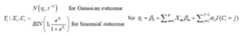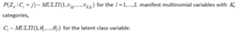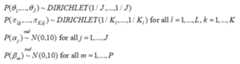- Notifications
You must be signed in to change notification settings - Fork0
umich-biostatistics/lcra
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation
A user-friendly interface for doing joint Bayesian latent class andregression analysis with binary and continuous outcomes.
Simply specify the regression model and number of classes and lcrapredicts class membership for each observation and accounts foruncertainty in class membership in the estimation of the regressionparameters.
This is the only package available for joint latent class and regressionanalysis. Other packages use a sequential procedure, where latentclasses are determined prior to fitting the usual regression model. Theonly Bayesian alternative will require you to program the model by hand,which can be time consuming.
ThisR package provides a user-friendly interface for fitting Bayesianjoint latent class and regression models. Using the standard R syntax,the user can specify the form of the regression model and the desirednumber of latent classes.
The technical details of the model implemented here are described inElliott, Michael R., Zhao, Zhangchen, Mukherjee, Bhramar, Kanaya, Alka,Needham, Belinda L., “Methods to account for uncertainty in latent classassignments when using latent classes as predictors in regressionmodels, with application to acculturation strategy measures” (2020) Inpress at Epidemiology.
JAGS is the new default Gibbs sampler for the package. Install JAGShere:JAGS Download.
This package uses the R interface for JAGS and WinBUGS. The packageR2WinBUGS in turn depends on the standalone Windows programWinBUGS,andrjags depends on the standalone program JAGS. Follow this link:JAGS Download for JAGS download andinstallation instructions.
Once you’ve chosen between JAGS or WinBUGS, open R and run:
If the devtools package is not yet installed, install it first:
install.packages('devtools')library(devtools)
# install lcra from Github:install_github('umich-biostatistics/lcra') library(lcra)
inits=list(list(theta= c(0.33,0.33,0.34),beta= rep(0,length=3),alpha= rep(0,length=2),tau=0.5,true= rep(1,length= nrow(express))))fit= lcra(formula=y~x1+x2,family="gaussian",data=express,nclasses=3,inits=inits,manifest= paste0("Z",1:5),n.chains=1,n.iter=500)# use coda to analyze sampleslibrary(coda)summary(fit)plot(fit)
NOTE: WinBUGS throws an error on Windows machines if you are not runningas administrator. The error reads: Error in file(con, “wb”) : cannotopen the connection.
Ignore the error. Once WinBUGS is finished drawing samples, follow thepromp and return to R.
The following priors are the default and cannot be altered by the user:
Please note also that the reference category for latent classes in theoutcome model output is always the Jth latent class in the output, andthe bugs output is defined by the Latin equivalent of the modelparameters (beta, alpha, tau, pi, theta). Also, the bugs output includesthe variable true, which corresponds to the MCMC draws of C_i, i =1,…,n, as well as the MCMC draws of the deviance (DIC) statistic.Finally the bugs output for pi is stored in a three dimensional arraycorresponding to (class, variable, category), where category is indexedby 1 through maximum K_l; for variables where the number of categoriesis less than maximum K_l, these cells will be set to NA. The parametersoutputed by the lcra function currently are not user definable.
The main function for the joint model islcra(). Use?lcra for the Rhelp file.
Here is an example analysis on simulated data with continuous anddiscrete outcomes:
# quick exampleinits=list(list(theta= c(0.33,0.33,0.34),beta= rep(0,length=3),alpha= rep(0,length=2),tau=0.5,true= rep(1,length= nrow(express))))fit= lcra(formula=y~x1+x2,family="gaussian",data=express,nclasses=3,inits=inits,manifest= paste0("Z",1:5),n.chains=1,n.iter=500)data('paper_sim')# Set initial valuesinits=list(list(theta= c(0.33,0.33,0.34),beta= rep(0,length=3),alpha= rep(0,length=2),tau=0.5,true= rep(1,length=100)),list(theta= c(0.33,0.33,0.34),beta= rep(0,length=3),alpha= rep(0,length=2),tau=0.5,true= rep(1,length=100)),list(theta= c(0.33,0.33,0.34),beta= rep(0,length=3),alpha= rep(0,length=2),tau=0.5,true= rep(1,length=100)) )# Fit model 1fit.gaus_paper= lcra(formula=Y~X1+X2,family="gaussian",data=paper_sim,nclasses=3,manifest= paste0("Z",1:10),inits=inits,n.chains=3,n.iter=5000)# Model 1 resultslibrary(coda)summary(fit.gaus_paper)plot(fit.gaus_paper)# simulated exampleslibrary(gtools)# for Dirichel distribution# with binary responsen<-500X1<- runif(n,2,8)X2<- rbinom(n,1,.5)Cstar<- rnorm(n,.25*X1-.75*X2,1)C<-1* (Cstar<=.8)+2* ((Cstar>.8)& (Cstar<=1.6))+3* (Cstar>1.6)pi1<- rdirichlet(10, c(5,4,3,2,1))pi2<- rdirichlet(10, c(1,3,5,3,1))pi3<- rdirichlet(10, c(1,2,3,4,5))Z1<-(C==1)*t(rmultinom(n,1,pi1[1,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[1,]))%*%c(1:5)+(C==3)*t(rmultinom(n,1,pi3[1,]))%*%c(1:5)Z2<-(C==1)*t(rmultinom(n,1,pi1[2,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[2,]))%*%c(1:5)+(C==3)*t(rmultinom(n,1,pi3[2,]))%*%c(1:5)Z3<-(C==1)*t(rmultinom(n,1,pi1[3,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[3,]))%*%c(1:5)+(C==3)*t(rmultinom(n,1,pi3[3,]))%*%c(1:5)Z4<-(C==1)*t(rmultinom(n,1,pi1[4,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[4,]))%*%c(1:5)+(C==3)*t(rmultinom(n,1,pi3[4,]))%*%c(1:5)Z5<-(C==1)*t(rmultinom(n,1,pi1[5,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[5,]))%*%c(1:5)+(C==3)*t(rmultinom(n,1,pi3[5,]))%*%c(1:5)Z6<-(C==1)*t(rmultinom(n,1,pi1[6,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[6,]))%*%c(1:5)+(C==3)*t(rmultinom(n,1,pi3[6,]))%*%c(1:5)Z7<-(C==1)*t(rmultinom(n,1,pi1[7,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[7,]))%*%c(1:5)+(C==3)*t(rmultinom(n,1,pi3[7,]))%*%c(1:5)Z8<-(C==1)*t(rmultinom(n,1,pi1[8,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[8,]))%*%c(1:5)+(C==3)*t(rmultinom(n,1,pi3[8,]))%*%c(1:5)Z9<-(C==1)*t(rmultinom(n,1,pi1[9,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[9,]))%*%c(1:5)+(C==3)*t(rmultinom(n,1,pi3[9,]))%*%c(1:5)Z10<-(C==1)*t(rmultinom(n,1,pi1[10,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[10,]))%*%c(1:5)+(C==3)*t(rmultinom(n,1,pi3[10,]))%*%c(1:5)Z<- cbind(Z1,Z2,Z3,Z4,Z5,Z6,Z7,Z8,Z9,Z10)Y<- rbinom(n,1, exp(-1-.1*X1+X2+2*(C==1)+1*(C==2))/ (1+ exp(1-.1*X1+X2+2*(C==1)+1*(C==2))))mydata=data.frame(Y,X1,X2,Z1,Z2,Z3,Z4,Z5,Z6,Z7,Z8,Z9,Z10)inits=list(list(theta= c(0.33,0.33,0.34),beta= rep(0,length=3),alpha= rep(0,length=2),true= rep(1,length= nrow(mydata))))fit= lcra(formula=Y~X1+X2,family="binomial",data=mydata,nclasses=3,inits=inits,manifest= paste0("Z",1:10),n.chains=1,n.iter=1000)summary(fit)plot(fit)# with continuous responsen<-500X1<- runif(n,2,8)X2<- rbinom(n,1,.5)Cstar<- rnorm(n,.25*X1-.75*X2,1)C<-1* (Cstar<=.8)+2*((Cstar>.8)& (Cstar<=1.6))+3*(Cstar>1.6)pi1<- rdirichlet(10, c(5,4,3,2,1))pi2<- rdirichlet(10, c(1,3,5,3,1))pi3<- rdirichlet(10, c(1,2,3,4,5))pi4<- rdirichlet(10, c(1,1,1,1,1))Z1<-(C==1)*t(rmultinom(n,1,pi1[1,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[1,]))%*%c(1:5)+(C==3)* t(rmultinom(n,1,pi3[1,]))%*%c(1:5)+(C==4)*t(rmultinom(n,1,pi4[1,]))%*%c(1:5)Z2<-(C==1)*t(rmultinom(n,1,pi1[2,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[2,]))%*%c(1:5)+(C==3)* t(rmultinom(n,1,pi3[2,]))%*%c(1:5)+(C==4)*t(rmultinom(n,1,pi4[2,]))%*%c(1:5)Z3<-(C==1)*t(rmultinom(n,1,pi1[3,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[3,]))%*%c(1:5)+(C==3)* t(rmultinom(n,1,pi3[3,]))%*%c(1:5)+(C==4)*t(rmultinom(n,1,pi4[3,]))%*%c(1:5)Z4<-(C==1)*t(rmultinom(n,1,pi1[4,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[4,]))%*%c(1:5)+(C==3)* t(rmultinom(n,1,pi3[4,]))%*%c(1:5)+(C==4)*t(rmultinom(n,1,pi4[4,]))%*%c(1:5)Z5<-(C==1)*t(rmultinom(n,1,pi1[5,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[5,]))%*%c(1:5)+(C==3)* t(rmultinom(n,1,pi3[5,]))%*%c(1:5)+(C==4)*t(rmultinom(n,1,pi4[5,]))%*%c(1:5)Z6<-(C==1)*t(rmultinom(n,1,pi1[6,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[6,]))%*%c(1:5)+(C==3)* t(rmultinom(n,1,pi3[6,]))%*%c(1:5)+(C==4)*t(rmultinom(n,1,pi4[6,]))%*%c(1:5)Z7<-(C==1)*t(rmultinom(n,1,pi1[7,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[7,]))%*%c(1:5)+(C==3)* t(rmultinom(n,1,pi3[7,]))%*%c(1:5)+(C==4)*t(rmultinom(n,1,pi4[7,]))%*%c(1:5)Z8<-(C==1)*t(rmultinom(n,1,pi1[8,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[8,]))%*%c(1:5)+(C==3)* t(rmultinom(n,1,pi3[8,]))%*%c(1:5)+(C==4)*t(rmultinom(n,1,pi4[8,]))%*%c(1:5)Z9<-(C==1)*t(rmultinom(n,1,pi1[9,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[9,]))%*%c(1:5)+(C==3)* t(rmultinom(n,1,pi3[9,]))%*%c(1:5)+(C==4)*t(rmultinom(n,1,pi4[9,]))%*%c(1:5)Z10<-(C==1)*t(rmultinom(n,1,pi1[10,]))%*%c(1:5)+(C==2)* t(rmultinom(n,1,pi2[10,]))%*%c(1:5)+(C==3)* t(rmultinom(n,1,pi3[10,]))%*%c(1:5)+(C==4)*t(rmultinom(n,1,pi4[10,]))%*%c(1:5)Z<- cbind(Z1,Z2,Z3,Z4,Z5,Z6,Z7,Z8,Z9,Z10)Y<- rnorm(n,10-.5*X1+2*X2+2*(C==1)+1*(C==2),1)mydata=data.frame(Y,X1,X2,Z1,Z2,Z3,Z4,Z5,Z6,Z7,Z8,Z9,Z10)inits=list(list(theta= c(0.33,0.33,0.34),beta= rep(0,length=3),alpha= rep(0,length=2),true= rep(1,length= nrow(mydata)),tau=0.5))fit= lcra(formula=Y~X1+X2,family="gaussian",data=mydata,nclasses=3,inits=inits,manifest= paste0("Z",1:10),n.chains=1,n.iter=1000)summary(fit)plot(fit)
“Methods to account for uncertainty in latent class assignments whenusing latent classes as predictors in regression models, withapplication to acculturation strategy measures” (2020) In press atEpidemiology.
About
Joint latent class and regression analysis using MCMC/ Michael Elliot
Resources
Uh oh!
There was an error while loading.Please reload this page.