Uh oh!
There was an error while loading.Please reload this page.
- Notifications
You must be signed in to change notification settings - Fork87
bayesplot R package for plotting Bayesian models
License
stan-dev/bayesplot
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation
bayesplot is an R package providing an extensive library of plottingfunctions for use after fitting Bayesian models (typically with MCMC).The plots created bybayesplot are ggplot objects, which means that aftera plot is created it can be further customized using various functions fromtheggplot2 package.
Currentlybayesplot offers a variety of plots of posterior draws,visual MCMC diagnostics, graphical posterior (or prior) predictive checking,and general plots of posterior (or prior) predictive distributions.
The idea behindbayesplot is not only to provide convenient functionalityfor users, but also a common set of functions that can be easily used bydevelopers working on a variety of packages for Bayesian modeling, particularly(but not necessarily) those powered byRStan.
If you are just getting started withbayesplot we recommend starting withthe tutorialvignettes,the examples throughout the packagedocumentation,and the paperVisualization in Bayesian workflow:
- Gabry J, Simpson D, Vehtari A, Betancourt M, Gelman A (2019). Visualization in Bayesian workflow.J. R. Stat. Soc. A, 182: 389-402. doi:10.1111/rssa.12378.(journal version,arXiv preprint,code on GitHub)
- mc-stan.org/bayesplot (online documentation, vignettes)
- Ask a question (Stan Forums on Discourse)
- Open an issue (GitHub issues for bug reports, feature requests)
We are always looking for new contributors! SeeCONTRIBUTING.md for details and/or reach out via the issue tracker.
- Install from CRAN:
install.packages("bayesplot")- Install latest development version from GitHub (requiresdevtools package):
if (!require("devtools")) { install.packages("devtools")}devtools::install_github("stan-dev/bayesplot",dependencies=TRUE,build_vignettes=FALSE)
This installation won't include the vignettes (they take some time to build), but all of the vignettes areavailable online atmc-stan.org/bayesplot/articles.
Some quick examples using MCMC draws obtained from therstanarmandrstan packages.
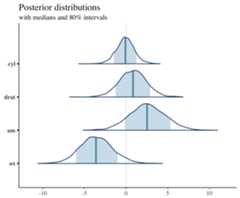
library("bayesplot")library("rstanarm")library("ggplot2")fit<- stan_glm(mpg~.,data=mtcars)posterior<- as.matrix(fit)plot_title<- ggtitle("Posterior distributions","with medians and 80% intervals")mcmc_areas(posterior,pars= c("cyl","drat","am","wt"),prob=0.8)+plot_title
color_scheme_set("red")ppc_dens_overlay(y=fit$y,yrep= posterior_predict(fit,draws=50))
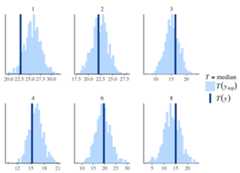
# also works nicely with pipinglibrary("dplyr")color_scheme_set("brightblue")fit %>% posterior_predict(draws=500) %>% ppc_stat_grouped(y=mtcars$mpg,group=mtcars$carb,stat="median")
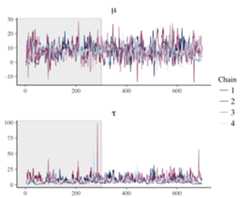
# with rstan demo modellibrary("rstan")fit2<- stan_demo("eight_schools",warmup=300,iter=700)posterior2<- extract(fit2,inc_warmup=TRUE,permuted=FALSE)color_scheme_set("mix-blue-pink")p<- mcmc_trace(posterior2,pars= c("mu","tau"),n_warmup=300,facet_args=list(nrow=2,labeller=label_parsed))p+ facet_text(size=15)
# scatter plot also showing divergencescolor_scheme_set("darkgray")mcmc_scatter( as.matrix(fit2),pars= c("tau","theta[1]"),np= nuts_params(fit2),np_style= scatter_style_np(div_color="green",div_alpha=0.8))
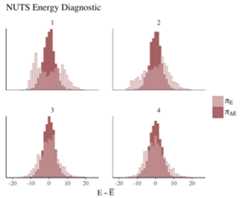
color_scheme_set("red")np<- nuts_params(fit2)mcmc_nuts_energy(np)+ ggtitle("NUTS Energy Diagnostic")
# another example with rstanarmcolor_scheme_set("purple")fit<- stan_glmer(mpg~wt+ (1|cyl),data=mtcars)ppc_intervals(y=mtcars$mpg,yrep= posterior_predict(fit),x=mtcars$wt,prob=0.5)+ labs(x="Weight (1000 lbs)",y="MPG",title="50% posterior predictive intervals\nvs observed miles per gallon",subtitle="by vehicle weight" )+ panel_bg(fill="gray95",color=NA)+ grid_lines(color="white")
About
bayesplot R package for plotting Bayesian models
Topics
Resources
License
Contributing
Uh oh!
There was an error while loading.Please reload this page.
Stars
Watchers
Forks
Sponsor this project
Uh oh!
There was an error while loading.Please reload this page.
Uh oh!
There was an error while loading.Please reload this page.