Before You Start
- In the following example, we demonstrate key package functionalityusing the test data set that is included in the package
- You can follow along because all test data and associated CSV inputfiles are contained in the package.
- Additional examples are also available underExampleVignettes
Components of the test dataset
- Paired-end short read amplicon data
- Files: S1_R1.fastq.gz, S1_R2.fastq.gz, S2_R1.fastq.gz,S2_R1.fastq.gz
- The files must be zipped and end in eitherR1.fastq.gz orR2.fastq.gz and each sample must have both R1 and R2files.
- New row for each unique sample
- Samples entered twice if samples contain two pooled metabarcodes, asin the test data template
- New row for each unique barcode and associated primer sequence
- Optional
CutadaptandDADA2parameters
- Taxonomy databases
- UNITE fungal database (abridged version)
- oomyceteDB
Format of the paired-end read files
- The package takes forward and reverse Illumina short read sequencedata.
- Format of file names To avoid errors, the onlycharacters that are acceptable in sample names are letters and numbers.Characters can be separated by underscores, but no other symbols. Thefiles must end with the suffixR1.fastq.gz orR2.fastq.gz.
Format of metadata file
- The format of the CSV file namedmetadata.csv issimple.
- A template ishere.
- The only tworequired columns (with these headers)are:
- Column 1.sample_name
- Column 2.primer_info (applicable options: ‘rps10’,‘its’, ‘r16S’, ‘other1’, other2’)
- Column 1.sample_name
Additional columns
- Other columns should be pasted after these two columns.
- They can be referenced later during the analysis steps and save astep of loading metadata later.
Notes
- S1 and S2 are come from a rhododendron rhizobiome dataset and are arandom subset of the full set of reads.
- S1 and S2 are included twice in the ‘metadata.csv’ sheet. This isbecause these two samples contain pooled metabarcodes (ITS1 andrps10).
- To demultiplex and run both analyses in tandem, include the samesample twice under sample_name, and then change the primer_name.
Here is what themetadata.csv looks like for this testdataset:
| sample_name | primer_name | organism |
|---|---|---|
| S1 | rps10 | Cry |
| S2 | rps10 | Cin |
| S1 | its | Cry |
| S2 | its | Cin |
Format of primer and parameters file
Primer sequence information and user-defined parameters are placed inprimerinfo_params.csv
To simplify how functions are called, user will provideparameters within this input file.
We recommend using the template linkedhere.
Remember to add any additional optional
DADA2parameters you want to use.The only threerequired columns (with theseheaders) are:
- Column 1.sample_name
- Column 2.forward (primer sequence)
- Column 3.reverse(primer sequence)
| primer_name | forward | reverse | already_trimmed | minCutadaptlength | multithread | verbose | maxN | maxEE_forward | maxEE_reverse | truncLen_forward | truncLen_reverse | truncQ | minLen | maxLen | minQ | trimLeft | trimRight | rm.lowcomplex | minOverlap | maxMismatch | min_asv_length |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rps10 | GTTGGTTAGAGYARAAGACT | ATRYYTAGAAAGAYTYGAACT | FALSE | 100 | TRUE | FALSE | 1.00E+05 | 5 | 5 | 0 | 0 | 5 | 150 | Inf | 0 | 0 | 0 | 0 | 15 | 0 | 50 |
| its | CTTGGTCATTTAGAGGAAGTAA | GCTGCGTTCTTCATCGATGC | FALSE | 50 | TRUE | FALSE | 1.00E+05 | 5 | 5 | 0 | 0 | 5 | 50 | Inf | 0 | 0 | 0 | 0 | 15 | 0 | 50 |
For more info on parameter specifics, seeDocumentation
Reference Database Format
- For now, the package is compatible with the following databases:
- oomycetedb from:https://grunwaldlab.github.io/OomyceteDB/
- SILVA 16S database with species assignments:https://www.arb-silva.de/
- UNITE database fromhttps://unite.ut.ee/repository.php
- A user can select up to two other databases, but will first need toreformat headers exactly like the SILVA database headers. SeeDocumentation
- oomycetedb from:https://grunwaldlab.github.io/OomyceteDB/
Databases are downloaded from these sources and then placed in thefrom the user-specified data folder where raw data files and csv filesare located.
Additional Notes
- Computer specifications may be a limiting factor.
- If you are using the SILVA or UNITE databases for taxonomicassignment steps, an ordinary personal computer (unless it hassufficient RAM) may not have enough memory for the taxonomic assignmentsteps, even with few samples.
- The test databases and reads are subsetted and the following exampleshould run on a personal computer with at least 16 GB of RAM.
- If computer crashes during the taxonomic assignment step, you willneed to switch to a computer with sufficient memory.
- You must ensure that you have enough storage to save intermediatefiles in a temporary directory (default) or user-specified directorybefore proceeding.
Installation
Dependencies:
First installCutadapt program followingthe instructions here:https://cutadapt.readthedocs.io/en/stable/installation.html
Let’s locate where theCutadaptexecutable is.
You must do this from aTerminal window:
#If you installed with pip or pipx, or homebrew, run this command from a Terminal windowwhich cutadaptcutadapt--versionIf you followed the cutadapt installation instructions to create aconda environment calledCutadapt (changeto whatever you named your environment), to install it in.
Open up aTerminal window and type thesecommands:
Second, make sure the following R packages are installed:
demulticoder(Available throughCRAN)DADA2(Latest version is 3.20)- To install, follow these instructions:https://www.bioconductor.org/packages/release/bioc/html/dada2.html
- To install, follow these instructions:https://www.bioconductor.org/packages/release/bioc/html/dada2.html
phyloseqmetacoder(Available throughCRAN)
Reorganize data tables and set-up data directory structure
The sample names, primer sequences, and other metadata arereorganized in preparation for runningCutadapt to remove primers.
output<-prepare_reads( data_directory=system.file("extdata", package="demulticoder"), output_directory=tempdir(),# Change to your desired output directory path or leave as is", overwrite_existing=TRUE)#> Existing files found in the output directory. Overwriting existing files.#>Rows:2Columns:25#>──Column specification────────────────────────────────────────────────────────#>Delimiter: ","#>chr (3): primer_name, forward, reverse#>dbl (18): minCutadaptlength, maxN, maxEE_forward, maxEE_reverse, truncLen_fo...#>lgl (4): already_trimmed, count_all_samples, multithread, verbose#>#>ℹ Use `spec()` to retrieve the full column specification for this data.#>ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.#>Rows:2Columns:25#>──Column specification────────────────────────────────────────────────────────#>Delimiter: ","#>chr (3): primer_name, forward, reverse#>dbl (18): minCutadaptlength, maxN, maxEE_forward, maxEE_reverse, truncLen_fo...#>lgl (4): already_trimmed, count_all_samples, multithread, verbose#>#>ℹ Use `spec()` to retrieve the full column specification for this data.#>ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.#>Rows:4Columns:3#>──Column specification────────────────────────────────────────────────────────#>Delimiter: ","#>chr (3): sample_name, primer_name, organism#>#>ℹ Use `spec()` to retrieve the full column specification for this data.#>ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.#> Creating output directory: /tmp/RtmpyNk8MG/demulticoder_run/prefiltered_sequences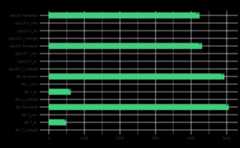
Remove primers with Cutadapt
Before runningCutadapt,pleaseensure that you have installed it
cut_trim(output, cutadapt_path="/usr/bin/cutadapt",#CHANGE LOCATION TO YOUR LOCAL INSTALLATION overwrite_existing=TRUE)#> Running cutadapt 3.5 for its sequence data
#> Running cutadapt 3.5 for rps10 sequence data
ASV inference step
Raw reads will be merged and ASVs will be inferred
make_asv_abund_matrix(output, overwrite_existing=TRUE)#> 80608 total bases in 307 reads from 2 samples will be used for learning the error rates.#> Sample 1 - 163 reads in 84 unique sequences.#> Sample 2 - 144 reads in 96 unique sequences.#> 82114 total bases in 307 reads from 2 samples will be used for learning the error rates.#> Sample 1 - 163 reads in 128 unique sequences.#> Sample 2 - 144 reads in 119 unique sequences.
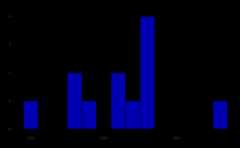
#> 91897 total bases in 327 reads from 2 samples will be used for learning the error rates.#> Sample 1 - 145 reads in 107 unique sequences.#> Sample 2 - 182 reads in 133 unique sequences.#> 91567 total bases in 327 reads from 2 samples will be used for learning the error rates.#> Sample 1 - 145 reads in 114 unique sequences.#> Sample 2 - 182 reads in 170 unique sequences.

#> $its#> [1] "/tmp/RtmpyNk8MG/demulticoder_run/asvabund_matrixDADA2_its.RData"#>#> $rps10#> [1] "/tmp/RtmpyNk8MG/demulticoder_run/asvabund_matrixDADA2_rps10.RData"Taxonomic assignment step
Using the core assignTaxonomy function fromDADA2, taxonomic assignments will be givento ASVs.
assign_tax(output,asv_abund_matrix, retrieve_files=TRUE, overwrite_existing=TRUE)#> samplename_barcode input filtered denoisedF denoisedR merged nonchim#> 1 S1_its 299 163 146 141 132 132#> 2 S2_its 235 144 113 99 99 99#> samplename_barcode input filtered denoisedF denoisedR merged nonchim#> 1 S1_rps10 196 145 145 145 145 145#> 2 S2_rps10 253 182 181 181 181 181Format of output matrices
- The default output is a CSV file per metabarcode with the inferredASVs and sequences, taxonomic assignments, and bootstrap supportsprovided by
DADA2. These data can then beused as input for downstream steps. - To view these files, locate the files with the prefixfinal_asv_abundance_matrix in your specified outputdirectory.
- In this example, it will be in the home directory withindemulticoder_test.
- You can then open your matrices using Excel
- We recommend checking the column with the ASV sequences to make surethere aren’t truncated sequences *It is important to also review thecolumn showing the taxonomic assignments of each ASV and theirassociated bootstrap supports.
We can also load the matrices into our R environment and view thenfrom R Studio.
We do that here with therps10 matrix
rps10_matrix_file<-file.path(output$directory_paths$output_directory,"final_asv_abundance_matrix_rps10.csv")rps10_matrix<-read.csv(rps10_matrix_file, header=TRUE)head(rps10_matrix)#> asv_id#> 1 asv_1#> 2 asv_2#> 3 asv_3#> 4 asv_4#> 5 asv_5#> sequence#> 1 GAAAATCTTTGTGTCGGTGGTTCAAATCCACCTCCAGACAAATTAATTTTTTAAAACTTATGTTTATATTAAGAATTACATTTAAATCTATTAAAAAAATAAATAACATTAAACCTATTTTATTAAATCAAAAAAATTTAAATAAATTTAATAATATTAAAATAAATGGAATATTTAAAACAAAAAATAAAAATAAAATTTTTACAGTATTAAAATCACCACATGTTAATAAAAAAGCACGTGAACATTTTATTTATAAAAATTTTACACAAAAAGTTGAAATTAAATGTTTAAATATTTTTCAATTATTAAATTTTTTAATTTTGATTAAAAAAATCTTAACAGAAAATTTTATTATTACTACAAAAATAATTAAACAATAATTAATATATGCTTATAGCTTAATGGATAAAGCATTAGATTGCGGATCTACAAAATGAA#> 2 GAAAATCTTTGTGTCGGTGGTTCAAGTCCACCTCCAGACAAAAATAATAACTATGTATATTTTAAGAATAACTTTTAAATCAATACAAAAAATAAATAATATTAAAAAAAATTTATTAAAATTAAAAAAAATAAATAAATTTAAAAATATTCAAATAAAAGGAATATTTAAAATAAAAGATAAAAATAAAATTTTTACTTTATTAAAATCACCTCACGTAAATAAAAAATCTCGTGAACATTTTATTTATAAAAATTATACTCAAAAAATAGATGTAAAATTTTCAAATATTATTGAATTATTTAATTTTATAATACTTATTAAAAAAGTTTTAACTAAAAATTTTATAATAAATTTTAAAATTATAAAATATAATAAAAAAAAAATGCTTATAGCTTAATGGATAAAGCGTTAGATTGCGGATCTATAAAATGAA#> 3 GAAAATCTTTGTGTCGGTGGTTCAAATCCGCCTCCAAACAAAAATATAAATTATTTTATATGTATATTTTAAGAATAAGTTTTAAATCTGTATCAAAAATAGATAAAATTAAAAAAGAATTATCAAGATTAAAAAAAATATATAATTTTAAAAATATAACAATTAATGGTATTTTTAAAACAAAAAATAAAGATAAAATTTTTACATTATTAAAATCACCTCATGTTAATAAAAAATCTCGCGAACATTTTATTTATAAAAATTATGTACAAAAAATAGATTTAAATTTTATTAATATTTTTCAATTAATAAATTTTTTAATAATTTTAAAAAAAAAATTATCAAAAAATGTATTAATAAATGTAAAAATAATTAAAAAATAAAAAAATATGCTTATAGCTTAATGGATAAAGCGTTAGATTGCGGATCTATAAAATGAA#> 4 GAAAATCTTTGTGTCGGTGGTTCAAGTCCGCCTCCAAACATCTTATAAAAATGTATGTATATTTTACGAATTAGTTTTAAATCAGTAGAAAAAATAAATGAATTAAAAAAAAATATCTCAAAAATAAAAAAAATATATAATTTTAAAAATTTAAAAGTAAATGGTATTTTTAGAACAAAAAATAAAAATAAAATTTTTACATTATTAAAATCACCCCATGTTAATAAAAAATCACGTGAACATTTTATATATAAAAATTATATTCAAAAAATTGATTTAAATTTTTCAAATTTTTTTCAATTATTAAATTTTTTAGTAATTTTAAAAAAAATATTATCTAAAGATTTTTTAATCAATATCAAAATTATTAAAAAAAATAAAAAAAATATGCTTATAGCTTAATGGATAAAGCGTTAGATTGCGGATCTATAAAATGAA#> 5 GAAAATCTTTGTGTCGGTGGTTCAAGTCCGCCTCCAAACATCTTATAAAAATGTATGTATATTTTACGAATTAGTTTTAAATCAGTAGAAAAAATAAATGAATTAAAAAAAAATATCTCAAAAATAAAAAAAATATATAATTTTAAAAATTTAAAAGTAAATGGTATTTTTAGAACAAAAAATAAAAATAAAATTTTTACATTATTAAAATCACCCCATGTTAATAAAAAATCACGTGAACATTTTATATATAAAAATTATATTCAAAAAATTGATTTAAATTTTTCAAATTTTTTTCAATTATTAAATTTTTTAGTAATTTTAAAAAAAACATTATCTAAAGATTTTTTAATCAATATTAAAATTATTAAAAAAAATAAAAAAATATGCTTATAGCTTAATGGATAAAGCGTTAGATTGCGGATCTATAAAATGAA#> dada2_tax#> 1 Stramenopila--100--Kingdom;Oomycota--100--Phylum;Oomycetes--100--Class;Peronosporales--26--Order;Peronosporaceae--26--Family;Phytophthora--23--Genus;Phytophthora_sp.kununurra--13--Species;oomycetedb_650--13--Reference#> 2 Stramenopila--100--Kingdom;Oomycota--100--Phylum;Oomycetes--100--Class;Pythiales--100--Order;Pythiaceae--100--Family;Globisporangium--100--Genus;Globisporangium_cylindrosporum--28--Species;oomycetedb_41--28--Reference#> 3 Stramenopila--100--Kingdom;Oomycota--100--Phylum;Oomycetes--100--Class;Pythiales--93--Order;Pythiaceae--93--Family;Pythium--86--Genus;Pythium_aphanidermatum--22--Species;oomycetedb_727--22--Reference#> 4 Stramenopila--100--Kingdom;Oomycota--100--Phylum;Oomycetes--100--Class;Pythiales--100--Order;Pythiaceae--100--Family;Pythium--100--Genus;Pythium_dissotocum--58--Species;oomycetedb_750--28--Reference#> 5 Stramenopila--100--Kingdom;Oomycota--100--Phylum;Oomycetes--100--Class;Pythiales--100--Order;Pythiaceae--100--Family;Pythium--100--Genus;Pythium_oopapillum--64--Species;oomycetedb_771--64--Reference#> S1_rps10 S2_rps10#> 1 87 0#> 2 0 71#> 3 0 63#> 4 58 0#> 5 0 47We can also inspect the first rows of theITS1 matrix
its_matrix_file<-file.path(output$directory_paths$output_directory,"final_asv_abundance_matrix_its.csv")its_matrix<-read.csv(its_matrix_file, header=TRUE)head(its_matrix)#> asv_id#> 1 asv_1#> 2 asv_2#> 3 asv_3#> 4 asv_4#> 5 asv_5#> 6 asv_6#> sequence#> 1 AAAAAGTCGTAACAAGGTTTCCGTAGGTGAACCTGCGGAAGGATCATTAGTGAATCTTCAAAGTCGGCTCGTCGGATTGTGCTGGTGGAGACACATGTGCACGTCTACGAGTCGCAAACCCACACACCTGTGCATCTATGACTCTGAGTGCCGCTTTGCATGGCCCCTTGATTTGGGCCTGGCGCTCGAGTACTTTCACACACTCTCGAATGTAATGGAATGTCTTGTTGTGCATAACGTACAAACAGAAACAACTTTCAACAACGGATCTCTTGGCTCTC#> 2 AAAAAGTCGTAACAAGGTTTCCGTAGGTGAACCTGCGGAAGGATCATTAGTGAATCTTCAAAGTCGGCTCGTCAGATTGTGCTGGTGGAGACACATGTGCACGTCTACGAGTCGCAAACCCACACACCTGTGCATCTATGACTCTGAGTGCCGCTTTGCATGGCCCCTTGATTTGGGCCTGGCGCTCGAGTACTTTCACACACTCTCGAATGTAATGGAATGTCTTCTTGTGCATAACGTACAAACAGAAACAACTTTCAACAACGGATCTCTTGGCTCTC#> 3 AAAAAGTCGTAACAAGGTTTCCGTAGGTGAACCTGCGGAAGGATCATTAGTGAATCTTCAAAGTCGGCTCGTCGGATTGTGCTGGTGGAGACACATGTGCACGTCTACGAGTCGCAAACCCACACACCTGTGCATCTATGACTCTGAGTGCCGCTTTGCATGGCCCCTTGATTTGGGCCTGGCGCTCGAGTACTTTCACACACTCTCGAATGTAATGGAATGACTTGTTGTGCATAACGTACAAACAGAAACAACTTTCAACAACGGATCTCTTGGCTCTC#> 4 AAGTCGTAACAAGGTTTCCGTAGGTGAACCTGCGGAAGGATCATTAACGAGTTCAACTGGGGTCTGTGGGGATTCGATGCTGGCCTCCGGGCATGTGCTCGTCCCCCGGACCTCACATCCACTCACAACCCCTGTGCATCATGAGAGGTGTGGTCCTCCGTCATGGGAGCAGCTCCCCTCCGGGCGTGTTTGTACACCCCGGGCGGTCGAGCCGGGACTGCACTTCGACGCCTTTACACAAACCTTTGAATCAGTGATGTAGAATGTCATGGCCAGCGATGGTCGAACTTTAAATACAACTTTCAACAACGGATCTCTTGGCTCTC#> 5 AAAAAGTCGTAACAAGGTTTCCGTAGGTGAACCTGCGGAAGGATCATTAGTGAATCTTCAAAGTCGGCTCGTCGGATTGTGCTGGTGGAGACACATGTGCACATCTACGAGTCGCAAACCCACACACCTGTGCATCTATGACTCTGAGTGCCGCTTTGCATGGCCCCTTGATTTGGGCCTGGCGCTCGAGTACTTTCACACACTCTCGAATGTAATGGAATGTCTTGTTGTGCATAACGTACAAACAGAAACAACTTTCAACAACGGATCTCTTGGCTCTC#> 6 AAGTCGTAACAAGGTTTCCGTAGGTGAACCTGCGGAAGGATCATTAAAAATGAAGCCGGGAAACCGGTCCTTCTAACCCTTGATTATCTTAATTTGTTGCTTTGGTGGGCCGCGCAAGCACCGGCTTCGGCTGGATCGTGCCCGCCAGAGGACCACAACTCTGTATCAAATGTCGTCTGAGTACTATATAATAGTTAAAACTTTCAACAACGGATCTCTTGGTTCTG#> dada2_tax#> 1 Fungi--100--Kingdom;Basidiomycota--100--Phylum;Agaricomycetes--100--Class;Sebacinales--100--Order;unidentified--92--Family;unidentified--92--Genus;Sebacinales_sp--92--Species#> 2 Fungi--100--Kingdom;Basidiomycota--100--Phylum;Agaricomycetes--100--Class;Sebacinales--100--Order;unidentified--95--Family;unidentified--95--Genus;Sebacinales_sp--95--Species#> 3 Fungi--100--Kingdom;Basidiomycota--100--Phylum;Agaricomycetes--100--Class;Sebacinales--100--Order;unidentified--96--Family;unidentified--96--Genus;Sebacinales_sp--96--Species#> 4 Fungi--100--Kingdom;Basidiomycota--69--Phylum;Agaricomycetes--66--Class;Hymenochaetales--26--Order;Repetobasidiaceae--11--Family;Rickenella--11--Genus;Rickenella_sp--11--Species#> 5 Fungi--100--Kingdom;Basidiomycota--100--Phylum;Agaricomycetes--100--Class;Sebacinales--100--Order;unidentified--92--Family;unidentified--92--Genus;Sebacinales_sp--92--Species#> 6 Fungi--100--Kingdom;Ascomycota--96--Phylum;Leotiomycetes--72--Class;Helotiales--72--Order;Helotiales_fam_Incertae_sedis--44--Family;Chlorencoelia--41--Genus;Chlorencoelia_sp--41--Species#> S1_its S2_its#> 1 60 0#> 2 46 0#> 3 0 35#> 4 21 0#> 5 0 21#> 6 0 13Reformat ASV matrix astaxmap andphyloseq objects after optional filteringof low abundance ASVs
objs<-convert_asv_matrix_to_objs(output, minimum_bootstrap=0, save_outputs=TRUE)Objects can now be used for downstream data analysis
Here we make heattrees usingtaxmapobject.
First we make a heat tree to examine fungal community compositionusing ourITS1 data.
objs$taxmap_its%>%filter_taxa(!grepl(x=taxon_names,"NA", ignore.case=TRUE))%>%metacoder::heat_tree(node_label=taxon_names, node_size=n_obs, node_color=n_obs, node_color_axis_label="ASV count", node_size_axis_label="Total Abundance of Taxa", layout="da", initial_layout="re")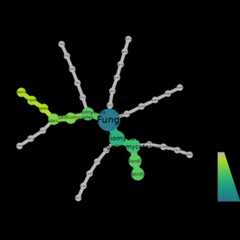
Now we make a heat tree to showcase oomycete community compositionusing ourrps10 data
objs$taxmap_rps10%>%filter_taxa(!grepl(x=taxon_names,"NA", ignore.case=TRUE))%>%metacoder::heat_tree(node_label=taxon_names, node_size=n_obs, node_color=n_obs, node_color_axis_label="ASV count", node_size_axis_label="Total Abundance of Taxa", layout="da", initial_layout="re")
We can also do a variety of analyses, if we convert tophyloseq object
Here we demonstrate how to make a stacked bar plot of the relativeabundance of taxa by sample for the ITS1-barcoded samples
data<-objs$phyloseq_its%>%phyloseq::transform_sample_counts(function(x){x/sum(x)})%>%phyloseq::psmelt()%>%dplyr::filter(Abundance>0.02)%>%dplyr::arrange(Genus)abund_plot<-ggplot2::ggplot(data,ggplot2::aes(x=Sample, y=Abundance, fill=Genus))+ggplot2::geom_bar(stat="identity", position="stack", color="black", size=0.2)+ggplot2::scale_fill_viridis_d()+ggplot2::theme_minimal()+ggplot2::labs( y="Relative Abundance", title="Relative abundance of taxa by sample", fill="Genus")+ggplot2::theme( axis.text.x=ggplot2::element_text(angle=90, hjust=1, vjust=0.5, size=14), panel.grid.major=ggplot2::element_blank(), panel.grid.minor=ggplot2::element_blank(), legend.position="top", legend.text=ggplot2::element_text(size=14), legend.title=ggplot2::element_text(size=14), strip.text=ggplot2::element_text(size=14), strip.background=ggplot2::element_blank())+ggplot2::guides( fill=ggplot2::guide_legend( reverse=TRUE, keywidth=1, keyheight=1, title.position="top", title.hjust=0.5,# Center the legend title label.theme=ggplot2::element_text(size=10)))print(abund_plot)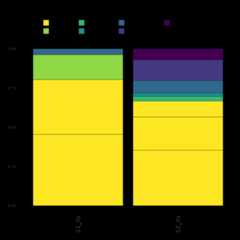
Finally, we demonstrate how to make a stacked bar plot of therelative abundance of taxa by sample for the rps10-barcoded samples
data<-objs$phyloseq_rps10%>%phyloseq::transform_sample_counts(function(x){x/sum(x)})%>%phyloseq::psmelt()%>%dplyr::filter(Abundance>0.02)%>%dplyr::arrange(Genus)abund_plot<-ggplot2::ggplot(data,ggplot2::aes(x=Sample, y=Abundance, fill=Genus))+ggplot2::geom_bar(stat="identity", position="stack", color="black", size=0.2)+ggplot2::scale_fill_viridis_d()+ggplot2::theme_minimal()+ggplot2::labs( y="Relative Abundance", title="Relative abundance of taxa by sample", fill="Genus")+ggplot2::theme( axis.text.x=ggplot2::element_text(angle=90, hjust=1, vjust=0.5, size=14), panel.grid.major=ggplot2::element_blank(), panel.grid.minor=ggplot2::element_blank(), legend.position="top", legend.text=ggplot2::element_text(size=14), legend.title=ggplot2::element_text(size=14),# Adjust legend title size strip.text=ggplot2::element_text(size=14), strip.background=ggplot2::element_blank())+ggplot2::guides( fill=ggplot2::guide_legend( reverse=TRUE, keywidth=1, keyheight=1, title.position="top", title.hjust=0.5,# Center the legend title label.theme=ggplot2::element_text(size=10)# Adjust the size of the legend labels))print(abund_plot)