA-N0.000description1
- VOOINLQYUZOREH-SRVKXCTJSA-NMet-Gln-LeuChemical compoundCC(C)C[C@@H](C(=O)O)NC(=O)[C@H](CCC(=O)N)NC(=O)[C@H](CCSC)NVOOINLQYUZOREH-SRVKXCTJSA-N0.000description1
- HHCOOFPGNXKFGR-HJGDQZAQSA-NMet-Gln-ThrChemical compound[H]N[C@@H](CCSC)C(=O)N[C@@H](CCC(N)=O)C(=O)N[C@@H]([C@@H](C)O)C(O)=OHHCOOFPGNXKFGR-HJGDQZAQSA-N0.000description1
- LRALLISKBZNSKN-BQBZGAKWSA-NMet-Gly-SerChemical compoundCSCC[C@H](N)C(=O)NCC(=O)N[C@@H](CO)C(O)=OLRALLISKBZNSKN-BQBZGAKWSA-N0.000description1
- SXWQMBGNFXAGAT-FJXKBIBVSA-NMet-Gly-ThrChemical compoundCSCC[C@H](N)C(=O)NCC(=O)N[C@@H]([C@@H](C)O)C(O)=OSXWQMBGNFXAGAT-FJXKBIBVSA-N0.000description1
- VBGGTAPDGFQMKF-AVGNSLFASA-NMet-Lys-MetChemical compoundCSCC[C@H](N)C(=O)N[C@@H](CCCCN)C(=O)N[C@@H](CCSC)C(O)=OVBGGTAPDGFQMKF-AVGNSLFASA-N0.000description1
- WXUUEPIDLLQBLJ-DCAQKATOSA-NMet-Met-GlnChemical compoundCSCC[C@@H](C(=O)N[C@@H](CCSC)C(=O)N[C@@H](CCC(=O)N)C(=O)O)NWXUUEPIDLLQBLJ-DCAQKATOSA-N0.000description1
- VWWGEKCAPBMIFE-SRVKXCTJSA-NMet-Met-MetChemical compoundCSCC[C@H](N)C(=O)N[C@@H](CCSC)C(=O)N[C@@H](CCSC)C(O)=OVWWGEKCAPBMIFE-SRVKXCTJSA-N0.000description1
- ZACMJPCWVSLCNS-JYJNAYRXSA-NMet-Phe-MetChemical compoundCSCC[C@H](N)C(=O)N[C@H](C(=O)N[C@@H](CCSC)C(O)=O)CC1=CC=CC=C1ZACMJPCWVSLCNS-JYJNAYRXSA-N0.000description1
- UFWIBTONFRDIAS-UHFFFAOYSA-NNaphthaleneChemical compoundC1=CC=CC2=CC=CC=C21UFWIBTONFRDIAS-UHFFFAOYSA-N0.000description1
- PVNIIMVLHYAWGP-UHFFFAOYSA-NNiacinChemical compoundOC(=O)C1=CC=CN=C1PVNIIMVLHYAWGP-UHFFFAOYSA-N0.000description1
- 244000061176Nicotiana tabacumSpecies0.000description1
- 235000002637Nicotiana tabacumNutrition0.000description1
- KAGCQPSEVAETCA-JYJNAYRXSA-NPhe-Gln-LeuChemical compoundCC(C)C[C@@H](C(=O)O)NC(=O)[C@H](CCC(=O)N)NC(=O)[C@H](CC1=CC=CC=C1)NKAGCQPSEVAETCA-JYJNAYRXSA-N0.000description1
- FQUUYTNBMIBOHS-IHRRRGAJSA-NPhe-Met-SerChemical compoundCSCC[C@@H](C(=O)N[C@@H](CO)C(=O)O)NC(=O)[C@H](CC1=CC=CC=C1)NFQUUYTNBMIBOHS-IHRRRGAJSA-N0.000description1
- NJJBATPLUQHRBM-IHRRRGAJSA-NPhe-Pro-SerChemical compoundC1C[C@H](N(C1)C(=O)[C@H](CC2=CC=CC=C2)N)C(=O)N[C@@H](CO)C(=O)ONJJBATPLUQHRBM-IHRRRGAJSA-N0.000description1
- ODGNUUUDJONJSC-UFYCRDLUSA-NPhe-Pro-TyrChemical compoundC1C[C@H](N(C1)C(=O)[C@H](CC2=CC=CC=C2)N)C(=O)N[C@@H](CC3=CC=C(C=C3)O)C(=O)OODGNUUUDJONJSC-UFYCRDLUSA-N0.000description1
- 108020005120Plant DNAProteins0.000description1
- 108700001094Plant GenesProteins0.000description1
- DBALDZKOTNSBFM-FXQIFTODSA-NPro-Ala-AsnChemical compound[H]N1CCC[C@H]1C(=O)N[C@@H](C)C(=O)N[C@@H](CC(N)=O)C(O)=ODBALDZKOTNSBFM-FXQIFTODSA-N0.000description1
- OLTFZQIYCNOBLI-DCAQKATOSA-NPro-Cys-LysChemical compoundC1C[C@H](NC1)C(=O)N[C@@H](CS)C(=O)N[C@@H](CCCCN)C(=O)OOLTFZQIYCNOBLI-DCAQKATOSA-N0.000description1
- LZHHZYDPMZEMRX-STQMWFEESA-NPro-Tyr-GlyChemical compound[H]N1CCC[C@H]1C(=O)N[C@@H](CC1=CC=C(O)C=C1)C(=O)NCC(O)=OLZHHZYDPMZEMRX-STQMWFEESA-N0.000description1
- FIDNSJUXESUDOV-JYJNAYRXSA-NPro-Tyr-ValChemical compound[H]N1CCC[C@H]1C(=O)N[C@@H](CC1=CC=C(O)C=C1)C(=O)N[C@@H](C(C)C)C(O)=OFIDNSJUXESUDOV-JYJNAYRXSA-N0.000description1
- 108010009736Protein HydrolysatesProteins0.000description1
- 241000725643Respiratory syncytial virusSpecies0.000description1
- 108010016634Seed Storage ProteinsProteins0.000description1
- ZUGXSSFMTXKHJS-ZLUOBGJFSA-NSer-Ala-AlaChemical compound[H]N[C@@H](CO)C(=O)N[C@@H](C)C(=O)N[C@@H](C)C(O)=OZUGXSSFMTXKHJS-ZLUOBGJFSA-N0.000description1
- TUYBIWUZWJUZDD-ACZMJKKPSA-NSer-Cys-GlnChemical compoundOC[C@H](N)C(=O)N[C@@H](CS)C(=O)N[C@H](C(O)=O)CCC(N)=OTUYBIWUZWJUZDD-ACZMJKKPSA-N0.000description1
- BQWCDDAISCPDQV-XHNCKOQMSA-NSer-Gln-ProChemical compoundC1C[C@@H](N(C1)C(=O)[C@H](CCC(=O)N)NC(=O)[C@H](CO)N)C(=O)OBQWCDDAISCPDQV-XHNCKOQMSA-N0.000description1
- 230000024932T cell mediated immunityEffects0.000description1
- PXQUBKWZENPDGE-CIQUZCHMSA-NThr-Ala-IleChemical compoundCC[C@H](C)[C@@H](C(=O)O)NC(=O)[C@H](C)NC(=O)[C@H]([C@@H](C)O)NPXQUBKWZENPDGE-CIQUZCHMSA-N0.000description1
- YJCVECXVYHZOBK-KNZXXDILSA-NThr-Ile-ProChemical compoundCC[C@H](C)[C@@H](C(=O)N1CCC[C@@H]1C(=O)O)NC(=O)[C@H]([C@@H](C)O)NYJCVECXVYHZOBK-KNZXXDILSA-N0.000description1
- BVOVIGCHYNFJBZ-JXUBOQSCSA-NThr-Leu-AlaChemical compound[H]N[C@@H]([C@@H](C)O)C(=O)N[C@@H](CC(C)C)C(=O)N[C@@H](C)C(O)=OBVOVIGCHYNFJBZ-JXUBOQSCSA-N0.000description1
- RFKVQLIXNVEOMB-WEDXCCLWSA-NThr-Leu-GlyChemical compoundC[C@H]([C@@H](C(=O)N[C@@H](CC(C)C)C(=O)NCC(=O)O)N)ORFKVQLIXNVEOMB-WEDXCCLWSA-N0.000description1
- PCMDGXKXVMBIFP-VEVYYDQMSA-NThr-Met-AsnChemical compound[H]N[C@@H]([C@@H](C)O)C(=O)N[C@@H](CCSC)C(=O)N[C@@H](CC(N)=O)C(O)=OPCMDGXKXVMBIFP-VEVYYDQMSA-N0.000description1
- COYHRQWNJDJCNA-NUJDXYNKSA-NThr-Thr-ThrChemical compoundC[C@@H](O)[C@H](N)C(=O)N[C@@H]([C@@H](C)O)C(=O)N[C@@H]([C@@H](C)O)C(O)=OCOYHRQWNJDJCNA-NUJDXYNKSA-N0.000description1
- NOXKHHXSHQFSGJ-FQPOAREZSA-NTyr-Ala-ThrChemical compoundC[C@@H](O)[C@@H](C(O)=O)NC(=O)[C@H](C)NC(=O)[C@@H](N)CC1=CC=C(O)C=C1NOXKHHXSHQFSGJ-FQPOAREZSA-N0.000description1
- HNERGSKJJZQGEA-JYJNAYRXSA-NTyr-Met-MetChemical compoundCSCC[C@@H](C(=O)N[C@@H](CCSC)C(=O)O)NC(=O)[C@H](CC1=CC=C(C=C1)O)NHNERGSKJJZQGEA-JYJNAYRXSA-N0.000description1
- WURLIFOWSMBUAR-SLFFLAALSA-NTyr-Phe-ProChemical compoundC1C[C@@H](N(C1)C(=O)[C@H](CC2=CC=CC=C2)NC(=O)[C@H](CC3=CC=C(C=C3)O)N)C(=O)OWURLIFOWSMBUAR-SLFFLAALSA-N0.000description1
- BQASAMYRHNCKQE-IHRRRGAJSA-NTyr-Val-CysChemical compoundCC(C)[C@@H](C(=O)N[C@@H](CS)C(=O)O)NC(=O)[C@H](CC1=CC=C(C=C1)O)NBQASAMYRHNCKQE-IHRRRGAJSA-N0.000description1
- 241000700605VirusesSpecies0.000description1
- 238000009825accumulationMethods0.000description1
- 238000012271agricultural productionMethods0.000description1
- 108010087924alanylprolineProteins0.000description1
- 230000004075alterationEffects0.000description1
- BFNBIHQBYMNNAN-UHFFFAOYSA-Nammonium sulfateChemical compoundN.N.OS(O)(=O)=OBFNBIHQBYMNNAN-UHFFFAOYSA-N0.000description1
- 229910052921ammonium sulfateInorganic materials0.000description1
- 235000011130ammonium sulphateNutrition0.000description1
- 229960000723ampicillinDrugs0.000description1
- AVKUERGKIZMTKX-NJBDSQKTSA-NampicillinChemical compoundC1([C@@H](N)C(=O)N[C@H]2[C@H]3SC([C@@H](N3C2=O)C(O)=O)(C)C)=CC=CC=C1AVKUERGKIZMTKX-NJBDSQKTSA-N0.000description1
- 230000003321amplificationEffects0.000description1
- 238000004458analytical methodMethods0.000description1
- 230000005875antibody responseEffects0.000description1
- 239000000427antigenSubstances0.000description1
- 102000036639antigensHuman genes0.000description1
- 108010077245asparaginyl-prolineProteins0.000description1
- 230000001580bacterial effectEffects0.000description1
- 238000010170biological methodMethods0.000description1
- 210000000234capsidAnatomy0.000description1
- 239000005018caseinSubstances0.000description1
- BECPQYXYKAMYBN-UHFFFAOYSA-Ncasein, tech.Chemical compoundNCCCCC(C(O)=O)N=C(O)C(CC(O)=O)N=C(O)C(CCC(O)=N)N=C(O)C(CC(C)C)N=C(O)C(CCC(O)=O)N=C(O)C(CC(O)=O)N=C(O)C(CCC(O)=O)N=C(O)C(C(C)O)N=C(O)C(CCC(O)=N)N=C(O)C(CCC(O)=N)N=C(O)C(CCC(O)=N)N=C(O)C(CCC(O)=O)N=C(O)C(CCC(O)=O)N=C(O)C(COP(O)(O)=O)N=C(O)C(CCC(O)=N)N=C(O)C(N)CC1=CC=CC=C1BECPQYXYKAMYBN-UHFFFAOYSA-N0.000description1
- 235000021240caseinsNutrition0.000description1
- 235000013339cerealsNutrition0.000description1
- 230000008859changeEffects0.000description1
- RCTYPNKXASFOBE-UHFFFAOYSA-MchloromercuryChemical compound[Hg]ClRCTYPNKXASFOBE-UHFFFAOYSA-M0.000description1
- GVPFVAHMJGGAJG-UHFFFAOYSA-Lcobalt dichlorideChemical compound[Cl-].[Cl-].[Co+2]GVPFVAHMJGGAJG-UHFFFAOYSA-L0.000description1
- 238000004891communicationMethods0.000description1
- 239000002299complementary DNASubstances0.000description1
- 238000007796conventional methodMethods0.000description1
- 229910000365copper sulfateInorganic materials0.000description1
- ARUVKPQLZAKDPS-UHFFFAOYSA-Lcopper(II) sulfateChemical compound[Cu+2].[O-][S+2]([O-])([O-])[O-]ARUVKPQLZAKDPS-UHFFFAOYSA-L0.000description1
- 108010016616cysteinylglycineProteins0.000description1
- 230000034994deathEffects0.000description1
- 230000007812deficiencyEffects0.000description1
- 230000018109developmental processEffects0.000description1
- 230000009025developmental regulationEffects0.000description1
- 238000010586diagramMethods0.000description1
- 230000029087digestionEffects0.000description1
- 235000019301disodium ethylene diamine tetraacetateNutrition0.000description1
- 238000005516engineering processMethods0.000description1
- 239000003623enhancerSubstances0.000description1
- 230000002708enhancing effectEffects0.000description1
- 238000009472formulationMethods0.000description1
- 230000030279gene silencingEffects0.000description1
- 238000012226gene silencing methodMethods0.000description1
- 108010050792gluteninProteins0.000description1
- 108010010147glycylglutamineProteins0.000description1
- 239000003501hydroponicsSubstances0.000description1
- GRRNUXAQVGOGFE-NZSRVPFOSA-Nhygromycin BChemical compoundO[C@@H]1[C@@H](NC)C[C@@H](N)[C@H](O)[C@H]1O[C@H]1[C@H]2O[C@@]3([C@@H]([C@@H](O)[C@@H](O)[C@@H](C(N)CO)O3)O)O[C@H]2[C@@H](O)[C@@H](CO)O1GRRNUXAQVGOGFE-NZSRVPFOSA-N0.000description1
- 229940097277hygromycin bDrugs0.000description1
- 230000028993immune responseEffects0.000description1
- 230000036039immunityEffects0.000description1
- 230000003053immunizationEffects0.000description1
- 238000002649immunizationMethods0.000description1
- 229960000367inositolDrugs0.000description1
- CDAISMWEOUEBRE-GPIVLXJGSA-NinositolChemical compoundO[C@H]1[C@H](O)[C@@H](O)[C@H](O)[C@H](O)[C@@H]1OCDAISMWEOUEBRE-GPIVLXJGSA-N0.000description1
- 230000003993interactionEffects0.000description1
- 230000003834intracellular effectEffects0.000description1
- 150000002505ironChemical class0.000description1
- 108010057821leucylprolineProteins0.000description1
- 108010012058leucyltyrosineProteins0.000description1
- 239000007791liquid phaseSubstances0.000description1
- 238000011177media preparationMethods0.000description1
- 229960002523mercuric chlorideDrugs0.000description1
- LWJROJCJINYWOX-UHFFFAOYSA-Lmercury dichlorideChemical compoundCl[Hg]ClLWJROJCJINYWOX-UHFFFAOYSA-L0.000description1
- 230000002503metabolic effectEffects0.000description1
- 230000037353metabolic pathwayEffects0.000description1
- 239000002207metaboliteSubstances0.000description1
- 108010005942methionylglycineProteins0.000description1
- 108010068488methionylphenylalanineProteins0.000description1
- 238000002493microarrayMethods0.000description1
- 238000003199nucleic acid amplification methodMethods0.000description1
- 108020004707nucleic acidsProteins0.000description1
- 102000039446nucleic acidsHuman genes0.000description1
- 150000007523nucleic acidsChemical class0.000description1
- 235000016709nutritionNutrition0.000description1
- 230000035764nutritionEffects0.000description1
- 230000035790physiological processes and functionsEffects0.000description1
- 230000008635plant growthEffects0.000description1
- 239000002244precipitateSubstances0.000description1
- 238000012545processingMethods0.000description1
- 108010031719prolyl-serineProteins0.000description1
- 108010079317prolyl-tyrosineProteins0.000description1
- 108010015796prolylisoleucineProteins0.000description1
- RADKZDMFGJYCBB-UHFFFAOYSA-Npyridoxal hydrochlorideNatural productsCC1=NC=C(CO)C(C=O)=C1ORADKZDMFGJYCBB-UHFFFAOYSA-N0.000description1
- 235000019171pyridoxine hydrochlorideNutrition0.000description1
- 239000011764pyridoxine hydrochlorideSubstances0.000description1
- 238000004445quantitative analysisMethods0.000description1
- 230000008929regenerationEffects0.000description1
- 238000011069regeneration methodMethods0.000description1
- 230000001105regulatory effectEffects0.000description1
- 230000001850reproductive effectEffects0.000description1
- 230000000241respiratory effectEffects0.000description1
- 230000004044responseEffects0.000description1
- 238000010839reverse transcriptionMethods0.000description1
- CDAISMWEOUEBRE-UHFFFAOYSA-Nscyllo-inosotolNatural productsOC1C(O)C(O)C(O)C(O)C1OCDAISMWEOUEBRE-UHFFFAOYSA-N0.000description1
- 238000000926separation methodMethods0.000description1
- 230000000405serological effectEffects0.000description1
- 238000004904shorteningMethods0.000description1
- 239000011780sodium chlorideSubstances0.000description1
- 239000011684sodium molybdateSubstances0.000description1
- 235000015393sodium molybdateNutrition0.000description1
- TVXXNOYZHKPKGW-UHFFFAOYSA-Nsodium molybdate (anhydrous)Chemical compound[Na+].[Na+].[O-][Mo]([O-])(=O)=OTVXXNOYZHKPKGW-UHFFFAOYSA-N0.000description1
- 238000007447staining methodMethods0.000description1
- 239000006228supernatantSubstances0.000description1
- 230000009885systemic effectEffects0.000description1
- DPJRMOMPQZCRJU-UHFFFAOYSA-Mthiamine hydrochlorideChemical compoundCl.[Cl-].CC1=C(CCO)SC=[N+]1CC1=CN=C(C)N=C1NDPJRMOMPQZCRJU-UHFFFAOYSA-M0.000description1
- 230000025366tissue developmentEffects0.000description1
- 231100000331toxicToxicity0.000description1
- 230000002588toxic effectEffects0.000description1
- 238000013518transcriptionMethods0.000description1
- 230000035897transcriptionEffects0.000description1
- 238000012795verificationMethods0.000description1
- 235000019158vitamin B6Nutrition0.000description1
- 239000011726vitamin B6Substances0.000description1
- 238000005406washingMethods0.000description1
Images
Landscapes
- Breeding Of Plants And Reproduction By Means Of Culturing (AREA)
Abstract
Description
Translated fromChinese技术领域technical field
本发明属于植物基因工程技术领域,具体涉及到水稻中同一胚乳特异性表达基因的不同长度启动子区域的分离克隆及表达模式鉴定。The invention belongs to the technical field of plant genetic engineering, and in particular relates to the isolation and cloning of different length promoter regions of the same endosperm-specific expression gene in rice and the identification of expression patterns.
背景技术Background technique
通过分子生物学以及基因工程的手段去研究和改良作物在农业生产中具有很好的应用前景。自1983年获得第一株转基因烟草以来(Zambryski et al.,1983),在近30年的时间里,植物基因工程的研究取得了突飞猛进的进展,已经成为现代生物学和育种学中一项重要的方法和技术。但是在其广泛运用的过程中也逐渐暴露了一些问题。其中之一就是现阶段研究中经常用组成型启动子来驱动外源基因表达,由于组成型启动子会使驱动的目的基因在植物各组织中恒定而持续的表达,从而过度消耗细胞内的物质和能量,并且不能从时间和空间上有效地调控目的基因的表达,所以有时会带来一些负面效应。如外源基因在整株植物中表达,产生的大量异源蛋白质或代谢产物在植物体内积累,打破了植物原有的代谢平衡,不利于产量和品质的提高;有些产物对植物并非必需甚至有毒,因而阻碍了植物的正常生长,甚至导致死亡(Karlowsikiand Hirsch,2003;Ehsani et al.,2003)。如利用组成型启动子表达病毒衣壳蛋白,就可能会引起病毒农壳转移,从而导致植物病毒新株系的产生(Robinson,1996),因此也引起了一些安全性方面的思考。另外,重复使用同一种组成型启动子驱动两个或两个以上的外源基因表达可能会引起基因沉默或共抑制现象(Kumpatlaet al.,1998),所以给多基因转化也带来了不少麻烦。因此,启动子在植物基因工程中的研究地位也越来越突出。研究者们通过寻找更为有效的组织、器官特异性表达启动子或者是诱导表达启动子来代替组成型启动子,以期更好地调控植物基因表达。诱导型启动子虽然优点突出,利用诱导型启动子获得转基因植株尽管也有一些报导,但到目前为止能用于生产实际的还比较少,而组织器官特异表达的启动子有望用于生产实际则有一些较成功的例子(Cai et al.,2007;Ye et al.,2009)。Researching and improving crops by means of molecular biology and genetic engineering has a good application prospect in agricultural production. Since the first transgenic tobacco was obtained in 1983 (Zambryski et al., 1983), in the past 30 years, the research on plant genetic engineering has made rapid progress, and has become an important task in modern biology and breeding. methods and techniques. However, some problems have gradually been exposed in the process of its wide application. One of them is that constitutive promoters are often used to drive the expression of exogenous genes in current research, because constitutive promoters will cause the driven target genes to express constantly and continuously in various plant tissues, thereby excessively consuming intracellular substances and energy, and cannot effectively regulate the expression of the target gene in time and space, so sometimes it will bring some negative effects. If the exogenous gene is expressed in the whole plant, a large amount of heterologous protein or metabolites will accumulate in the plant, breaking the original metabolic balance of the plant, which is not conducive to the improvement of yield and quality; some products are not necessary or even toxic to plants , thus hindering the normal growth of plants and even leading to death (Karlowsiki and Hirsch, 2003; Ehsani et al., 2003). If a constitutive promoter is used to express the viral capsid protein, it may cause the transfer of the viral capsid, thereby leading to the generation of new plant virus strains (Robinson, 1996), which also caused some safety considerations. In addition, repeated use of the same constitutive promoter to drive the expression of two or more foreign genes may cause gene silencing or co-suppression (Kumpatla et al., 1998), so it also brings a lot of benefits to multigene transformation. trouble. Therefore, the research status of promoters in plant genetic engineering is becoming more and more prominent. Researchers have replaced constitutive promoters by looking for more effective tissue- and organ-specific expression promoters or inducible expression promoters in order to better regulate plant gene expression. Although inducible promoters have outstanding advantages, although there are some reports on the use of inducible promoters to obtain transgenic plants, there are still relatively few that can be used for actual production so far, and the promoters with specific expression in tissues and organs are expected to be used in actual production. Some more successful examples (Cai et al., 2007; Ye et al., 2009).
所谓组织特异型启动子是指除具有一般启动子的结构外,通常还有增强子和沉默子的一般特性。在组织特异型启动子调控下,基因的表达常常只发生在某些特定的器官或组织部位,并常常表现出发育调节的特性。这些启动子的调控往往受到组织细胞生理状态和化学物理信号等物质的诱导,还受到发育阶段的调控,其表达是多种因子相互作用的结果。The so-called tissue-specific promoter refers to the general characteristics of enhancers and silencers in addition to the structure of general promoters. Under the regulation of tissue-specific promoters, the expression of genes often occurs only in some specific organs or tissue parts, and often shows the characteristics of developmental regulation. The regulation of these promoters is often induced by substances such as the physiological state of tissue cells and chemical and physical signals, and is also regulated by the developmental stage, and its expression is the result of the interaction of various factors.
果实和种子是植物的生殖器官,也是营养物质主要的储藏场所。利用果实或种子等器官特异性启动子调控基因表达,不仅可提高基因在这些部位的表达量,将生物能耗降到最低,利于表达产物的分离,而且可有目的地提高转基因植物果实或种子的营养或改善其品质。Sandhu等利用E8启动子(成熟果实中乙烯应答性基因的启动子)驱动呼吸道合胞病毒F抗原基因成功转化番茄植株,用果实特异表达抗原喂饲小鼠,可诱导小鼠产生特异的粘膜免疫反应、血清学抗体反应及TH1型细胞免疫反应(Sandhu et al.,2000)。Vasconcelos等用水稻胚乳特异性谷蛋白基因启动子驱动大豆铁蛋白基因,使转基因水稻谷粒中铁和锌的含量都有所增加,而且铁蛋白主要积累在胚乳中,不会在食品加工过程中丢失(Vasconcelos et al.,2003)。研究人员在一些单子叶植物的种子储藏蛋白中也分离了一些胚乳特异表达的顺式作用元件,最著名的GCN4元件被认为对维持胚乳特异表达活性是必须的(Wu et al.,1998)。但也有文献报导,GCN4元件仅仅是起到增强启动子在胚乳中的表达强度的作用,其并不能决定启动子胚乳特异表达的特性(Vickers et al.,2006)。另外,一些种子中特异表达的启动子如PsGNS2启动子(Buchner et al.,2002)、FAE1(Rossak et al.,2001)启动子等也有相关的报导。Fruits and seeds are the reproductive organs of plants and the main storage places for nutrients. Utilizing organ-specific promoters such as fruits or seeds to regulate gene expression can not only increase the expression of genes in these parts, minimize bioenergy consumption, facilitate the separation of expression products, but also can purposefully improve the expression of transgenic plant fruits or seeds. nutrition or improve its quality. Sandhu et al. used the E8 promoter (the promoter of ethylene-responsive genes in mature fruits) to drive the respiratory syncytial virus F antigen gene to successfully transform tomato plants, and fed mice with fruit-specific antigen expression, which could induce specific mucosal immunity in mice response, serological antibody response and TH1 type cellular immune response (Sandhu et al., 2000). Vasconcelos et al. used rice endosperm-specific glutenin gene promoter to drive soybean ferritin gene, which increased the content of iron and zinc in transgenic rice grains, and ferritin was mainly accumulated in endosperm and would not be lost during food processing (Vasconcelos et al., 2003). Researchers have also isolated some endosperm-specific cis-acting elements in some monocot seed storage proteins, and the most famous GCN4 element is considered to be necessary for maintaining endosperm-specific expression activity (Wu et al., 1998). However, it has also been reported in the literature that the GCN4 element only plays a role in enhancing the expression intensity of the promoter in the endosperm, and it cannot determine the specific expression characteristics of the promoter in the endosperm (Vickers et al., 2006). In addition, some promoters specifically expressed in seeds, such as PsGNS2 promoter (Buchner et al., 2002), FAE1 (Rossak et al., 2001) promoter, etc., have also been reported.
水稻是世界上最重要的粮食作物之一,是禾本科作物功能基因组学研究的模式植物。启动子是精确调控基因表达的“开关”。因此对水稻组织器官特异表达启动子的分离克隆及深入研究,不仅有助于阐明植物形态、发育、代谢途径等基础理论,还可以指导转基因育种,创造巨大的经济效益和社会效益,更好的为人类生产生活服务。本发明就是利用有关分子生物学方法,从水稻“明恢63”基因组中分离克隆了同一个胚乳特异表达基因上游不同长度的启动子区域,将不同长度的启动子融合报告基因GUS导入水稻“中花11”中,验证其表达模式,进而鉴定出控制表达量及表达模式区段。Rice is one of the most important food crops in the world, and it is a model plant for functional genomics research of gramineous crops. Promoters are "switches" that precisely regulate gene expression. Therefore, the isolation, cloning and in-depth study of rice tissue and organ-specific expression promoters will not only help to clarify basic theories such as plant morphology, development, and metabolic pathways, but also guide transgenic breeding and create huge economic and social benefits. Serving human production and life. The present invention utilizes relevant molecular biological methods to separate and clone the same endosperm-specific expression gene upstream promoter regions of different lengths from the genome of rice "Minghui 63", and introduce the promoter fusion reporter gene GUS of different lengths into rice "Minghui 63" In Hua 11", the expression pattern was verified, and the segment that controlled the expression level and expression pattern was identified.
发明内容Contents of the invention
本发明的目的在于克服现有可用于水稻基因工程研究的组织特异表达启动子数目的不足,从水稻“明恢63”(我国广泛应用的优良水稻恢复系)基因组中分离并鉴定出胚乳特异表达的启动子,有望将这些启动子用于水稻的基因工程改良。为了便于研究利用,我们创建了EnP3、EnP3-859、EnP3-678、EnP2-471、EnP3-292和EnP3-110共6个不同长度的启动子。将这些启动子融合报告基因β-葡糖醛酸酶基因(以下简称GUS基因)导入水稻“中花11”(中国农业科学院作物研究所商业经营品种)中,验证其表达模式,并进行表达量的测定,为日后利用该启动子奠定了基础。The purpose of the present invention is to overcome the deficiency in the number of tissue-specific expression promoters available for rice genetic engineering research, to isolate and identify endosperm-specific expression promoters from the genome of rice "Minghui 63" (an excellent rice restorer line widely used in my country) It is expected that these promoters will be used for genetic engineering improvement of rice. In order to facilitate research and utilization, we created six promoters of different lengths, namely EnP3, EnP3-859, EnP3-678, EnP2-471, EnP3-292 and EnP3-110. These promoters were fused with the reporter gene β-glucuronidase gene (hereinafter referred to as the GUS gene) into the rice "Zhonghua 11" (commercially operated variety of the Crop Research Institute of the Chinese Academy of Agricultural Sciences), and the expression pattern was verified, and the expression level was measured. The determination of the promoter laid the foundation for the future use of the promoter.
本发明是这样实现的(本发明的技术路线见图1):The present invention is realized like this (technical path of the present invention sees Fig. 1):
利用华中农业大学作物遗传改良国家重点实验室水稻全生育期表达谱芯片数据库CREP(http://crep.ncpgr.cn)(Wang et al.,2010)及反转录PCR(以下简称RT-PCR)等方法,从水稻“明恢63”(我国广泛推广应用的优良水稻恢复系)基因组中分离并鉴定出具有胚乳特异性表达的启动子。申请人将其命名为EnP3、EnP3-859、EnP3-678、EnP3-471、EnP3-292和EnP3-110。所述的启动子EnP3序列,它是序列表SEQ ID NO:1所示的序列。通过分析该启动子驱动GUS报告基因在转基因水稻中的表达模式发现:该启动子仅在转化植株的胚乳表达,并且随着胚乳发育阶段的不断进行,表达量呈现下降趋势;在其他组织如叶片、叶鞘、茎杆、根、花、颖壳及胚中均检测不到表达。所述的启动子EnP3-859、EnP3-678、EnP2-471和EnP3-292序列,分别是序列表SEQ ID NO:2、SEQ ID NO:3、SEQ ID NO:4、SEQ ID NO:5所示的序列,它们分别是由启动子EnP3截短的核心区859bp、678bp、471bp及292bp的序列。这4个片段均具有独立的启动基因表达的功能,并且在转化植株中的表达模式与EnP3相同,即仅在胚乳表达,不过表达量各有区别。所述的启动子EnP3-110序列,它是序列表SEQ ID NO:6所示的序列,是由EnP3-292进一步截短的110bp的序列,分析其驱动GUS报告基因在转基因水稻中表达情况发现,这个区段的表达模式发生了显著的改变:在叶片、叶鞘、茎杆及颖壳这些绿色组织中可以检测到表达,但是在根、花及种子(包括胚和胚乳)中却检测不到表达,成为一个短的绿色组织特异表达启动子。Using the CREP (http://crep.ncpgr.cn) (Wang et al., 2010) and reverse transcription PCR (hereinafter referred to as RT-PCR) ) and other methods, isolated and identified an endosperm-specific expression promoter from the genome of rice "Minghui 63" (an excellent rice restorer line widely used in my country). Applicants named them EnP3, EnP3-859, EnP3-678, EnP3-471, EnP3-292 and EnP3-110. The promoter EnP3 sequence is the sequence shown in SEQ ID NO: 1 in the sequence table. By analyzing the expression pattern of the promoter-driven GUS reporter gene in transgenic rice, it was found that the promoter was only expressed in the endosperm of transformed plants, and the expression level showed a downward trend as the endosperm development continued; in other tissues such as leaves , leaf sheaths, stems, roots, flowers, glumes and embryos, no expression was detected. The promoters EnP3-859, EnP3-678, EnP2-471 and EnP3-292 sequences are respectively listed in the sequence table SEQ ID NO: 2, SEQ ID NO: 3, SEQ ID NO: 4, and SEQ ID NO: 5 The sequences shown are the 859bp, 678bp, 471bp and 292bp sequences of the core region truncated by the promoter EnP3 respectively. These four fragments all have the function of independently promoting gene expression, and the expression pattern in transformed plants is the same as that of EnP3, that is, only expressed in the endosperm, but the expression levels are different. The promoter EnP3-110 sequence, which is the sequence shown in the sequence table SEQ ID NO: 6, is a 110bp sequence further truncated by EnP3-292, and it is found that it drives the expression of the GUS reporter gene in transgenic rice , the expression pattern of this segment has changed significantly: expression can be detected in green tissues such as leaves, leaf sheaths, stems and glumes, but not in roots, flowers and seeds (including embryos and endosperms) expression, as a short green tissue-specific expression promoter.
本发明的具体步骤是:Concrete steps of the present invention are:
首先在华中农业大学作物遗传改良国家重点实验室水稻全生育期表达谱芯片数据库CREP(http://crep.ncpgr.cn)(Wang et al.,2010)上发现了一个可能是胚乳特异表达的候选基因,在美国国立生物研究所网站NCBI(http://www.ncbi.nlm.nih.gov/)上获得该基因的序列,如SEQ ID NO:7所示,共459bp,以这459bp的序列设计引物来做RT-PCR反应从而进一步确定其表达谱,RT-PCR结果显示(见图2):该基因仅在胚乳中表达。在此基础上用特异性引物以PCR的方法从水稻“明恢63”基因组中扩增得到一个被命名为EnP3的启动子候选片段,将该启动子候选片段EnP3与报告基因GUS编码序列构建成融合基因并装载到双元Ti载体DX2181(载体图及多克隆位点等信息见图3)上,装配成EnP3-GUS载体(见图4),再通过农杆菌介导的遗传转化方法,将EnP3-GUS载体转入水稻受体“中花11”中,同时转化空载体DX2181作为阴性对照,转化花椰菜花叶病毒35S启动子驱动GUS(CaMV35S-GUS)作为阳性对照。获得转基因植株后通过细织化学染色及GUS活性定量分析,考察该启动子在胚乳发育各时期的表达变化,进而验证和克隆该启动子。检测结果表明:该启动子仅在转化植株的胚乳表达,并且随着胚乳发育阶段的不断进行,表达量呈现下降趋势;在其他组织如叶片、叶鞘、茎杆、根、花、颖壳及胚中均检测不到表达。采用片段缺失的方法,我们构建了5个来源于EnP3的5’端缺失片段连接GUS的表达载体,同样通过农杆菌介导的遗传转化方法将其转入水稻受体“中花11”中,分析这5个片段的表达模式,结果显示:来源于启动子EnP3的4个区段(EnP3-859、EnP3-678、EnP3-471和EnP3-292)均具有独立的启动基因表达的功能,并且在转化植株中的表达模式与EnP3相同,即仅在胚乳表达,不过表达量各有区别;由EnP3-292进一步截短的EnP3-110区段的表达模式发生了显著的改变:在叶片、叶鞘、茎杆及颖壳这些绿色组织中可以检测到表达,但是在根、花及种子(包括胚和胚乳)中却检测不到表达,成为一个短的绿色组织特异表达启动子。Firstly, a protein that may be endosperm-specific expression was found on the CREP (http://crep.ncpgr.cn) (Wang et al., 2010) of the rice expression profile chip database of the State Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University. Candidate gene, obtain the sequence of this gene on the website NCBI of National Institute of Biology (http://www.ncbi.nlm.nih.gov/), as shown in SEQ ID NO: 7, a total of 459bp, with this 459bp Primers were designed for the sequence to do RT-PCR reaction to further confirm its expression profile, and the RT-PCR results showed (see Figure 2): the gene was only expressed in the endosperm. On this basis, a candidate promoter fragment named EnP3 was amplified from the rice "Minghui 63" genome by PCR with specific primers, and the candidate promoter fragment EnP3 and the reporter gene GUS coding sequence were constructed into The fusion gene was loaded onto the binary Ti vector DX2181 (see Figure 3 for the vector map and multiple cloning sites, etc.), assembled into an EnP3-GUS vector (see Figure 4), and then through the Agrobacterium-mediated genetic transformation method, the The EnP3-GUS vector was transformed into the rice recipient "Zhonghua 11", and the empty vector DX2181 was transformed as a negative control, and the cauliflower mosaic virus 35S promoter-driven GUS (CaMV35S-GUS) was transformed as a positive control. After the transgenic plants were obtained, chemical staining and GUS activity quantitative analysis were carried out to investigate the expression changes of the promoter at various stages of endosperm development, and then to verify and clone the promoter. The test results showed that the promoter was only expressed in the endosperm of transformed plants, and the expression level showed a downward trend as the endosperm development continued; in other tissues such as leaves, leaf sheaths, stems, roots, flowers, glumes and embryos No expression was detected in any of the . Using the method of fragment deletion, we constructed 5 expression vectors with 5'-end deletion fragments derived from EnP3 linked to GUS, and transferred them into the rice recipient "Zhonghua 11" through the genetic transformation method mediated by Agrobacterium. Analysis of the expression patterns of these 5 fragments showed that the 4 segments (EnP3-859, EnP3-678, EnP3-471 and EnP3-292) derived from the promoter EnP3 all have the function of independently promoting gene expression, and The expression pattern in transformed plants is the same as that of EnP3, that is, it is only expressed in the endosperm, but the expression levels are different; the expression pattern of the EnP3-110 segment further truncated by EnP3-292 has changed significantly: in leaves, leaf sheaths Expression can be detected in green tissues such as , stem and glume, but no expression can be detected in roots, flowers and seeds (including embryo and endosperm), and it becomes a short green tissue-specific expression promoter.
本发明的优点在于:The advantages of the present invention are:
(1)本发明鉴定了胚乳特异表达的启动子EnP3及控制其表达量及表达模式的核心区域,为基因工程和分子育种提供了新的特异表达的启动子资源。(1) The present invention identifies the endosperm-specific expression promoter EnP3 and the core region controlling its expression level and expression pattern, and provides new specific expression promoter resources for genetic engineering and molecular breeding.
(2)本发明可以直接应用于水稻胚乳特异表达的启动子的鉴定和克隆。(2) The present invention can be directly applied to the identification and cloning of the rice endosperm-specific expression promoter.
附图说明Description of drawings
序列表SEQ ID NO:1,公开了本发明克隆的的水稻胚乳特异表达启动子EnP3的核苷酸序列,长度为1040bp。Sequence Listing SEQ ID NO: 1 discloses the nucleotide sequence of the rice endosperm-specific expression promoter EnP3 cloned in the present invention, with a length of 1040bp.
序列表SEQ ID NO:2是从所述的SEQ ID NO:1核苷酸序列的克隆得到的作为另一个启动子应用的核苷酸序列,长度为859bp。Sequence listing SEQ ID NO: 2 is a nucleotide sequence used as another promoter obtained from the cloning of the SEQ ID NO: 1 nucleotide sequence, and the length is 859bp.
序列表SEQ ID NO:3是从所述的SEQ ID NO:1核苷酸序列的克隆得到的作为另一个启动子应用的核苷酸序列,长度为678bp。Sequence Listing SEQ ID NO: 3 is a nucleotide sequence used as another promoter obtained from the cloning of the nucleotide sequence of SEQ ID NO: 1, and the length is 678bp.
序列表SEQ ID NO:4是从所述的SEQ ID NO:1核苷酸序列的克隆得到的作为另一个启动子应用的核苷酸序列,长度为471bp。Sequence Listing SEQ ID NO: 4 is a nucleotide sequence used as another promoter obtained from the cloning of the SEQ ID NO: 1 nucleotide sequence, with a length of 471bp.
序列表SEQ ID NO:5是从所述的SEQ ID NO:1核苷酸序列的克隆得到的作为另一个启动子应用的核苷酸序列,长度为292bp。Sequence Listing SEQ ID NO: 5 is a nucleotide sequence used as another promoter obtained from the cloning of the SEQ ID NO: 1 nucleotide sequence, and the length is 292bp.
序列表SEQ ID NO:6是从所述的SEQ ID NO:1核苷酸序列的克隆得到的作为另一个启动子应用的核苷酸序列,长度为110bp。Sequence Listing SEQ ID NO: 6 is a nucleotide sequence obtained from the cloning of the nucleotide sequence of SEQ ID NO: 1 and used as another promoter, with a length of 110bp.
序列表SEQ ID NO:7是本发明克隆的水稻胚乳特异表达启动子EnP3在基因组中所驱动的基因(Genebank登录号为GI:31415924)的核苷酸序列,长度为459bp。Sequence listing SEQ ID NO: 7 is the nucleotide sequence of the gene (Genebank accession number is GI: 31415924) driven by the rice endosperm-specific expression promoter EnP3 cloned in the present invention in the genome, and the length is 459bp.
序列表SEQ ID NO:8是本发明克隆的水稻胚乳特异表达启动子EnP3在基因组中所驱动的基因(Genebank登录号为GI:31415924)编码的氨基酸序列,长度为134个氨基酸。Sequence Listing SEQ ID NO: 8 is the amino acid sequence encoded by the gene (Genebank accession number GI: 31415924) driven by the rice endosperm-specific expression promoter EnP3 cloned in the present invention in the genome, with a length of 134 amino acids.
图1:实现本发明的技术路线图。Figure 1: A technical roadmap for realizing the present invention.
图2:利用RT-PCR的方法来分析EnP3在基因组中所驱动的基因(GI:31415924)的表达模式。上下列分别表示的为内参水稻Actin1基因和目的基因(GI:31415924)在各个组织中的表达情况。Figure 2: Using RT-PCR to analyze the expression pattern of the gene (GI: 31415924) driven by EnP3 in the genome. The upper and lower columns respectively represent the expression of the internal reference rice Actin1 gene and the target gene (GI: 31415924) in each tissue.
图3:表示双元载体DX2181结构示意图。该载体是在pCAMBIA1380的基础上改造而来,在多克隆位点处以相反的方向分别构建了一个GUS基因和EGFP基因。Figure 3: Schematic representation of the structure of the binary vector DX2181. The vector was transformed on the basis of pCAMBIA1380, and a GUS gene and EGFP gene were constructed in opposite directions at the multiple cloning site.
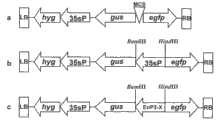
图4:构建好的以DX2181为骨架的表达载体简图。a为阴性对照,即DX2181空载体,无启动子驱动GUS基因表达;b为阳性对照,即用CaMV35S启动子来驱动GUS基因表达;c为EnP3及其系列缺失片段来驱动GUS基因表达。LB、RB分别为DX2181中T-DNA的左边界和右边界;hyg为潮霉素抗性筛选基因;gus为报告基因GUS(β-葡萄糖苷酸酶);egfp为报告基因EGFP(增强的绿色荧光蛋白);MCS表示多克隆位点;35sP表示CaMV35S启动子(花椰菜花叶病毒35S启动子);EnP3-X表示EnP3及其系列缺失片段;箭头方向表示基因或启动子表达的方向。Figure 4: Schematic diagram of the constructed expression vector with DX2181 as the backbone. a is the negative control, that is, the DX2181 empty vector, without a promoter to drive the expression of the GUS gene; b is the positive control, that is, the CaMV35S promoter is used to drive the expression of the GUS gene; c is EnP3 and its series of deletion fragments to drive the expression of the GUS gene. LB and RB are the left and right borders of T-DNA in DX2181, respectively; hyg is the hygromycin resistance screening gene; gus is the reporter gene GUS (β-glucuronidase); egfp is the reporter gene EGFP (enhanced green Fluorescent protein); MCS indicates the multiple cloning site; 35sP indicates the CaMV35S promoter (cauliflower mosaic virus 35S promoter); EnP3-X indicates EnP3 and its series of deletion fragments; the direction of the arrow indicates the direction of gene or promoter expression.
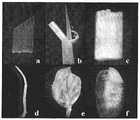
图5:由EnP3驱动GUS基因的转化植株的各个组织的组织化学染色。a为叶片;b为叶鞘;c为茎杆;d为根;e为花;f为种子(含胚与胚乳)。Fig. 5: Histochemical staining of various tissues of transformed plants with GUS gene driven by EnP3. a is the leaf; b is the sheath; c is the stem; d is the root; e is the flower; f is the seed (embryo and endosperm).
图6:由EnP3及其系列缺失片段驱动GUS基因的转化植株的成熟种子的组织化学染色。a、b、c、d、e、f分别表示的是启动子片段EnP3、EnP3-859、EnP3-678、EnP3-471、EnP3-292和EnP3-110的表达情况,图中En表示胚乳,Em表示胚。Figure 6: Histochemical staining of mature seeds of transformed plants with GUS gene driven by EnP3 and its serial deletion fragments. a, b, c, d, e, f represent the expression of promoter fragments EnP3, EnP3-859, EnP3-678, EnP3-471, EnP3-292 and EnP3-110 respectively, in the figure En represents endosperm, Em Indicates embryo.
图7:由EnP3-110驱动GUS基因的转化植株的各个组织的组织化学染色。a为叶片;b为叶鞘;c为茎杆;d为根;e为花;f为种子(含胚与胚乳)。Fig. 7: Histochemical staining of various tissues of transformed plants with GUS gene driven by EnP3-110. a is the leaf; b is the sheath; c is the stem; d is the root; e is the flower; f is the seed (embryo and endosperm).
图8:由EnP3-110驱动GUS基因的转化植株的各个组织的GUS活性定量检测,DX2181表示阴性对照。Figure 8: Quantitative detection of GUS activity in various tissues of transformed plants driven by EnP3-110 GUS gene, DX2181 represents the negative control.
图9:由EnP3及其系列缺失片段驱动GUS基因的转化植株的胚乳在不同发育时期的GUS活性定量检测。图例中的DAF表示开花后天数(days after flowering);DX2181表示阴性对照。Figure 9: Quantitative detection of GUS activity in endosperm of transformed plants driven by EnP3 and its serial deletion fragments of GUS gene at different developmental stages. DAF in the legend indicates the days after flowering (days after flowering); DX2181 indicates the negative control.
具体实施方式Detailed ways
实施例1:胚乳特异表达候选基因(Genebank登录号为GI:31415924)的表达谱验证Example 1: Verification of the expression profile of the endosperm-specific expression candidate gene (Genebank accession number is GI: 31415924)
材料准备:本实施例所用材料是籼稻品种(Oryza sativa ssp.Indica)明恢63(我国广泛应用的优良水稻恢复系),种植方法是水培。营养液成分如下所示:1.44mM NH4NO3,0.3mM NaH2PO4,0.5mM K2SO4,1.0mM CaCl2,1.6mM MgSO4,0.17mM NaSiO3,50μm Fe-EDTA,0.06μM(NH4)6Mo7O24,15μM H3BO3,8μMMnCl2,0.12μM CuSO4,0.12μM ZnSO4,29μM FeCl3,40.5μM Citric acid,pH5.5(Yoshida et al.,1976)。在孕穗期取叶片、叶鞘、茎杆、根、穗,受精14天后取胚乳。RNA提取采用Trizol Reagent(Invitrogen,Carisbad,CA,USA)方法。总RNA经1.4%琼脂糖胶电泳检测和浓度测定确认质量合格(18S、28S两条主带清晰无拖尾,OD260/OD280=1.8-2.0)后方可进行后续试验。将各个组织的RNA反转录成cDNA,置于-20℃冰箱保存。反转录使用的试剂盒是Promega公司的M-MLV,按照产品说明书操作即可。Material preparation: The material used in this example is the indica rice variety (Oryza sativa ssp.Indica) Minghui 63 (an excellent rice restorer line widely used in my country), and the planting method is hydroponics. The composition of the nutrient solution is as follows: 1.44mM NH4 NO3 , 0.3mM NaH2 PO4 , 0.5mM K2 SO4 , 1.0mM CaCl2 , 1.6mM MgSO4 , 0.17mM NaSiO3 , 50μm Fe-EDTA, 0.06μM (NH4 )6 Mo7 O24 , 15 μM H3 BO3 , 8 μM nCl2 , 0.12 μM CuSO4 , 0.12 μM ZnSO4 , 29 μM FeCl3 , 40.5 μM Citric acid, pH 5.5 (Yoshida et al., 1976). Take leaves, leaf sheaths, stems, roots and ears at the booting stage, and take endosperm 14 days after fertilization. RNA was extracted using the Trizol Reagent (Invitrogen, Carisbad, CA, USA) method. The total RNA was detected by 1.4% agarose gel electrophoresis and concentration determination to confirm that the quality was qualified (the two main bands of 18S and 28S were clear without tailing, OD260/OD280=1.8-2.0) before subsequent tests could be carried out. The RNA of each tissue was reverse-transcribed into cDNA and stored in a -20°C refrigerator. The kit used for reverse transcription is M-MLV from Promega, and it can be operated according to the product instructions.
首先在华中农业大学作物遗传改良国家重点实验室水稻全生育期表达谱芯片数据库CREP(http://crep.ncpgr.cn)(Wang et al.,2010)中发现了一个可能是胚乳特异表达的候选基因,所对应的探针号是Os.13536.1.S1_at,芯片显示在胚乳发育的7,14,21天均高量表达,在其他组织无表达或表达量很低。该探针所表征的基因在TIGR上的登录号是LOC_Os03g55734,在NCBI上的基因号为GI:31415924,基因功能注释为“可能的醇溶谷蛋白(putative prolamin)”。基因大小为459bp,mRNA大小为459bp,编码134个氨基酸(见序列表SEQ ID NO:8)。设计能扩增部分mRNA的RT-PCR引物(YRJTZ3F:AAGCATCTCCCATACGCTGT,YRJTZ3R:TCAACAACAACCATAAGGAAAGA,扩增大小为430bp),以水稻“明恢63”的叶片、叶鞘、茎杆、根、穗、胚乳的RNA反转录产物为模板,以水稻Actin1为内参(扩增引物为actin1F:GCCACACTGTCCCCATCTAT,actin1R:GCGACCACCTTGATCTTCAT)。PCR反应条件:94℃ 5min,94℃ 30sec,55.5℃ 30sec,72℃ 50sec,30个循环,72℃ 7min。PCR产物经0.8%琼脂糖胶电泳检测。检测结果见附图2。结果显示,候选基因GI:31415924在胚乳中表达,在其他组织不表达。Firstly, a protein that may be endosperm-specific expression was found in the CREP (http://crep.ncpgr.cn) (Wang et al., 2010) of the rice expression profile chip database of the State Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University. The candidate gene, corresponding to the probe number is Os.13536.1.S1_at, the microarray showed that it was highly expressed on the 7th, 14th and 21st days of endosperm development, and had no expression or very low expression in other tissues. The accession number of the gene characterized by this probe is LOC_Os03g55734 on TIGR, the gene number on NCBI is GI:31415924, and the gene function is annotated as "possible prolamin (putative prolamin)". The size of the gene is 459bp, the size of the mRNA is 459bp, encoding 134 amino acids (see the sequence listing SEQ ID NO: 8). RT-PCR primers (YRJTZ3F: AAGCATCTCCCATACGCTGT, YRJTZ3R: TCAACAACAACCATAAGGAAAGA, 430 bp) capable of amplifying part of mRNA were designed, and the RNA of leaves, leaf sheaths, stems, roots, panicles, and endosperms of rice "Minghui 63" were used for reverse expression. The transcript was used as a template, and rice Actin1 was used as an internal reference (amplification primers were actin1F: GCCACACTGTCCCCCATCTAT, actin1R: GCGACCACCTTGATCTTCAT). PCR reaction conditions: 94°C for 5min, 94°C for 30sec, 55.5°C for 30sec, 72°C for 50sec, 30 cycles, 72°C for 7min. PCR products were detected by 0.8% agarose gel electrophoresis. The test results are shown in Figure 2. The results showed that the candidate gene GI:31415924 was expressed in endosperm but not in other tissues.
实施例2:胚乳特异表达启动子EnP3候选片段及相应缺失片段的获得(相应分子生物学常规操作参考《分子克隆实验指南(第二版)》(J.萨姆布鲁克等,1996))Example 2: Acquisition of endosperm-specific expression promoter EnP3 candidate fragments and corresponding deletion fragments (refer to "Molecular Cloning Experiment Guide (Second Edition)" (J. Sambrook et al., 1996) for routine molecular biology operations)
明恢63基因组DNA的抽提:在明恢63分蘖盛期取新鲜叶片用于抽提其基因组DNA,具体方法为Murray报道的十六烷基三甲基溴化铵(Cetyltrimethyl Ammonium Bromide,以下简称CTAB)抽提法(Murray and Thompson.1980),抽提出的DNA完全溶解后置于-20℃冰箱保存。Extraction of Minghui 63 genomic DNA: Take fresh leaves at the peak tillering stage of Minghui 63 to extract its genomic DNA. CTAB) extraction method (Murray and Thompson.1980), the extracted DNA was completely dissolved and stored in a -20°C refrigerator.
在生物信息学网站NCBI(http://www.ncbi.nlm.nih.gov/)上提取候选基因GI:31415924的上游序列,具体来讲是-976至+54(转录起始点为+1)区间共计1130bp作为启动子候选片段。将其命名为EnP3。利用PCR法,以抽提的明恢63基因组DNA为模板,通过设计特异引物(EnP3F:AAGCTTGGAGCATTTGTAGGAATGCC;EnP3R:GGATCCTGTTGACAGCGTATGGGAGA)来扩增EnP3,为了后续构建载体的方便,左右引物分别添加HindIII、BamHI酶切位点(下划线表示)及相应的保护碱基(阴影表示)。PCR反应条件:94℃ 5min,94℃ 1min,66℃ 50sec,72℃ 1min30sec,30个循环,72℃ 7min。Extract the upstream sequence of the candidate gene GI: 31415924 on the bioinformatics website NCBI (http://www.ncbi.nlm.nih.gov/), specifically -976 to +54 (transcription start point is +1) A total of 1130bp intervals were used as promoter candidate fragments. Name it EnP3. Using the PCR method, using the extracted Minghui 63 genomic DNA as a template, by designing specific primers (EnP3F:AAGCTT GGAGCATTTGTAGGAATGCC; EnP3R:GGATCC TGTTGACAGCGTATGGGAGA) to amplify EnP3, for the convenience of subsequent vector construction, the left and right primers were added with HindIII, BamHI restriction sites (underlined) and corresponding protective bases (shaded). PCR reaction conditions: 94°C for 5min, 94°C for 1min, 66°C for 50sec, 72°C for 1min30sec, 30 cycles, 72°C for 7min.
取EnP3的部分PCR产物用0.8%琼脂糖电泳检测。剩余的PCR产物用于做TA克隆。使用的试剂盒为Promega公司的T-Vctor系统。反应体系为5.0μl,具体如下:EnP3PCR产物1.9μl、T-Vctor 0.3μl、2倍缓冲液(以下简写为buffer)2.5μl、T4 ligase 0.3μl。Part of the PCR product of EnP3 was detected by 0.8% agarose electrophoresis. The remaining PCR products were used for TA cloning. The kit used is the T-Vctor system of Promega Company. The reaction system was 5.0 μl, specifically as follows: 1.9 μl of EnP3 PCR product, 0.3 μl of T-Vctor, 2.5 μl of 2-fold buffer (abbreviated as buffer hereinafter), and 0.3 μl of T4 ligase.
连接产物于16℃低温水浴10h,然后电转化(1800伏,电击2-3秒)入大肠杆菌DH5α(该菌株为商业化的大肠杆菌菌株),37℃复苏40min,涂含氨苄霉素(Amp)、异丙基-β-D-硫代半乳糖苷(IPTG)、5-溴-4-氯-3-吲哚-β-D-半乳糖苷(X-gal)的平板,37℃培养过夜,挑选白斑用含Amp的LB培养基来扩大培养。抽提所摇菌的质粒,用HindIII和BamHI双酶切来筛选阳性克隆。酶切体系为20.0μl,具体如下:质粒3.0μl、10×K buffer2.0μl、HindIII0.1μl、BamHI0.1μl、ddH2O14.8μl,37℃反应4h,酶切产物用0.8%琼脂糖电泳检测。挑选阳性克隆用ABI3730测序仪进行测序。测序结果显示来源于明恢63基因组的EnP3序列(序列表SEQ ID NO:1所示)与NCBI上报道日本晴的序列并非是100%完全匹配,存在少量差异,最终大小为1040bp,后续5’缺失片段的设计均以明恢63基因组的序列为准。The ligation product was placed in a low-temperature water bath at 16°C for 10 hours, then electrotransformed (1800 volts, electric shock for 2-3 seconds) into Escherichia coli DH5α (this strain is a commercial Escherichia coli strain), recovered at 37°C for 40 minutes, and coated with ampicillin (Amp ), isopropyl-β-D-thiogalactoside (IPTG), 5-bromo-4-chloro-3-indole-β-D-galactoside (X-gal), cultured at 37°C Overnight, pick the white spot and use LB medium containing Amp to expand the culture. Extract the plasmid of the shaken bacteria, and use HindIII and BamHI double digestion to screen positive clones. The enzyme digestion system is 20.0 μl, specifically as follows: 3.0 μl of plasmid, 2.0 μl of 10×K buffer, 0.1 μl of HindIII, 0.1 μl of BamHI, 14.8 μl of ddH2 O, react at 37°C for 4 hours, and detect the digested products by 0.8% agarose electrophoresis . Positive clones were selected for sequencing with an ABI3730 sequencer. Sequencing results show that the EnP3 sequence derived from the Minghui 63 genome (shown in the sequence table as SEQ ID NO: 1) is not 100% completely matched with the sequence reported by Nipponbare on NCBI, and there are a few differences. The final size is 1040bp, and the subsequent 5' deletion The fragments were designed based on the genome sequence of Minghui 63.
5个5’缺失片段与全长启动子共用同一个右引物(EnP3R:GGATCCTGTTGACAGCGTATGGGAGA),左引物根据它们最终的扩增片段大小分别命名为EnP3F-859(AAGCTTTTGGAATTGCGCGAAGTTAG)、EnP3F-678(AAGCTTGCTAAATGTTACTTCCTCCG)、EnP3F-471(CAAGCTTGTCGTGACTTATGTAACACG)、EnP3F-292(AAGCTTGCACCAATGCACCATTGATC)和EnP3F-110(AAGCTTCCTATAAATAATCCCCTAGAGC)。在每条左引物5’端都引入HindIII酶切位点(以下划线表示)及相应的保护碱基(以阴影表示)。PCR模板为测序正确的TA-EnP3质粒。得到扩增产物之后,后续操作按照构建TA-EnP3的方法进行。The five 5' deletions share the same right primer with the full-length promoter (EnP3R:GGATCC TGTTGACAGCGTATGGGAGA), the left primers were named EnP3F-859 according to their final amplified fragment size (AAGCTTTTGGAATTGCGCGAAGTTAG ), EnP3F-678(AAGCTT GCTAAATGTTACTTCCTCCG), EnP3F-471 (CAAGCTT GTCGTGACTTATGTAACACG), EnP3F-292 (AAGCTT GCACCAATGCACCATTGATC) and EnP3F-110 (AAGCTT CCTATAAATAATCCCCTAGAGC). A HindIII restriction site (indicated by underline) and corresponding protective bases (indicated by shadow) were introduced at the 5' end of each left primer. The PCR template is the correctly sequenced TA-EnP3 plasmid. After the amplified product was obtained, subsequent operations were performed according to the method for constructing TA-EnP3.
实施例3:胚乳特异表达启动子EnP3候选片段及相应缺失片段的转化载体构建(相应分子生物学常规操作参考J.萨姆布鲁克等,《分子克隆实验指南(第二版)》,科学出版社(1996)。Example 3: Construction of transformation vectors for endosperm-specific expression promoter EnP3 candidate fragments and corresponding deletion fragments (refer to J. Sambrook et al., "Molecular Cloning Experiment Guide (Second Edition)" for the corresponding molecular biology routine operations, Science Press (1996).
(1)酶切载体DX2181。DX2181载体是在pCAMBIA1380(该载体是澳大利亚CAMBIA(Center forthe Application of Molecular Biology to International Agriculture,CAMBIA)实验室在全世界公开交流使用的载体)的基础上改造而来,在多克隆位点处以相反的方向分别构建了一个GUS基因和EGFP基因,载体图及多克隆位点等信息见图3。用HindIII、BamHI双酶切DX2181,酶切产物用UNIQ-10柱式DNA胶回收试剂盒(上海生工生物工程技术服务有限公司生产)回收,电泳检测其酶切完整性,置于-20℃冰箱保存。(1) Digest vector DX2181. The DX2181 vector was transformed on the basis of pCAMBIA1380 (the vector used by the Australian CAMBIA (Center for the Application of Molecular Biology to International Agriculture, CAMBIA) laboratory in open communication all over the world), and at the multiple cloning site with the opposite Direction A GUS gene and EGFP gene were respectively constructed, and the vector map and multiple cloning sites and other information are shown in Figure 3. Digest DX2181 with HindIII and BamHI, recover the digested product with UNIQ-10 Column DNA Gel Recovery Kit (produced by Shanghai Sangon Bioengineering Technology Service Co., Ltd.), detect the integrity of the enzyme digestion by electrophoresis, and store at -20 °C Store in the refrigerator.
(2)酶切EnP3及相应缺失片段的TA克隆质粒。用HindIII、BamHI双酶切经过测序为正确的EnP3及相应缺失片段的TA克隆质粒,酶切产物用0.8%琼脂糖电泳分离,然后目标带用UNIQ-10柱式DNA胶回收试剂盒回收,电泳检测其酶切完整性,置于-20℃冰箱保存。(2) TA cloning plasmids digested with EnP3 and corresponding deletion fragments. Use HindIII and BamHI to double-digest the TA cloning plasmid that has been sequenced as the correct EnP3 and the corresponding missing fragment. The digested product is separated by 0.8% agarose electrophoresis, and then the target band is recovered with UNIQ-10 column DNA gel recovery kit, electrophoresis Check the integrity of the enzyme digestion and store in a -20°C refrigerator.
(3)将酶切回收的EnP3及相应缺失片段构建到载体DX2181上(见附图4),电转化入大肠杆菌DH5α(该菌株为商业化的大肠杆菌菌株)。酶切检测与测序后将构建好的载体导入农杆碱型的根癌农杆菌EHA105菌株(该菌株为商业化的农杆菌菌株),构成可用于转化的工程菌株。然后通过农杆菌介导的遗传转化法(林拥军等,2002)转化水稻品种“中花11”。(3) The recovered EnP3 and the corresponding deletion fragment were constructed on the vector DX2181 (see Figure 4), and electrotransformed into Escherichia coli DH5α (this strain is a commercial Escherichia coli strain). After enzyme digestion detection and sequencing, the constructed vector was introduced into the Agrobacterium tumefaciens EHA105 strain of the Agrobacterium base type (this strain is a commercial Agrobacterium strain) to form an engineering strain that can be used for transformation. Then the rice variety "Zhonghua 11" was transformed by the Agrobacterium-mediated genetic transformation method (Lin Yongjun et al., 2002).
实施例4:农杆菌介导的遗传转化Example 4: Agrobacterium-mediated genetic transformation
农杆菌介导的遗传转化方法主要参照华中农业大学作物遗传改良国家重点实验室发表的“农杆菌介导的遗传转化操作手册”所示的方法(林拥军等,2002)。转化受体为水稻品种“中花11”的成熟种子所诱导产生的胚性愈伤组织。经过预培养、侵染、共培养、筛选得到具有潮霉素抗性的愈伤,再经过分化、生根、炼苗和移栽,得到转基因植株。本发明的遗传转化的主要步骤、培养基及其配制的方法如下所述:The Agrobacterium-mediated genetic transformation method mainly refers to the method shown in the "Agrobacterium-mediated Genetic Transformation Manual" published by the State Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University (Lin Yongjun et al., 2002). The transformation recipient is the embryogenic callus induced by the mature seeds of the rice variety "Zhonghua 11". The hygromycin-resistant callus is obtained through precultivation, infection, cocultivation and screening, and then through differentiation, rooting, seedling hardening and transplanting to obtain transgenic plants. The main steps of the genetic transformation of the present invention, the culture medium and the preparation method thereof are as follows:
(1)试剂和溶液缩写(1) Abbreviation of reagents and solutions
本发明中培养基配制过程中所涉及到得的主要试剂的名称及缩写表示如下:6-BA(6-BenzylaminoPurine,6-苄基腺嘌呤);CN(Carbenicillin,羧苄青霉素);KT(Kinetin,激动素);NAA(Napthalene acetic acid,萘乙酸);IAA(Indole-3-acetic acid,吲哚乙酸);2,4-D(2,4-Dichlorophenoxyacetic acid,2,4-二氯苯氧乙酸);AS(Acetosringone,乙酰丁香酮);CH(CaseinEnzymatic Hydrolysate,水解酪蛋白);HN(Hygromycin B,潮霉素);DMSO(Dimethyl Sulfoxide,二甲基亚砜);N6max(N6大量元素成分溶液);N6mix(N6微量元素成分溶液);MSmax(MS大量元素成分溶液);MSmix(MS微量元素成分溶液)The title and the abbreviation of the main reagent involved in the medium preparation process in the present invention are as follows: 6-BA (6-BenzylaminoPurine, 6-benzyl adenine); CN (Carbenicillin, carbenicillin); KT (Kinetin , kinetin); NAA (Napthalene acetic acid, naphthalene acetic acid); IAA (Indole-3-acetic acid, indole acetic acid); 2,4-D (2,4-Dichlorophenoxyacetic acid, 2,4-dichlorophenoxy Acetic acid); AS (Acetosringone, acetylsyringone); CH (CaseinEnzymatic Hydrolysate, hydrolyzed casein); HN (Hygromycin B, hygromycin); DMSO (Dimethyl Sulfoxide, dimethyl sulfoxide); N6 max (N6 Major element composition solution); N6 mix (N6 trace element composition solution); MSmax (MS macroelement composition solution); MSmix (MS trace element composition solution)
(2)主要溶液配方(2) Main solution formula
1)N6培养基大量元素母液(按照10倍浓缩液(10X)配制):1)N6 medium macroelement mother solution (prepared according to 10 times concentrated solution (10X)):
硝酸钾(KNO3) 28.3gPotassium nitrate (KNO3 ) 28.3g
磷酸二氢钾(KH2PO4) 4.0gPotassium dihydrogen phosphate (KH2 PO4 ) 4.0g
硫酸铵((NH4)2SO4) 4.63gAmmonium sulfate ((NH4 )2 SO4 ) 4.63g
硫酸镁(MgSO4·7H2O) 1.85gMagnesium sulfate (MgSO4 ·7H2 O) 1.85g
氯化钙(CaCl2·2H2O) 1.66gCalcium chloride (CaCl2 2H2 O) 1.66g
将上述试剂逐一溶解,然后室温下用蒸馏水定容至1000ml。Dissolve the above reagents one by one, and then dilute to 1000ml with distilled water at room temperature.
2)N6培养基微量元素母液(按照100倍浓缩液(100X)配制:2) N6 medium trace element mother solution (prepared according to 100 times concentrated solution (100X):
碘化钾(KI) 0.08gPotassium iodide (KI) 0.08g
硼酸(H3BO3) 0.16gBoric acid (H3 BO3 ) 0.16g
硫酸锰(MnSO4·4H2O) 0.44gManganese sulfate (MnSO4 4H2 O) 0.44g
硫酸锌(ZnSO4·7H2O) 0.15gZinc sulfate (ZnSO4 ·7H2 O) 0.15g
将上述试剂在室温下溶解并用蒸馏水定容至1000ml。The above reagents were dissolved at room temperature and made up to 1000ml with distilled water.
3)铁盐(Fe2-EDTA)贮存液(按照100X浓缩液配制):3) Iron salt (Fe2 -EDTA) stock solution (prepared according to 100X concentrated solution):
将3.73克乙二胺四乙酸二钠(Na2EDTA·2H2O)和2.78克FeSO4·7H2O分别溶解,混合并用蒸馏水定容至1000ml,至70℃温浴2小时,4℃保存备用。Dissolve 3.73 g of disodium ethylenediamine tetraacetate (Na2 EDTA·2H2 O) and 2.78 g of FeSO4 ·7H2 O respectively, mix and dilute to 1000 ml with distilled water, incubate at 70°C for 2 hours, store at 4°C for later use .
4)维生素贮存液(按照100X浓缩液配制):4) Vitamin storage solution (prepared according to 100X concentrated solution):
烟酸(Nicotinic acid) 0.1gNiacin (Nicotinic acid) 0.1g
维生素B1(Thiamine HCl) 0.1gVitamin B1 (Thiamine HCl) 0.1g
维生素B6(Pyridoxine HCl) 0.1gVitamin B6 (Pyridoxine HCl) 0.1g
甘氨酸(Glycine) 0.2gGlycine 0.2g
肌醇(Inositol) 10gInositol 10g
加蒸馏水定容至1000ml,4℃保存备用。Add distilled water to make up to 1000ml, and store at 4°C for later use.
5)MS培养基大量元素母液(按照10X浓缩液配制):5) MS medium macroelement mother solution (prepared according to 10X concentrated solution):
硝酸铵(NH4NO3) 16.5gAmmonium nitrate (NH4 NO3 ) 16.5g
硝酸钾(KNO3) 19.0gPotassium nitrate (KNO3 ) 19.0g
磷酸二氢钾(KH2PO4) 1.7gPotassium dihydrogen phosphate (KH2 PO4 ) 1.7g
硫酸镁(MgSO4·7H2O) 3.7gMagnesium sulfate (MgSO4 ·7H2 O) 3.7g
氯化钙(CaCl2·2H2O) 4.4gCalcium chloride (CaCl2 2H2 O) 4.4g
将上述试剂在室温下溶解,并用蒸馏水定容至1000ml。Dissolve the above reagents at room temperature and make up to 1000ml with distilled water.
6)MS培养基微量元素母液(按照100X浓缩液配制):6) MS medium trace element mother solution (prepared according to 100X concentrated solution):
硫酸锰(MnSO4·4H2O) 2.23gManganese sulfate (MnSO4 4H2 O) 2.23g
硫酸锌(ZnSO4·7H2O) 0.86gZinc sulfate (ZnSO4 ·7H2 O) 0.86g
硼酸(H3BO3) 0.62gBoric acid (H3 BO3 ) 0.62g
碘化钾(KI) 0.083gPotassium iodide (KI) 0.083g
钼酸钠(Na2MoO4·2H2O) 0.025gSodium molybdate (Na2 MoO4 2H2 O) 0.025g
硫酸铜(CuSO4·5H2O) 0.0025gCopper sulfate (CuSO4 5H2 O) 0.0025g
氯化钴(CoCl2·6H2O) 0.0025gCobalt chloride (CoCl2 6H2 O) 0.0025g
将上述试剂在室温下溶解,并用蒸馏水定容至1000ml。Dissolve the above reagents at room temperature and make up to 1000ml with distilled water.
7)2,4-D贮存液(1mg/ml)的配制:7) Preparation of 2,4-D stock solution (1mg/ml):
称取2,4-D 100mg,用1ml 1N氢氧化钾溶解5分钟,然后加10ml蒸馏水溶解完全后定容至100ml,于室温下保存。Weigh 100mg of 2,4-D, dissolve it in 1ml 1N potassium hydroxide for 5 minutes, then add 10ml of distilled water to dissolve completely, dilute to 100ml, and store at room temperature.
8)6-BA贮存液(1mg/ml)的配制:8) Preparation of 6-BA stock solution (1mg/ml):
称取6-BA 100mg,用1ml 1N氢氧化钾溶解5分钟,然后加10ml蒸馏水溶解完全后定容至100ml,室温保存。Weigh 100mg of 6-BA, dissolve it with 1ml 1N potassium hydroxide for 5 minutes, then add 10ml of distilled water to dissolve completely, then dilute to 100ml, and store at room temperature.
9)萘乙酸(NAA)贮存液(1mg/ml)的配制:9) Preparation of naphthaleneacetic acid (NAA) stock solution (1mg/ml):
称取NAA 100mg,用1ml 1N氢氧化钾溶解5分钟,然后加10ml蒸馏水溶解完全后定容至100ml,4℃保存备用。Weigh 100mg of NAA, dissolve it with 1ml 1N potassium hydroxide for 5 minutes, then add 10ml of distilled water to dissolve completely, then dilute to 100ml, store at 4°C for later use.
10)吲哚乙酸(IAA)贮存液(1mg/ml)的配制:10) Preparation of indole acetic acid (IAA) stock solution (1mg/ml):
称取IAA 100mg,用1ml 1N氢氧化钾溶解5分钟,然后加10ml蒸馏水溶解完全后定容至100ml,4℃保存备用。Weigh 100mg of IAA, dissolve it with 1ml 1N potassium hydroxide for 5 minutes, then add 10ml of distilled water to dissolve completely, then dilute to 100ml, store at 4°C for later use.
11)葡萄糖贮存液(0.5g/ml)的配制:11) Preparation of glucose storage solution (0.5g/ml):
称取葡萄糖125g,然后用蒸馏水溶解定容至250ml,灭菌后4℃保存备用。Weigh 125g of glucose, then dissolve it in distilled water to 250ml, and store it at 4°C after sterilization.
12)AS贮存液的配制:12) Preparation of AS stock solution:
称取AS 0.392g,加入DMSO 10ml溶解,分装至1.5ml离心管内,4℃保存备用。Weigh 0.392g of AS, add 10ml of DMSO to dissolve, dispense into 1.5ml centrifuge tubes, and store at 4°C for later use.
13)1N氢氧化钾贮存液配制:13) Preparation of 1N potassium hydroxide stock solution:
称取氢氧化钾5.6g,用蒸馏水溶解定容至100ml,室温保存备用。Weigh 5.6g of potassium hydroxide, dissolve it in distilled water to 100ml, and store it at room temperature for later use.
(3)用于水稻遗传转化的培养基配方(3) Medium formula for genetic transformation of rice
1)诱导培养基1) Induction medium
N6max母液(取已经制备好的10X浓缩液,下同) 100mlN6max mother liquor (take the prepared 10X concentrated solution, the same below) 100ml
N6mix母液(取已经制备好的100X浓缩液,下同) 10mlN6mix mother solution (take the prepared 100X concentrated solution, the same below) 10ml
Fe2+EDTA贮存液(取已经制备好的100X浓缩液,下同) 10mlFe2+ EDTA stock solution (take the prepared 100X concentrated solution, the same below) 10ml
维生素贮存液(取已经制备好的100X浓缩液,下同) 10mlVitamin storage solution (take the prepared 100X concentrated solution, the same below) 10ml
2,4-D贮存液(取上述制备好的) 2.5ml2,4-D stock solution (take the prepared above) 2.5ml
脯氨酸(Proline) 0.3gProline (Proline) 0.3g
CH 0.6gCH 0.6g
蔗糖 30gSucrose 30g
Phytagel 3gPhytagel 3g
加蒸馏水至900ml,1N氢氧化钾调节pH值到5.9,煮沸并定容至1000ml,分装到50ml三角瓶(25ml/瓶),封口后按常规方法灭菌(例如121℃下灭菌25分钟,下述的培养基灭菌方法与本培养基的灭菌方法相同)。Add distilled water to 900ml, adjust the pH value to 5.9 with 1N potassium hydroxide, boil and set the volume to 1000ml, dispense into 50ml Erlenmeyer flasks (25ml/bottle), seal and sterilize according to conventional methods (for example, sterilize at 121°C for 25 minutes , the following medium sterilization method is the same as the sterilization method of this medium).
2)继代培养基2) subculture medium
N6max母液(10X) 100mlN6max mother liquor (10X) 100ml
N6mix母液(100X) 10mlN6mix mother solution (100X) 10ml
Fe2+EDTA贮存液(100X) 10mlFe2+ EDTA stock solution (100X) 10ml
维生素贮存液(100X) 10mlVitamin stock solution (100X) 10ml
2,4-D贮存液 2.0ml2,4-D stock solution 2.0ml
脯氨酸 0.5gProline 0.5g
CH 0.6gCH 0.6g
蔗糖 30gSucrose 30g
Phytagel 3gPhytagel 3g
加蒸馏水至900ml,1N氢氧化钾调节pH值到5.9,煮沸并定容至1000ml,分装到50ml三角瓶(25ml/瓶),封口,按上述方法灭菌。Add distilled water to 900ml, adjust the pH value to 5.9 with 1N potassium hydroxide, boil and set the volume to 1000ml, dispense into 50ml Erlenmeyer flasks (25ml/bottle), seal, and sterilize as above.
3)预培养基3) Pre-medium
N6max母液(10X) 12.5mlN6max mother solution (10X) 12.5ml
N6mix母液(100X) 1.25mlN6mix mother solution (100X) 1.25ml
Fe2+EDTA贮存液(100X) 2.5mlFe2+ EDTA stock solution (100X) 2.5ml
维生素贮存液(100X) 2.5mlVitamin stock solution (100X) 2.5ml
2,4-D贮存液 0.75ml2,4-D stock solution 0.75ml
CH 0.15gCH 0.15g
蔗糖 5gSucrose 5g
琼脂粉 1.75gAgar powder 1.75g
加蒸馏水至250ml,1N氢氧化钾调节pH值到5.6,封口,按上述方法灭菌。Add distilled water to 250ml, adjust the pH value to 5.6 with 1N potassium hydroxide, seal, and sterilize as above.
使用前加热溶解培养基并加入5ml葡萄糖贮存液和250微升AS贮存液,分装倒入培养皿中(25ml/皿)。Heat to dissolve the culture medium before use, add 5ml of glucose stock solution and 250 microliters of AS stock solution, and pour them into Petri dishes (25ml/dish).
4)共培养基4) Co-culture medium
N6max母液(10X) 12.5mlN6max mother solution (10X) 12.5ml
N6mix母液(100X) 1.25mlN6mix mother solution (100X) 1.25ml
Fe2+EDTA贮存液(100X) 2.5mlFe2+E DTA stock solution (100X) 2.5ml
维生素贮存液(100X) 2.5mlVitamin stock solution (100X) 2.5ml
2,4-D贮存液 0.75ml2,4-D stock solution 0.75ml
CH 0.2gCH 0.2g
蔗糖 5gSucrose 5g
琼脂粉 1.75gAgar powder 1.75g
加蒸馏水至250ml,1N氢氧化钾调节pH值到5.6,封口,按上述方法灭菌。Add distilled water to 250ml, adjust the pH value to 5.6 with 1N potassium hydroxide, seal, and sterilize as above.
使用前加热溶解培养基并加入5ml葡萄糖贮存液和250微升AS贮存液,分装倒入培养皿中(25ml/每皿)。Heat to dissolve the culture medium before use, add 5ml of glucose stock solution and 250 microliters of AS stock solution, and pour them into Petri dishes (25ml/dish).
5)悬浮培养基5) Suspension medium
N6max母液(10X) 5mlN6max mother solution (10X) 5ml
N6mix母液(100X) 0.5mlN6mix mother solution (100X) 0.5ml
Fe2+EDTA贮存液(100X) 0.5mlFe2+ EDTA stock solution (100X) 0.5ml
维生素贮存液(100X) 1mlVitamin stock solution (100X) 1ml
2,4-D贮存液 0.2ml2,4-D stock solution 0.2ml
CH 0.08gCH 0.08g
蔗糖 2gSucrose 2g
加蒸馏水至100ml,调节pH值到5.4,分装到两个100ml的三角瓶中,封口,按上述方法灭菌。Add distilled water to 100ml, adjust the pH value to 5.4, divide into two 100ml Erlenmeyer flasks, seal, and sterilize according to the above method.
使用前加入1ml无菌葡萄糖贮存液和100微升AS贮存液。Add 1 ml of sterile glucose stock solution and 100 μl of AS stock solution before use.
6)选择培养基6) Select medium
N6max母液(10X) 25mlN6max mother solution (10X) 25ml
N6mix母液(100X) 2.5mlN6mix mother solution (100X) 2.5ml
Fe2+EDTA贮存液(100X) 2.5mlFe2+ EDTA stock solution (100X) 2.5ml
维生素贮存液(100X) 2.5mlVitamin stock solution (100X) 2.5ml
2,4-D贮存液 0.625ml2,4-D stock solution 0.625ml
CH 0.15gCH 0.15g
蔗糖 7.5gSucrose 7.5g
琼脂粉 1.75gAgar powder 1.75g
加蒸馏水至250ml,调节pH值到6.0,封口,按上述方法灭菌。Add distilled water to 250ml, adjust the pH value to 6.0, seal, and sterilize according to the above method.
使用前溶解培养基,加入250微升HN(50mg/ml)和400微升CN(250mg/ml)分装倒入培养皿中(25ml/皿)。Dissolve the medium before use, add 250 microliters of HN (50mg/ml) and 400 microliters of CN (250mg/ml) and pour them into petri dishes (25ml/dish).
(注:第一次选择培养基羧苄青霉素浓度为400mg/升,第二次及以后选择培养基羧苄青霉素浓度为250mg/l)。(Note: the concentration of carbenicillin in the selection medium for the first time is 400 mg/liter, and the concentration of carbenicillin in the selection medium for the second time and later is 250 mg/l).
7)预分化培养基7) Pre-differentiation medium
N6max母液(10X) 25mlN6max mother solution (10X) 25ml
N6mix母液(100X) 2.5mlN6mix mother solution (100X) 2.5ml
Fe2+EDTA贮存液(100X) 2.5mlFe2+ EDTA stock solution (100X) 2.5ml
维生素贮存液(100X) 2.5mlVitamin stock solution (100X) 2.5ml
6-BA贮存液 0.5ml6-BA stock solution 0.5ml
KT贮存液 0.5mlKT stock solution 0.5ml
NAA贮存液 50微升NAA stock solution 50 microliters
IAA贮存液 50微升IAA stock solution 50 microliters
CH 0.15gCH 0.15g
蔗糖 7.5gSucrose 7.5g
琼脂粉 1.75gAgar powder 1.75g
加蒸馏水至250ml,1N氢氧化钾调节pH值到5.9,封口,按上述方法灭菌。Add distilled water to 250ml, adjust the pH value to 5.9 with 1N potassium hydroxide, seal, and sterilize as above.
使用前溶解培养基,250微升HN(50mg/ml)250微升CN(250毫g/ml),分装倒入培养皿中(25ml/皿)。Dissolve the medium before use, 250 microliters of HN (50 mg/ml) and 250 microliters of CN (250 mg/ml), and pour them into petri dishes (25ml/dish).
8)分化培养基8) Differentiation medium
N6max母液(10X) 100mlN6max mother solution (10X) 100ml
N6mix母液(100X) 10mlN6mix mother solution (100X) 10ml
Fe2+EDTA贮存液(100X) 10mlFe2+ EDTA stock solution (100X) 10ml
维生素贮存液(100X) 10mlVitamin stock solution (100X) 10ml
6-BA贮存液 2ml6-BA stock solution 2ml
KT贮存液 2mlKT stock solution 2ml
NAA贮存液 0.2mlNAA stock solution 0.2ml
IAA贮存液 0.2mlIAA stock solution 0.2ml
CH 1gCH 1g
蔗糖 30gSucrose 30g
Phytagel 3gPhytagel 3g
加蒸馏水至900ml,1N氢氧化钾调节pH值到6.0。Add distilled water to 900ml, and adjust the pH value to 6.0 with 1N potassium hydroxide.
煮沸并用蒸馏水定容至1000ml,分装到50ml三角瓶(50ml/瓶),封口,按上述方法灭菌。Boil and dilute to 1000ml with distilled water, dispense into 50ml Erlenmeyer flasks (50ml/bottle), seal, and sterilize as above.
9)生根培养基9) Rooting medium
MSmax母液(10X) 50mlMSmax mother solution (10X) 50ml
MSmix母液(100X) 5mlMSmix mother solution (100X) 5ml
Fe2+EDTA贮存液(100X) 5mlFe2+ EDTA stock solution (100X) 5ml
维生素贮存液(100X) 5mlVitamin stock solution (100X) 5ml
蔗糖 20gSucrose 20g
Phytagel 3gPhytagel 3g
加蒸馏水至900ml,用1N氢氧化钾调节pH值到5.8。Add distilled water to 900ml, and adjust the pH value to 5.8 with 1N potassium hydroxide.
煮沸并用蒸馏水定容至1000ml,分装到生根管中(25ml/管),封口,按上述方法灭菌。Boil and dilute to 1000ml with distilled water, pack into rooting tubes (25ml/tube), seal, and sterilize according to the above method.
(4)农杆菌介导的遗传转化步骤(4) Agrobacterium-mediated genetic transformation step
1)愈伤诱导1) Callus induction
a.将成熟的中花11水稻种子去壳,然后依次用70%的乙醇处理1分钟,0.15%氯化汞(HgCl2)种子表面消毒15分钟;a. Ripe Zhonghua 11 rice seeds were dehulled, then treated with 70% ethanol for 1 minute, and 0.15% mercuric chloride (HgCl2 ) seed surface was sterilized for 15 minutes;
b.用灭菌水洗种子4-5次;b. Wash the seeds 4-5 times with sterilized water;
c.将种子放在诱导培养基上;c. placing the seeds on the induction medium;
d.将接种后的培养基置于黑暗处培养4周,温度25±1℃。d. Culture the inoculated culture medium in a dark place for 4 weeks at a temperature of 25±1°C.
2)愈伤继代2) Callus subculture
挑选亮黄色、紧实且相对干燥的胚性愈伤,放于继代培养基上黑暗下培养2周,温度25±1℃。Select bright yellow, compact and relatively dry embryogenic calli, and place them on the subculture medium for 2 weeks in the dark at a temperature of 25±1°C.
3)预培养3) Pre-culture
挑选紧实且相对干燥的胚性愈伤,放于预培养基上黑暗下培养2周,温度25±1℃。Select compact and relatively dry embryogenic callus, put it on the pre-medium and culture it in the dark for 2 weeks at a temperature of 25±1°C.
4)农杆菌培养4) Agrobacterium culture
a.在带有对应抗性选择的LA培养基上预培养农杆菌EHA105(该菌株来自CAMBIA公司商业化的农杆菌菌株)两天,温度28℃;a. Pre-cultivate Agrobacterium EHA105 (this bacterial strain is from the commercialized Agrobacterium strain of CAMBIA Company) on the LA medium with corresponding resistance selection, at a temperature of 28° C.;
b.将农杆菌转移至悬浮培养基里,28℃摇床上培养2-3小时。b. Transfer the Agrobacterium to the suspension medium and culture on a shaker at 28°C for 2-3 hours.
5)农杆菌侵染5) Agrobacterium infection
a.将预培养的愈伤转移至灭好菌的瓶子内;a. transfer the pre-cultured callus to the bottle of sterilized bacteria;
b.调节农杆菌的悬浮液至OD6000.8-1.0;b. adjust the suspension of Agrobacterium to OD600 0.8-1.0;
c.将愈伤在农杆菌悬浮液中浸泡30分钟;c. Soak the callus in the Agrobacterium suspension for 30 minutes;
d.转移愈伤至灭菌好的滤纸上吸干;然后放在共培养基上培养3天,温度19-20℃。d. Transfer the callus to the sterilized filter paper and blot it dry; then culture it on the co-culture medium for 3 days at a temperature of 19-20°C.
6)愈伤洗涤和选择培养6) Callus washing and selection culture
a.灭菌水洗涤愈伤至看不见农杆菌;a. Wash the callus with sterilized water until the Agrobacterium cannot be seen;
b.浸泡在含400毫克/L羧苄青霉素(CN)的灭菌水中30分钟;b. Soak in sterilized water containing 400 mg/L carbenicillin (CN) for 30 minutes;
c.转移愈伤至灭菌好的滤纸上吸干;c. transfer the callus to dry on the sterilized filter paper;
d.转移愈伤至选择培养基上选择培养2-3次,每次2周。d. Transfer the callus to the selection medium for selection and culture for 2-3 times, each time for 2 weeks.
7)分化7) differentiation
a.将抗性愈伤转移至预分化培养基上于黑暗处培养5-7天;a. Transfer the resistant callus to the pre-differentiation medium and cultivate it in the dark for 5-7 days;
b.转移预分化培养的愈伤至分化培养基上,光照(1500-2000Lux)下培养,培养温度26℃。b. Transfer the pre-differentiation cultured callus to the differentiation medium, culture under light (1500-2000Lux), culture temperature 26°C.
8)生根8) Rooting
剪掉分化时产生的根;然后将其转移至生根培养基中光照(1500-2000Lux)下培养2-3周,培养温度26℃。Cut off the roots produced during differentiation; then transfer them to the rooting medium and cultivate them under light (1500-2000 Lux) for 2-3 weeks at a culture temperature of 26°C.
9)移栽9) Transplanting
洗掉根上的残留培养基,将具有良好根系的幼苗转入温室,同时在最初的几天保持水分湿润。Wash off the residual medium on the roots and transfer the seedlings with a good root system to the greenhouse while keeping them moist for the first few days.
实施例5:PCR法检测转基因阳性植株Embodiment 5: Detection of transgenic positive plants by PCR method
转化苗移入温室后,待其返青,然后分单株取1-2cm幼嫩叶片,抽提其基因组DNA为模板,用PCR法检测阳性植株。扩增片段为报告基因gus的部分片段,大小为699bp。引物序列为GUS-F:GGGCGAACAGTTCCTGATTA,GUS-R:AACGTATCCACGCCGTATTC。PCR反应条件:94℃ 5min,94℃ 50sec,57℃ 40sec,72℃ 50sec,30个循环,72℃ 7min。PCR产物经0.8%琼脂糖胶电泳检测。通过此法,可剔除阴性植株。After the transformed seedlings are moved into the greenhouse, wait for them to turn green, then take 1-2cm young leaves from individual plants, extract their genomic DNA as a template, and use PCR to detect positive plants. The amplified fragment is a partial fragment of the reporter gene gus, with a size of 699bp. The primer sequences are GUS-F: GGGCGAACAGTTCCTGATTA, GUS-R: AACGTATCCACGCCGTATTC. PCR reaction conditions: 94°C for 5min, 94°C for 50sec, 57°C for 40sec, 72°C for 50sec, 30 cycles, 72°C for 7min. PCR products were detected by 0.8% agarose gel electrophoresis. By this method, negative plants can be eliminated.
小量叶片基因组DNA的提取方法:取适量幼嫩叶片,加800ul 1.5×CTAB(1.5×CTAB配方:1.5%CTAB、75mM Tris-HCl、15mM EDTA及1.05M NaCl)研磨,转入1.5ml离心管中;65℃水浴30min;加入600μL氯仿/异戊醇(体积比为24∶1)上下颠倒数次(约15min),下层液相呈深绿色为止;室温下12000r/min离心10min;取500μL上清于一新1.5ml离心管,加入预冷的95%乙醇1mL,混匀后置-20℃,30min;室温下12000r/min离心10min,去上清,用75%乙醇浸洗沉淀,自然干燥;加入100μLddH2O溶解,备用。Extraction method of a small amount of genomic DNA from leaves: Take an appropriate amount of young leaves, add 800ul 1.5×CTAB (1.5×CTAB formula: 1.5% CTAB, 75mM Tris-HCl, 15mM EDTA and 1.05M NaCl) to grind, transfer to a 1.5ml centrifuge tube Medium; water bath at 65°C for 30 min; add 600 μL chloroform/isoamyl alcohol (volume ratio 24:1) and invert several times (about 15 min), until the lower liquid phase turns dark green; centrifuge at 12000 r/min for 10 min at room temperature; take 500 μL upper Clear in a new 1.5ml centrifuge tube, add 1mL of pre-cooled 95% ethanol, mix well, place at -20°C for 30min; centrifuge at 12000r/min for 10min at room temperature, remove the supernatant, soak the precipitate with 75% ethanol, and dry naturally ; Add 100 μL ddH2 O to dissolve, set aside.
实施例6:GUS组织染色法分析EnP3候选片段及相应缺失片段各种组织中的表达模式Example 6: GUS tissue staining method to analyze the expression patterns of EnP3 candidate fragments and corresponding deletion fragments in various tissues
分别取经PCR检测(参考实施例5)为阳性的EnP3-GUS转化植株(10株以上)的根、叶片、叶鞘、茎杆、颖壳、花、种子切成约0.5CM长度的适当大小,浸入约200μl的GUS染液,37℃过夜,然后用75%酒精脱色,观察是否有蓝色出现。染色液的配方参照Jefferson等报道的方法(Jefferson等,1987)。结果表明:EnP3候选片段仅在转化植株的胚乳表达,在其他组织如叶片、叶鞘、茎杆、根、花、颖壳及胚中均检测不到表达(见附图5)。这些结果证实,EnP3为一个胚乳特异表达的启动子。Get the roots, blades, leaf sheaths, stems, glumes, flowers, and seeds of positive EnP3-GUS transformed plants (more than 10 strains) through PCR detection (reference example 5) and cut them into appropriate sizes of about 0.5CM length. About 200μl of GUS staining solution, overnight at 37°C, then decolorize with 75% alcohol, and observe whether there is blue color. The formulation of the staining solution refers to the method reported by Jefferson et al. (Jefferson et al., 1987). The results showed that the EnP3 candidate fragment was only expressed in the endosperm of the transformed plant, and no expression could be detected in other tissues such as leaves, leaf sheaths, stems, roots, flowers, glumes and embryos (see Figure 5). These results confirmed that EnP3 is an endosperm-specific expression promoter.
用同样的办法分析了EnP3的系列缺失片段驱动下的GUS在各种组织中的表达模式(见图6)。结果表明:启动子EnP3-859、EnP3-678、EnP3-471、EnP3-292均具有独立的启动基因表达的功能,并且在转化植株中的表达模式与EnP3相同,即仅在胚乳表达,不过表达量各有区别。同时发现,由EnP3-292进一步截短的EnP3-110区段的表达模式发生了显著的改变:在叶片、叶鞘、茎杆及颖壳这些绿色组织中可以检测到表达,但是在根、花及种子(包括胚和胚乳)中却检测不到表达,成为一个短的绿色组织特异表达启动子(见图7)。In the same way, the expression pattern of GUS driven by the serial deletion of EnP3 in various tissues was analyzed (see FIG. 6 ). The results showed that the promoters EnP3-859, EnP3-678, EnP3-471, and EnP3-292 all had the function of independently promoting gene expression, and the expression pattern in transformed plants was the same as that of EnP3, that is, only expressed in the endosperm, but not Quantities vary. At the same time, it was found that the expression pattern of the EnP3-110 segment further truncated by EnP3-292 changed significantly: expression could be detected in green tissues such as leaves, leaf sheaths, stems and glumes, but not in roots, flowers and However, no expression was detected in seeds (including embryos and endosperms), and it became a short green tissue-specific expression promoter (see Figure 7).
实施例7:启动子EnP3及相应缺失片段GUS活性定量检测Example 7: Quantitative detection of promoter EnP3 and corresponding deletion fragment GUS activity
取EnP3及相应缺失片段经GUS组织化学染色为阳性(参考实施例6)植株及转空载体DX2181植株的T0代种子,统一播种。待其开花受精后7天、14天、21天时取胚乳组织,抽提其总蛋白,然后测定其GUS活性(见图9);同时取EnP3-110及空载体DX2181抽穗期的叶片、叶鞘、茎杆、穗子和根组织,同样抽提其总蛋白,然后测定其GUS活性(见图8)。具体方法参考Bradford及Jefferson等报道的方法(BradfordM,1976;Jefferson等,1987)。结果表明:在胚乳组织发育的不同时期,启动子片段(EnP3、EnP3-859、EnP3-678、EnP3-471和EnP3-292)和阴性对照DX2181相比,均有显著差异;启动子片段EnP3-110的主要绿色组织(叶片、叶鞘、茎杆、颖壳(实验测量时为整穗))和阴性对照DX2181相比,均有显著差异这和组织化学染色结果是吻合的,即EnP3、EnP3-859、EnP3-678、EnP3-471和EnP3-292为胚乳特异表达启动子,而EnP3-110为一绿色组织特异表达启动子。从胚乳组织的表达量上来看,全长启动子EnP3的表达量最高(在各个发育时期均是如此),并且随着胚乳发育阶段的不断进行,表达量呈现下降趋势。对EnP3-678、EnP3-471和EnP3-292这三个启动子来讲,它们的表达模式与EnP3相同,即:随着胚乳发育阶段的不断进行,表达量呈现下降趋势;并且随着启动子长度的逐渐缩短,对同一发育时期比较而言,表达量呈现下降趋势。与其他几个胚乳特异表达的启动子相比而言,EnP3-859的表达模式略有不同,即在胚乳发育前期(开花受精后7天及14天)表达量无明显变化,在后期有一个急剧下降的过程。绿色组织特异表达启动子EnP3-110在叶片、叶鞘中的表达量(以阴性对照DX2181的相应组织为基准)要强于在茎杆和穗子中的表达量。同时从图上可以发现胚乳中开花受精后7天的GUS活性从EnP3到EnP3-859以及EnP3-471到EnP3-292有一个急剧下降的现象;而开花受精后14天及21天的GUS活性的急剧下降则出现在EnP3-471到EnP3-292。从上面所有结果可以推测启动子EnP3是一个在胚乳发育前期高量表达的特异表达启动子;在胚乳发育早期1040-859及471-292这两个区段中可能存在控制其表达量的顺式作用元件;在胚乳发育的中后期471-292这个区段中可能存在控制其表达量的顺式作用元件。EnP3-110为一绿色组织特异表达启动子,而EnP3-292为一胚乳特异表达启动子,表达模式发生显著改变,说明292-110这个区段可能存在控制启动子表达模式的顺式作用元件。The T0 generation seeds of the plants whose EnP3 and corresponding deletion fragments were positive by GUS histochemical staining (reference example 6) and the plants transformed with the empty vector DX2181 were sown uniformly. After 7 days, 14 days, and 21 days after flowering and fertilization, the endosperm tissue was taken, its total protein was extracted, and then its GUS activity was measured (see Figure 9); at the same time, leaves, leaf sheaths, leaf sheaths, and Stem, ear and root tissues were also extracted for total protein, and then their GUS activity was determined (see Figure 8). For specific methods, refer to the methods reported by Bradford and Jefferson et al. (Bradford M, 1976; Jefferson et al., 1987). The results showed that: at different stages of endosperm tissue development, the promoter fragments (EnP3, EnP3-859, EnP3-678, EnP3-471 and EnP3-292) had significant differences compared with the negative control DX2181; the promoter fragment EnP3- Compared with the negative control DX2181, the main green tissues (leaves, leaf sheaths, stems, glumes (whole panicle) in the experiment) of 110 were significantly different from the results of histochemical staining, that is, EnP3, EnP3- 859, EnP3-678, EnP3-471 and EnP3-292 are endosperm-specific expression promoters, while EnP3-110 is a green tissue-specific expression promoter. From the perspective of endosperm tissue expression, the expression level of the full-length promoter EnP3 is the highest (this is true in all developmental stages), and the expression level shows a downward trend as the endosperm development stage continues. For the three promoters EnP3-678, EnP3-471 and EnP3-292, their expression patterns are the same as those of EnP3, that is: as the endosperm development stage continues, the expression level shows a downward trend; and as the promoter Compared with the same developmental period, the expression level showed a downward trend with the gradual shortening of the length. Compared with several other endosperm-specific expression promoters, the expression pattern of EnP3-859 is slightly different, that is, there is no obvious change in the expression level in the early stage of endosperm development (7 days and 14 days after flowering and fertilization), and there is a a steep decline. The expression of the green tissue-specific expression promoter EnP3-110 in leaves and leaf sheaths (based on the corresponding tissues of the negative control DX2181) was stronger than that in stems and ears. At the same time, it can be found from the figure that the GUS activity in the
参考文献:references:
1.林拥军等,农杆菌介导的牡丹江8号高效转基因体系的建立,作物学报,2002,28(3):294-300;1. Lin Yongjun et al., Establishment of high-efficiency transgenic system of Mudanjiang No. 8 mediated by Agrobacterium, Acta Crops Sinica, 2002, 28(3): 294-300;
2.J.萨姆布鲁克,E.F.弗里奇,T.曼尼阿蒂斯.分子克隆实验指南(第二版).科学出版社,1996;2.J. Sambrook, E.F. Fritsch, T. Maniartis. Molecular Cloning Experiment Guide (Second Edition). Science Press, 1996;
3.Bradford M,A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing theprinciple of protein-dye binding.Anal Biochem,1976,72:248-254;3. Bradford M, A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem, 1976, 72: 248-254;
4.Buchner et al.,Characterization of a tissue-specific and developmentally regulated beta-1,3-glucanase gene inpea(Pisum sativum).Plant Mol Biol,2002,49:171-186;4. Buchner et al., Characterization of a tissue-specific and developmentally regulated beta-1, 3-glucanase gene inpea (Pisum sativum). Plant Mol Biol, 2002, 49: 171-186;
5.Cai M et al.,A rice promoter containing both novel positive and negative cis-elements for regulation of greentissue-specific gene expression in transgenic plants.Plant Biotechnology Journal,2007,5:664-674;5. Cai M et al., A rice promoter containing both novel positive and negative cis-elements for regulation of greenissue-specific gene expression in transgenic plants. Plant Biotechnology Journal, 2007, 5: 664-674;
6.Ehsani P et al.Expression of anti human IL-4 and IL-6 scFvs in transgenic tobacco plants.Plant Mol Biol,2003,52:17-29;6. Ehsani P et al. Expression of anti human IL-4 and IL-6 scFvs in transgenic tobacco plants. Plant Mol Biol, 2003, 52: 17-29;
7.Jefferson et al.,GUS fusions:beta-glucuronidase as a sensitive and versatile gene fusion marker in higherplants.EMBO J,1987,6:3901-3907;7. Jefferson et al., GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higherplants. EMBO J, 1987, 6: 3901-3907;
8.Karlowski W M,Hirsch A M.The over-expression of an alfalfa RING-H2 gene induces pleiotropic effects onplant growth and development.Plant Mol Biol,2003,52:121-133;8. Karlowski W M, Hirsch A M. The over-expression of an alfalfa RING-H2 gene induces pleiotropic effects on plant growth and development. Plant Mol Biol, 2003, 52: 121-133;
9.Kumpatla S P et al.,Genome intruder scanning and modulation systems and transgene silencing.Trends inPlant Sciences,1998,3:96-104;9. Kumpatla S P et al., Genome intruder scanning and modulation systems and transgene silencing. Trends in Plant Sciences, 1998, 3: 96-104;
10.Murray and Thompson.Rapid isolation of high molecular weight plant DNA.Nucleic Acids Res.1980,8:4321-4325;10. Murray and Thompson. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8: 4321-4325;
11.Robinson D J.Environmental risk assessment of release of transgenic plants containing virus-derived inserts.Transgen Res,1996,5:359-362;11. Robinson D J. Environmental risk assessment of release of transgenic plants containing virus-derived inserts. Transgen Res, 1996, 5: 359-362;
12.Rossak et al.,Expression of the FAE1 gene and FAE1 promoter activity in developing seeds of Arabidopsisthaliana.Plant Mol Biol,2001,46:717-725;12. Rossak et al., Expression of the FAE1 gene and FAE1 promoter activity in developing seeds of Arabidopsisthaliana. Plant Mol Biol, 2001, 46: 717-725;
13.Sandhu et al.,Oral immunization of mice with transgenic tomato fruit expressing respiratory syncytial virus-Fprotein induces a systemic immune response.Transgenic Res,2000,9:127-135;13. Sandhu et al., Oral immunization of mice with transgenic tomato fruit expressing respiratory syncytial virus-Fprotein induces a systemic immune response. Transgenic Res, 2000, 9: 127-135;
14.Vasconcelos et al.,Enhanced iron and zinc accumulation in transgenic rice with the ferritin gene.Plant Sci,2003,164:371-378;14. Vasconcelos et al., Enhanced iron and zinc accumulation in transgenic rice with the ferritin gene. Plant Sci, 2003, 164: 371-378;
15.Vickers et al.,A novel cis-acting element,ESP,contributes to high-level endosperm-specific expression in anoat globulin promoter.Plant Mol Biol,2006,62:195-214;15.Vickers et al., A novel cis-acting element, ESP, contributes to high-level endosperm-specific expression in anoat globulin promoter. Plant Mol Biol, 2006, 62: 195-214;
16.Wang L et al.,A dynamic gene expression atlas covering the entire life cycle of rice.Plant J.2010,61:752-766;16.Wang L et al., A dynamic gene expression atlas covering the entire life cycle of rice. Plant J.2010, 61:752-766;
17.Wu et al.,The GCN4 motif in a rice glutelin gene is essential for endosperm specific gene expression and isactivated by Opaque-2 in transgenic rice plants.Plant J,1998,14:673-683;17. Wu et al., The GCN4 motif in a rice glutelin gene is essential for endosperm specific gene expression and is activated by Opaque-2 in transgenic rice plants. Plant J, 1998, 14: 673-683;
18.Ye RJ et al.,Development of inseet-resistant transgenic rice with CrylC*-free endosperm.Pest Managementscience,2009,65:1015-1020;18. Ye RJ et al., Development of inseet-resistant transgenic rice with CrylC*-free endosperm. Pest Managementscience, 2009, 65: 1015-1020;
19.Yoshida et al.,Laboratory Manual for Physiological Studies of Rice,3rd Edn.International Rice ResearchInstitute,Manila.1976;19. Yoshida et al., Laboratory Manual for Physiological Studies of Rice, 3rd Edn. International Rice Research Institute, Manila.1976;
20.Zambryski et al.,Ti plasmid vector for the introduction of DNA into plant cells without alteration of theirnormal regeneration capacity.EMBO J,1983,2:2143-2150。20. Zambryski et al., Ti plasma vector for the introduction of DNA into plant cells without alteration of their normal regeneration capacity. EMBO J, 1983, 2: 2143-2150.
序列表sequence listing
<110>华中农业大学<110> Huazhong Agricultural University
<120>水稻胚乳特异表达基因的启动子及表达模式鉴定<120> Identification of the promoter and expression pattern of rice endosperm-specific expression genes
<130><130>
<141>2010-04-15<141>2010-04-15
<160>8<160>8
<170>PatentIn version 3.1<170>PatentIn version 3.1
<210>1<210>1
<211>1040<211>1040
<212>DNA<212>DNA
<213>水稻(Oryza sativa)<213> Rice (Oryza sativa)
<220><220>
<221>promoter<221>promoter
<222>(1)..(1040)<222>(1)..(1040)
<223><223>
<400>1<400>1
ggagcatttg taggaatgcc acgtggcggc ttaggagcgt ttgtagaaag tttaatggat 60ggagcatttg taggaatgcc acgtggcggc ttaggagcgt ttgtagaaag tttaatggat 60
ttttagtata taatagaaga tttgtataag acagcgtact agctatacat gtacaaggga 120ttttagtata taatagaaga tttgtataag acagcgtact agctatacat gtacaaggga 120
gaagtgagga taacaaagct aagtatggcg caattctgat tttccataat tccattaact 180gaagtgagga taacaaagct aagtatggcg caattctgat tttccataat tccattaact 180
tttggaattg cgcgaagtta gtttttcccc cctttttttc acgttttcat ttataaagta 240tttggaattg cgcgaagtta gtttttcccc cctttttttc acgttttcat ttataaagta 240
acagtgttcg atccttccat ttcctgttaa gaactcaatg tacatgccaa actacaaata 300acagtgttcg atccttccat ttcctgttaa gaactcaatg tacatgccaa actacaaata 300
tgtatgtatt ctattaaagc acttttactt gaaaaaaaaa ctctaacgta acaatcacct 360tgtatgtatt ctattaaagc acttttactt gaaaaaaaaa ctctaacgta acaatcacct 360
aggctaaatg ttacttcctc cgtttcacaa tataagtcat tctagcattt cccacattca 420aggctaaatg ttacttcctc cgtttcacaa tataagtcat tctagcattt cccacattca 420
tattgatgtt aatgaatctc gacatctaga ttcattaaca ttaatatgaa tgtgggaaat 480tattgatgtt aatgaatctc gacatctaga ttcattaaca ttaatatgaa tgtgggaaat 480
gctagaatgt ctagattcat taacatcaat atgaatgtgg gaaatgctag aatgacttac 540gctagaatgt ctagattcat taacatcaat atgaatgtgg gaaatgctag aatgacttac 540
attgtgaaac ggagggagta tatatgtaag tcgtgactta tgtaacacgc tcttaaaatt 600attgtgaaac ggagggagta tatatgtaag tcgtgactta tgtaacacgc tcttaaaatt 600
ggacaaatta tatatatgct tataatcaaa atactacttc acccatctaa aagaatattg 660ggacaaatta tatatatgct tataatcaaa atactacttc acccatctaa aagaatattg 660
ttagaataca aatttagttt aacatagtgc cgagccaata tggtatatct ttatcatcag 720ttagaataca aatttagttt aacatagtgc cgagccaata tggtatatct ttatcatcag 720
aatttgtttc cacgcaattc tcaataaagc accaatgcac cattgatcaa acacctttat 780aatttgtttc cacgcaattc tcaataaagc accaatgcac cattgatcaa acacctttat 780
atctacagca tcatctcgcc acgttcttcc aaaatattgg ttgaacaaat gtccaaatac 840atctacagca tcatctcgcc acgttcttcc aaaatattgg ttgaacaaat gtccaaatac 840
acctcatgac tcattgtttc tagcttactt gacctccacc aggtagtttt gcaaaaatta 900acctcatgac tcattgtttc tagcttactt gacctccacc aggtagtttt gcaaaaatta 900
aatccttttt ggcgtgtaat attgctatta cctataaata atcccctaga gcaattgtta 960aatccttttt ggcgtgtaat attgctatta cctataaata atcccctaga gcaattgtta 960
tctcatcacc ctcaacatat aatcttctac atttacacca tctagtaatc ttgttaagca 1020tctcatcacc ctcaacatat aatcttctac atttacacca tctagtaatc ttgttaagca 1020
tctcccatac gctgtcaaca 1040tctcccatac gctgtcaaca 1040
<210>2<210>2
<211>859<211>859
<212>DNA<212>DNA
<213>水稻(Oryza sativa)<213> Rice (Oryza sativa)
<220><220>
<221>promoter<221>promoter
<222>(1)..(859)<222>(1)..(859)
<223><223>
<400>2<400>2
ttggaattgc gcgaagttag tttttccccc ctttttttca cgttttcatt tataaagtaa 60ttggaattgc gcgaagttag tttttccccc ctttttttca cgttttcatt tataaagtaa 60
cagtgttcga tccttccatt tcctgttaag aactcaatgt acatgccaaa ctacaaatat 120cagtgttcga tccttccatt tcctgttaag aactcaatgt acatgccaaa ctacaaatat 120
gtatgtattc tattaaagca cttttacttg aaaaaaaaac tctaacgtaa caatcaccta 180gtatgtattc tattaaagca cttttacttg aaaaaaaaac tctaacgtaa caatcaccta 180
ggctaaatgt tacttcctcc gtttcacaat ataagtcatt ctagcatttc ccacattcat 240ggctaaatgt tacttcctcc gtttcacaat ataagtcatt ctagcatttc ccacattcat 240
attgatgtta atgaatctcg acatctagat tcattaacat taatatgaat gtgggaaatg 300attgatgtta atgaatctcg acatctagat tcattaacat taatatgaat gtgggaaatg 300
ctagaatgtc tagattcatt aacatcaata tgaatgtggg aaatgctaga atgacttaca 360ctagaatgtc tagattcatt aacatcaata tgaatgtggg aaatgctaga atgacttaca 360
ttgtgaaacg gagggagtat atatgtaagt cgtgacttat gtaacacgct cttaaaattg 420ttgtgaaacg gagggagtat atatgtaagt cgtgacttat gtaacacgct cttaaaattg 420
gacaaattat atatatgctt ataatcaaaa tactacttca cccatctaaa agaatattgt 480gacaaattat atatatgctt ataatcaaaa tactacttca cccatctaaa agaatattgt 480
tagaatacaa atttagttta acatagtgcc gagccaatat ggtatatctt tatcatcaga 540tagaatacaa atttagttta acatagtgcc gagccaatat ggtatatctt tatcatcaga 540
atttgtttcc acgcaattct caataaagca ccaatgcacc attgatcaaa cacctttata 600atttgtttcc acgcaattct caataaagca ccaatgcacc attgatcaaa cacctttata 600
tctacagcat catctcgcca cgttcttcca aaatattggt tgaacaaatg tccaaataca 660tctacagcat catctcgcca cgttcttcca aaatattggt tgaacaaatg tccaaataca 660
cctcatgact cattgtttct agcttacttg acctccacca ggtagttttg caaaaattaa 720cctcatgact cattgtttct agcttacttg acctccacca ggtagttttg caaaaattaa 720
atcctttttg gcgtgtaata ttgctattac ctataaataa tcccctagag caattgttat 780atcctttttg gcgtgtaata ttgctattac ctataaataa tcccctagag caattgttat 780
ctcatcaccc tcaacatata atcttctaca tttacaccat ctagtaatct tgttaagcat 840ctcatcaccc tcaacatata atcttctaca tttacaccat ctagtaatct tgttaagcat 840
ctcccatacg ctgtcaaca 859ctcccatacg ctgtcaaca 859
<210>3<210>3
<211>678<211>678
<212>DNA<212>DNA
<213>水稻(Oryza sativa)<213> Rice (Oryza sativa)
<220><220>
<221>promoter<221>promoter
<222>(1)..(678)<222>(1)..(678)
<223><223>
<400>3<400>3
gctaaatgtt acttcctccg tttcacaata taagtcattc tagcatttcc cacattcata 60gctaaatgtt acttcctccg tttcacaata taagtcattc tagcatttcc cacattcata 60
ttgatgttaa tgaatctcga catctagatt cattaacatt aatatgaatg tgggaaatgc 120ttgatgttaa tgaatctcga catctagatt cattaacatt aatatgaatg tgggaaatgc 120
tagaatgtct agattcatta acatcaatat gaatgtggga aatgctagaa tgacttacat 180tagaatgtct agattcatta acatcaatat gaatgtggga aatgctagaa tgacttacat 180
tgtgaaacgg agggagtata tatgtaagtc gtgacttatg taacacgctc ttaaaattgg 240tgtgaaacgg agggagtata tatgtaagtc gtgacttatg taacacgctc ttaaaattgg 240
acaaattata tatatgctta taatcaaaat actacttcac ccatctaaaa gaatattgtt 300acaaattata tatatgctta taatcaaaat actacttcac ccatctaaaa gaatattgtt 300
agaatacaaa tttagtttaa catagtgccg agccaatatg gtatatcttt atcatcagaa 360agaatacaaa tttagtttaa catagtgccg agccaatatg gtatatcttt atcatcagaa 360
tttgtttcca cgcaattctc aataaagcac caatgcacca ttgatcaaac acctttatat 420tttgtttcca cgcaattctc aataaagcac caatgcacca ttgatcaaac acctttatat 420
ctacagcatc atctcgccac gttcttccaa aatattggtt gaacaaatgt ccaaatacac 480ctacagcatc atctcgccac gttcttccaa aatattggtt gaacaaatgt ccaaatacac 480
ctcatgactc attgtttcta gcttacttga cctccaccag gtagttttgc aaaaattaaa 540ctcatgactc attgtttcta gcttacttga cctccaccag gtagttttgc aaaaattaaa 540
tcctttttgg cgtgtaatat tgctattacc tataaataat cccctagagc aattgttatc 600tcctttttgg cgtgtaatat tgctattacc tataaataat cccctagagc aattgttatc 600
tcatcaccct caacatataa tcttctacat ttacaccatc tagtaatctt gttaagcatc 660tcatcaccct caacatataa tcttctacat ttacaccatc tagtaatctt gttaagcatc 660
tcccatacgc tgtcaaca 678tcccatacgc tgtcaaca 678
<210>4<210>4
<211>471<211>471
<212>DNA<212>DNA
<213>水稻(Oryza sativa)<213> Rice (Oryza sativa)
<220><220>
<221>promoter<221>promoter
<222>(1)..(471)<222>(1)..(471)
<223><223>
<400>4<400>4
gtcgtgactt atgtaacacg ctcttaaaat tggacaaatt atatatatgc ttataatcaa 60gtcgtgactt atgtaacacg ctcttaaaat tggacaaatt atatatatgc ttataatcaa 60
aatactactt cacccatcta aaagaatatt gttagaatac aaatttagtt taacatagtg 120aatactactt cacccatcta aaagaatatt gttagaatac aaatttagtt taacatagtg 120
ccgagccaat atggtatatc tttatcatca gaatttgttt ccacgcaatt ctcaataaag 180ccgagccaat atggtatatc tttatcatca gaatttgttt ccacgcaatt ctcaataaag 180
caccaatgca ccattgatca aacaccttta tatctacagc atcatctcgc cacgttcttc 240caccaatgca ccattgatca aacaccttta tatctacagc atcatctcgc cacgttcttc 240
caaaatattg gttgaacaaa tgtccaaata cacctcatga ctcattgttt ctagcttact 300caaaatattg gttgaacaaa tgtccaaata cacctcatga ctcattgttt ctagcttact 300
tgacctccac caggtagttt tgcaaaaatt aaatcctttt tggcgtgtaa tattgctatt 360tgacctccac caggtagttt tgcaaaaatt aaatcctttt tggcgtgtaa tattgctatt 360
acctataaat aatcccctag agcaattgtt atctcatcac cctcaacata taatcttcta 420acctataaat aatcccctag agcaattgtt atctcatcac cctcaacata taatcttcta 420
catttacacc atctagtaat cttgttaagc atctcccata cgctgtcaac a 471catttacacc atctagtaat cttgttaagc atctcccata cgctgtcaac a 471
<210>5<210>5
<211>292<211>292
<212>DNA<212>DNA
<213>水稻(Oryza sativa)<213> Rice (Oryza sativa)
<220><220>
<221>promoter<221>promoter
<222>(1)..(292)<222>(1)..(292)
<223><223>
<400>5<400>5
gcaccaatgc accattgatc aaacaccttt atatctacag catcatctcg ccacgttctt 60gcaccaatgc accattgatc aaacaccttt atatctacag catcatctcg ccacgttctt 60
ccaaaatatt ggttgaacaa atgtccaaat acacctcatg actcattgtt tctagcttac 120ccaaaatatt ggttgaacaa atgtccaaat acacctcatg actcattgtt tctagcttac 120
ttgacctcca ccaggtagtt ttgcaaaaat taaatccttt ttggcgtgta atattgctat 180ttgacctcca ccaggtagtt ttgcaaaaat taaatccttt ttggcgtgta atattgctat 180
tacctataaa taatccccta gagcaattgt tatctcatca ccctcaacat ataatcttct 240tacctataaa taatccccta gagcaattgt tatctcatca ccctcaacat ataatcttct 240
acatttacac catctagtaa tcttgttaag catctcccat acgctgtcaa ca 292acatttacac catctagtaa tcttgttaag catctcccat acgctgtcaa ca 292
<210>6<210>6
<211>110<211>110
<212>DNA<212>DNA
<213>水稻(Oryza sativa)<213> Rice (Oryza sativa)
<220><220>
<221>promoter<221>promoter
<222>(1)..(110)<222>(1)..(110)
<223><223>
<400>6<400>6
cctataaata atcccctaga gcaattgtta tctcatcacc ctcaacatat aatcttctac 60cctataaata atcccctaga gcaattgtta tctcatcacc ctcaacatat aatcttctac 60
atttacacca tctagtaatc ttgttaagca tctcccatac gctgtcaaca 110atttacacca tctagtaatc ttgttaagca tctcccatac gctgtcaaca 110
<210>7<210>7
<211>459<211>459
<212>DNA<212>DNA
<213>水稻(Oryza sativa)<213> Rice (Oryza sativa)
<220><220>
<221>gene<221> gene
<222>(1)..(459)<222>(1)..(459)
<223><223>
<220><220>
<221>promoter<221>promoter
<222>(1)..(459)<222>(1)..(459)
<223><223>
<220><220>
<221>CDS<221> CDS
<222>(55)..(459)<222>(55)..(459)
<223><223>
<400>7<400>7
ctacatttac accatctagt aatcttgtta agcatctccc atacgctgtc aaca atg 57ctacatttac accatctagt aatcttgtta agcatctccc atacgctgtc aaca atg 57
MetMet
1 1
gca tca tac aag atc ttg gtt gtc ttt gct ttg cta gct ctt tct gca 105gca tca tac aag atc ttg gtt gtc ttt gct ttg cta gct ctt tct gca 105
Ala Ser Tyr Lys Ile Leu Val Val Phe Ala Leu Leu Ala Leu Ser AlaAla Ser Tyr Lys Ile Leu Val Val Phe Ala Leu Leu Ala Leu Ser Ala
5 10 155 10 15
agt gca gct acc gca atc acc acc act ata cca tac ttc cca tca aca 153agt gca gct acc gca atc acc acc act ata cca tac ttc cca tca aca 153
Ser Ala Ala Thr Ala Ile Thr Thr Thr Ile Pro Tyr Phe Pro Ser ThrSer Ala Ala Thr Ala Ile Thr Thr Thr Ile Pro Tyr Phe Pro Ser Thr
20 25 3020 25 30
cta gca atg ggc acc atg aat ccc tgt aag ctg tac atg atg caa act 201cta gca atg ggc acc atg aat ccc tgt aag ctg tac atg atg caa act 201
Leu Ala Met Gly Thr Met Asn Pro Cys Lys Leu Tyr Met Met Gln ThrLeu Ala Met Gly Thr Met Asn Pro Cys Lys Leu Tyr Met Met Gln Thr
35 40 4535 40 45
ttg ggc atg ggt agc tac gca acc atg ttc atg tca caa cca att gct 249ttg ggc atg ggt agc tac gca acc atg ttc atg tca caa cca att gct 249
Leu Gly Met Gly Ser Tyr Ala Thr Met Phe Met Ser Gln Pro Ile AlaLeu Gly Met Gly Ser Tyr Ala Thr Met Phe Met Ser Gln Pro Ile Ala
50 55 60 6550 55 60 65
ctc ctg caa caa caa tgt tgc atg caa cta caa ggc atg ata cca cag 297ctc ctg caa caa caa tgt tgc atg caa cta caa ggc atg ata cca cag 297
Leu Leu Gln Gln Gln Cys Cys Met Gln Leu Gln Gly Met Ile Pro GlnLeu Leu Gln Gln Gln Cys Cys Met Gln Leu Gln Gly Met Ile Pro Gln
70 75 8070 75 80
tgc cat tgt ggt gct agt tgt caa atg atg cag aac atg caa aat gct 345tgc cat tgt ggt gct agt tgt caa atg atg cag aac atg caa aat gct 345
Cys His Cys Gly Ala Ser Cys Gln Met Met Gln Asn Met Gln Asn AlaCys His Cys Gly Ala Ser Cys Gln Met Met Gln Asn Met Gln Asn Ala
85 90 9585 90 95
att tgt ggt gga ctc ggg caa caa caa atg atg atg aag atg gtg atg 393att tgt ggt gga ctc ggg caa caa caa atg atg atg aag atg gtg atg 393
Ile Cys Gly Gly Leu Gly Gln Gln Gln Met Met Met Lys Met Val MetIle Cys Gly Gly Leu Gly Gln Gln Gln Met Met Met Lys Met Val Met
100 105 110100 105 110
caa ctg cca tat gtg tgc aac atg gca ccc gcc aac ttt caa ctc ttt 441caa ctg cca tat gtg tgc aac atg gca ccc gcc aac ttt caa ctc ttt 441
Gln Leu Pro Tyr Val Cys Asn Met Ala Pro Ala Asn Phe Gln Leu PheGln Leu Pro Tyr Val Cys Asn Met Ala Pro Ala Asn Phe Gln Leu Phe
115 120 125115 120 125
cct tat ggt tgt tgt tga 459cct tat ggt tgt tgt tga 459
Pro Tyr Gly Cys CysPro Tyr Gly Cys Cys
130130
<210>8<210>8
<211>134<211>134
<212>PRT<212>PRT
<213>水稻(Oryza sativa)<213> Rice (Oryza sativa)
<400>8<400>8
Met Ala Ser Tyr Lys Ile Leu Val Val Phe Ala Leu Leu Ala Leu SerMet Ala Ser Tyr Lys Ile Leu Val Val Phe Ala Leu Leu Ala Leu Ser
1 5 10 151 5 10 15
Ala Ser Ala Ala Thr Ala Ile Thr Thr Thr Ile Pro Tyr Phe Pro SerAla Ser Ala Ala Thr Ala Ile Thr Thr Thr Ile Pro Tyr Phe Pro Ser
20 25 3020 25 30
Thr Leu Ala Met Gly Thr Met Asn Pro Cys Lys Leu Tyr Met Met GlnThr Leu Ala Met Gly Thr Met Asn Pro Cys Lys Leu Tyr Met Met Gln
35 40 4535 40 45
Thr Leu Gly Met Gly Ser Tyr Ala Thr Met Phe Met Ser Gln Pro IleThr Leu Gly Met Gly Ser Tyr Ala Thr Met Phe Met Ser Gln Pro Ile
50 55 6050 55 60
Ala Leu Leu Gln Gln Gln Cys Cys Met Gln Leu Gln Gly Met Ile ProAla Leu Leu Gln Gln Gln Cys Cys Met Gln Leu Gln Gly Met Ile Pro
65 70 75 8065 70 75 80
Gln Cys His Cys Gly Ala Ser Cys Gln Met Met Gln Asn Met Gln AsnGln Cys His Cys Gly Ala Ser Cys Gln Met Met Gln Asn Met Gln Asn
85 90 9585 90 95
Ala Ile Cys Gly Gly Leu Gly Gln Gln Gln Met Met Met Lys Met ValAla Ile Cys Gly Gly Leu Gly Gln Gln Gln Met Met Met Lys Met Val
100 105 110100 105 110
Met Gln Leu Pro Tyr Val Cys Asn Met Ala Pro Ala Asn Phe Gln LeuMet Gln Leu Pro Tyr Val Cys Asn Met Ala Pro Ala Asn Phe Gln Leu
115 120 125115 120 125
Phe Pro Tyr Gly Cys CysPhe Pro Tyr Gly Cys Cys
130130
Claims (7)
Priority Applications (1)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| CN2010101510338ACN101831429B (en) | 2010-04-15 | 2010-04-15 | Identification of promoters and expression patterns of endosperm-specific genes in rice |
Applications Claiming Priority (1)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| CN2010101510338ACN101831429B (en) | 2010-04-15 | 2010-04-15 | Identification of promoters and expression patterns of endosperm-specific genes in rice |
Publications (2)
| Publication Number | Publication Date |
|---|---|
| CN101831429A CN101831429A (en) | 2010-09-15 |
| CN101831429Btrue CN101831429B (en) | 2012-04-18 |
Family
ID=42715649
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| CN2010101510338AExpired - Fee RelatedCN101831429B (en) | 2010-04-15 | 2010-04-15 | Identification of promoters and expression patterns of endosperm-specific genes in rice |
Country Status (1)
| Country | Link |
|---|---|
| CN (1) | CN101831429B (en) |
Families Citing this family (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN103725680B (en)* | 2013-12-27 | 2015-06-10 | 安徽省农业科学院水稻研究所 | Plant endosperm specificity expression promoter pENP3 and application thereof |
| CN105274115B (en)* | 2015-11-25 | 2018-08-28 | 中国科学院植物研究所 | A kind of specifically expressed promoter P-MYBLIKE02 of rice paddy seed back side aleurone and its application |
Family Cites Families (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN1298021A (en)* | 1999-12-02 | 2001-06-06 | 中国科学院上海植物生理研究所 | One-purpose expression promoter in rice endosperm tissue and its application |
| CN100510077C (en)* | 2006-11-03 | 2009-07-08 | 上海师范大学 | Rice high efficient expression starter and application thereof |
- 2010
- 2010-04-15CNCN2010101510338Apatent/CN101831429B/ennot_activeExpired - Fee Related
Also Published As
| Publication number | Publication date |
|---|---|
| CN101831429A (en) | 2010-09-15 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| JP5938444B2 (en) | How to increase rice production | |
| CN101358193B (en) | Identification and application of rice leaf senescence-specific promoter | |
| CN100445384C (en) | Identification and utilization of a rice promoter CPIP with bidirectional expression activity specifically induced by stress | |
| CN104120137A (en) | Gene OsNAP for regulating and controlling senescence and drought resistant ability of rice leaves and application of gene OsNAP | |
| ES2560806T3 (en) | Expression of transcription regulators that provide heat tolerance | |
| CN114276429A (en) | Breeding method of TaLRK-R transgenic wheat with resistance to sheath blight and stem rot and related biological materials | |
| US20090210969A1 (en) | Antiporter gene from porteresia coarctata for conferring stress tolerance | |
| CN103421813A (en) | Application of SN1 gene in controlling high temperature resistance and drought resistance of rice | |
| US8207401B2 (en) | A method of creating a plant improved in tolerance to iron deficiency | |
| CN102747099A (en) | Application of rice gene OsbZIP46 in heat resistance and cold resistance regulation | |
| CN102747098B (en) | Application of Modified Gene OsbZIP46CA1 in Controlling Drought Resistance of Rice | |
| CN105219773B (en) | Chimeric promoters and application thereof | |
| CN111574606B (en) | Wheat disease-resistant and heading regulation gene TaCOK and related biological material and application thereof | |
| CN110627887B (en) | Application of SlTLFP8 protein and related biomaterials in regulating tomato drought resistance | |
| CN106566825B (en) | The method for screening rice tissue specifically expressing cis-acting elements and its flanking sequence | |
| CN101831429B (en) | Identification of promoters and expression patterns of endosperm-specific genes in rice | |
| CN104558132B (en) | Peanut DELLA gene families and its encoding gene and application | |
| CN109112135A (en) | Application of the OsREP4 gene in control paddy drought resistance | |
| WO2018010459A1 (en) | Isolated promoter safes6 not expressed in endosperm and application of promoter safes6 | |
| US20110055977A1 (en) | Enhancement of Reproductive Heat Tolerance in Plants | |
| CN114805508B (en) | Rice heading stage gene DHD3 function and application | |
| CN102586246B (en) | Identification and utilization of rice drought inducible promoter OXHS4P | |
| CN112159465B (en) | DRN protein and related biological material and application thereof in improving regeneration efficiency of plant somatic cells | |
| CN101831428A (en) | Separation clone and expression mode identification of promotor region of rice endosperm special expression gene | |
| CN103421788B (en) | Separation and utilization of stress response paddy rice promoter OsSN1P |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| C06 | Publication | ||
| PB01 | Publication | ||
| C10 | Entry into substantive examination | ||
| SE01 | Entry into force of request for substantive examination | ||
| C14 | Grant of patent or utility model | ||
| GR01 | Patent grant | ||
| CF01 | Termination of patent right due to non-payment of annual fee | ||
| CF01 | Termination of patent right due to non-payment of annual fee | Granted publication date:20120418 |