- Notifications
You must be signed in to change notification settings - Fork0
Developing an R Package for Influenza Surveillance Simulation Study
License
vjoshy/DESA
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation
DESA provides a comprehensive framework for early epidemic detectionthrough school absenteeism surveillance. The package enables:
- Simulation: Generate realistic populations, epidemic spreadpatterns, and resulting school absenteeism data
- Detection: Implement surveillance models that raise timelyalerts for impending epidemics
- Evaluation: Assess and optimize detection models using metricsthat balance timeliness and accuracy
The methods implemented are based on research byVanderkruk etal. (2023)andWard etal. (2019).
Children typically have higher influenza infection rates than other agegroups and are encouraged to stay home when ill. This makes schoolabsenteeism data a valuable early indicator of community-wide epidemicarrival, potentially providing public health officials with crucial leadtime to implement mitigation strategies.
Traditional epidemic surveillance systems often rely onlaboratory-confirmed cases, which can introduce significant delays indetection. To address this limitation, Ward et al. (2019) investigatedthe use of school absenteeism data for early epidemic detection in theWellington-Dufferin-Guelph region of Ontario, Canada.
The existing system in this region used a simple threshold-basedapproach (raising an alarm when 10% of students were absent) to identifypotential outbreaks. Ward et al. developed detection models andintroduced two evaluation metrics:
- False Alert Rate (FAR): Measures the proportion of incorrectlyraised alarms
- Accumulated Days Delayed (ADD): Quantifies the timeliness ofcorrect alarms
Building on this foundation, Vanderkruk et al. (2023) developed a morenuanced evaluation framework calledAlert Time Quality (ATQ). Unlikethe binary classification of previous metrics, ATQ assesses alerts whereones that are raised at sub optimal times receive penalties proportionalto their deviation from the ideal alert time. This approach balancesboth accuracy and timeliness in a single metric.
The DESA package implements the methodologies from both papers, enablingusers to:
- Generate simulations of population structures based on regionalcensus data
- Apply various detection models to simulated epidemics
- Evaluate model performance using FAR, ADD, and ATQ-based metrics
You can install the development version of DESA fromGithub
#install.packages("devtools")library(devtools)install_github("vjoshy/DESA")
The ATQ package includes the following main functions:
catchment_sim()- Simulates catchment area data
elementary_pop()- Simulates elementary school populations
subpop_children()- Simulates households with children
subpop_noChildren()- Simulates households without children
simulate_households()- Combines household simulations
ssir()- Simulates epidemic using SSIR model
compile_epi()- Compiles absenteeism data
eval_metrics()- Evaluates alarm metrics
plot()andsummary()- Methods for visualizing and summarizing results
Additionally, the package implements S3 methods for generic functions:
plot()andsummary()- These methods are extended for objects returned by
ssir()andeval_metrics() plot()provides visualizations of epidemic simulations andmetric evaluationssummary()offers concise summaries of the simulation results andmetric assessments
- These methods are extended for objects returned by
These functions and methods work together to facilitate comprehensiveepidemic simulation and evaluation of detection models
Please see example below:
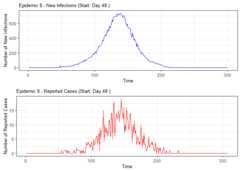
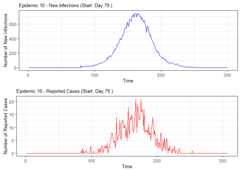
# Load the DESA packagelibrary(DESA)set.seed(69420)#Simulate number of elementary schools in each catchmentcatch_df<- catchment_sim(16,5,shape=4.68,rate=3.01)# Simulate elementary school populations for each catchment areaelementary_df<- elementary_pop(catch_df,shape=5.86,rate=0.01)# Simulate households with childrenhouse_children<- subpop_children(elementary_df,n=2,prop_parent_couple=0.7,prop_children_couple= c(0.3,0.5,0.2),prop_children_lone= c(0.4,0.4,0.2),prop_elem_age=0.6)# Simulate households without childrenhouse_nochildren<- subpop_noChildren(house_children,elementary_df,prop_house_size= c(0.2,0.3,0.25,0.15,0.1),prop_house_Children=0.3)# Combine household simulations and generate individual-level datasimulation<- simulate_households(house_children,house_nochildren)# Extract individual-level dataindividuals<-simulation$individual_sim# Simulate epidemic using SSIR (Stochastic Susceptible-Infectious-Recovered) modelepidemic<- ssir(nrow(individuals),T=300,alpha=0.298,inf_init=32,rep=10)# Summarize and plot the epidemic simulation resultssummary(epidemic)#> SSIR Epidemic Summary (Multiple Simulations):#> Number of simulations: 10#>#> Average total infected: 41065.5#> Average total reported cases: 826.9#> Average peak infected: 3036.4#>#> Model parameters:#> $N#> [1] 136784#>#> $T#> [1] 300#>#> $alpha#> [1] 0.298#>#> $inf_period#> [1] 4#>#> $inf_init#> [1] 32#>#> $report#> [1] 0.02#>#> $lag#> [1] 7#>#> $rep#> [1] 10plot(epidemic)
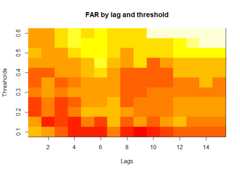
# Compile absenteeism data based on epidemic simulation and individual dataabsent_data<- compile_epi(epidemic,individuals)# Display structure of absenteeism datadplyr::glimpse(absent_data)#> Rows: 3,000#> Columns: 28#> $ Date <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,…#> $ ScYr <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1…#> $ pct_absent <dbl> 0.05096603, 0.05190044, 0.05079590, 0.04899765, 0.05083607…#> $ absent <dbl> 636, 661, 628, 627, 623, 624, 607, 603, 657, 673, 616, 667…#> $ absent_sick <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…#> $ new_inf <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…#> $ lab_conf <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…#> $ Case <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…#> $ sinterm <dbl> 0.01720158, 0.03439806, 0.05158437, 0.06875541, 0.08590610…#> $ costerm <dbl> 0.9998520, 0.9994082, 0.9986686, 0.9976335, 0.9963032, 0.9…#> $ window <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…#> $ ref_date <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…#> $ lag0 <dbl> 0.05096603, 0.05190044, 0.05079590, 0.04899765, 0.05083607…#> $ lag1 <dbl> NA, 0.05096603, 0.05190044, 0.05079590, 0.04899765, 0.0508…#> $ lag2 <dbl> NA, NA, 0.05096603, 0.05190044, 0.05079590, 0.04899765, 0.…#> $ lag3 <dbl> NA, NA, NA, 0.05096603, 0.05190044, 0.05079590, 0.04899765…#> $ lag4 <dbl> NA, NA, NA, NA, 0.05096603, 0.05190044, 0.05079590, 0.0489…#> $ lag5 <dbl> NA, NA, NA, NA, NA, 0.05096603, 0.05190044, 0.05079590, 0.…#> $ lag6 <dbl> NA, NA, NA, NA, NA, NA, 0.05096603, 0.05190044, 0.05079590…#> $ lag7 <dbl> NA, NA, NA, NA, NA, NA, NA, 0.05096603, 0.05190044, 0.0507…#> $ lag8 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, 0.05096603, 0.05190044, 0.…#> $ lag9 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, 0.05096603, 0.05190044…#> $ lag10 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 0.05096603, 0.0519…#> $ lag11 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 0.05096603, 0.…#> $ lag12 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 0.05096603…#> $ lag13 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 0.0509…#> $ lag14 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 0.…#> $ lag15 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA…# Evaluate alarm metrics for epidemic detectionalarm_metrics<- eval_metrics(absent_data,thres= seq(0.1,0.6,by=0.05))# Plot various alarm metricsplot(alarm_metrics$metrics,"FAR")# False Alert Rate
plot(alarm_metrics$metrics,"FATQ")# First Alert Time Quality
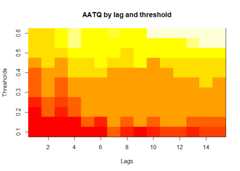
plot(alarm_metrics$metrics,"AATQ")# Average Alert Time Quality
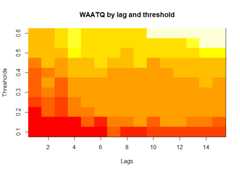
plot(alarm_metrics$metrics,"WAATQ")# Weighted Average Alert Time Quality
plot(alarm_metrics$metrics,"WFATQ")# Weighted First Alert Time Quality
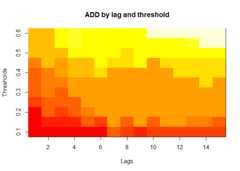
plot(alarm_metrics$metrics,"ADD")# Accumulated Delay Days
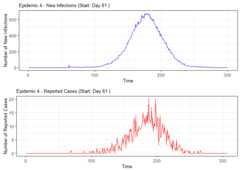
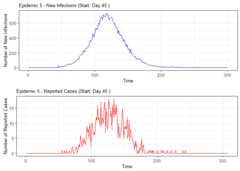
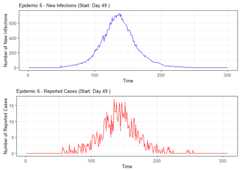
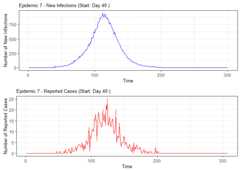
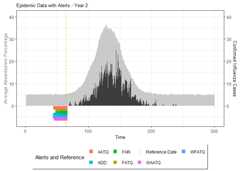
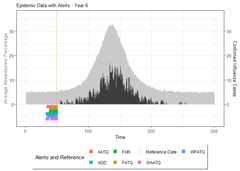
# Summarize alarm metricssummary(alarm_metrics$results)#> Alarm Metrics Summary#> =====================#>#> FAR :#> Mean: 0.5603#> Variance: 0.0191#> Optimal lag: 9#> Optimal threshold: 0.1#> Minimum value: 0.2111#>#> ADD :#> Mean: 25.831#> Variance: 75.0129#> Optimal lag: 1#> Optimal threshold: 0.1#> Minimum value: 6.1111#>#> AATQ :#> Mean: 0.542#> Variance: 0.028#> Optimal lag: 2#> Optimal threshold: 0.1#> Minimum value: 0.1861#>#> FATQ :#> Mean: 0.5451#> Variance: 0.0228#> Optimal lag: 9#> Optimal threshold: 0.1#> Minimum value: 0.1964#>#> WAATQ :#> Mean: 0.5288#> Variance: 0.0313#> Optimal lag: 1#> Optimal threshold: 0.15#> Minimum value: 0.1373#>#> WFATQ :#> Mean: 0.5474#> Variance: 0.0217#> Optimal lag: 4#> Optimal threshold: 0.1#> Minimum value: 0.2516#>#> Reference Dates and Model Selected Alert Dates:#> =====================#>#> year ref_date FAR ADD AATQ FATQ WAATQ WFATQ#> 1 1 53 NA NA NA NA NA NA#> 2 2 64 50 51 51 50 51 53#> 3 3 43 42 NA NA 42 NA NA#> 4 4 61 54 51 51 54 51 52#> 5 5 45 45 33 34 45 33 33#> 6 6 49 40 35 36 40 35 37#> 7 7 40 31 31 30 31 37 31#> 8 8 48 NA 35 41 NA 35 41#> 9 9 61 55 47 50 55 50 55#> 10 10 79 72 65 65 72 65 67# Generate and display plots for alarm metrics across epidemic yearsalarm_plots<- plot(alarm_metrics$plot_data)for(iin seq_along(alarm_plots)) { print(alarm_plots[[i]]) }
The final outputregion_metric will be a list of 6 matrices and 6 dataframes. The matrices describe the values of metrics for respective lagand thresholds.
An optimal lag and threshold value that minimizes each metric isselected and these optimal parameters are used to generate the 6 dataframes associated with the metrics. Simulated information like number oflab confirmed cases, number of students absent, etc, are also includedin the output.
About
Developing an R Package for Influenza Surveillance Simulation Study
Resources
License
Uh oh!
There was an error while loading.Please reload this page.
Stars
Watchers
Forks
Packages0
Uh oh!
There was an error while loading.Please reload this page.