- Notifications
You must be signed in to change notification settings - Fork40
An R Port of Stata's 'margins' Command
License
Unknown, MIT licenses found
Licenses found
leeper/margins
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation
| title | output |
|---|---|
Marginal Effects for Model Objects | github_document |
Themargins andprediction packages are a combined effort to port the functionality of Stata's (closed source)margins command to (open source) R. These tools provide ways of obtaining common quantities of interest from regression-type models.margins provides "marginal effects" summaries of models andprediction provides unit-specific and sample average predictions from models. Marginal effects are partial derivatives of the regression equation with respect to each variable in the model for each unit in the data; average marginal effects are simply the mean of these unit-specific partial derivatives over some sample. In ordinary least squares regression with no interactions or higher-order term, the estimated slope coefficients are marginal effects. In other cases and for generalized linear models, the coefficients are not marginal effects at least not on the scale of the response variable.margins therefore provides ways of calculating the marginal effects of variables to make these models more interpretable.
The major functionality of Stata'smargins command - namely the estimation of marginal (or partial) effects - is provided here through a single function,margins(). This is an S3 generic method for calculating the marginal effects of covariates included in model objects (like those of classes "lm" and "glm"). Users interested in generating predicted (fitted) values, such as the "predictive margins" generated by Stata'smargins command, should consider usingprediction() from the sibling project,prediction.
With the introduction of Stata'smargins command, it has become incredibly simple to estimate average marginal effects (i.e., "average partial effects") and marginal effects at representative cases. Indeed, in just a few lines of Stata code, regression results for almost any kind model can be transformed into meaningful quantities of interest and related plots:
. import delimited mtcars.csv. quietly reg mpg c.cyl##c.hp wt. margins, dydx(*)------------------------------------------------------------------------------ | Delta-method | dy/dx Std. Err. t P>|t| [95% Conf. Interval]-------------+---------------------------------------------------------------- cyl | .0381376 .5998897 0.06 0.950 -1.192735 1.26901 hp | -.0463187 .014516 -3.19 0.004 -.076103 -.0165343 wt | -3.119815 .661322 -4.72 0.000 -4.476736 -1.762894------------------------------------------------------------------------------. marginsplotStata'smargins command is incredibly robust. It works with nearly any kind of statistical model and estimation procedure, including OLS, generalized linear models, panel regression models, and so forth. It also represents a significant improvement over Stata's previous marginal effects command -mfx - which was subject to various well-known bugs. While other Stata modules have provided functionality for deriving quantities of interest from regression estimates (e.g.,Clarify), none has done so with the simplicity and genearlity ofmargins.
By comparison, R has no robust functionality in the base tools for drawing out marginal effects from model estimates (though the S3predict() methods implement some of the functionality for computing fitted/predicted values). The closest approximation ismodmarg, which does one-variable-at-a-time estimation of marginal effects is quite robust. Other than this relatively new package on the scene, no packages implement appropriate marginal effect estimates. Notably, several packages provide estimates of marginal effects for different types of models. Among these arecar,alr3,mfx,erer, among others. Unfortunately, none of these packages implement marginal effects correctly (i.e., correctly account for interrelated variables such as interaction terms (e.g.,a:b) or power terms (e.g.,I(a^2)) and the packages all implement quite different interfaces for different types of models.interflex,interplot, andplotMElm provide functionality simply for plotting quantities of interest from multiplicative interaction terms in models but do not appear to support general marginal effects displays (in either tabular or graphical form), whilevisreg provides a more general plotting function but no tabular output.interactionTest provides some additional useful functionality for controlling the false discovery rate when making such plots and interpretations, but is again not a general tool for marginal effect estimation.
Given the challenges of interpreting the contribution of a given regressor in any model that includes quadratic terms, multiplicative interactions, a non-linear transformation, or other complexities, there is a clear need for a simple, consistent way to estimate marginal effects for popular statistical models. This package aims to correctly calculate marginal effects that include complex terms and provide a uniform interface for doing those calculations. Thus, the package implements a single S3 generic method (margins()) that can be easily generalized for any type of model implemented in R.
Some technical details of the package are worth briefly noting. The estimation of marginal effects relies on numerical approximations of derivatives produced usingpredict() (actually, a wrapper aroundpredict() calledprediction() that is type-safe). Variance estimation, by default is provided using the delta method a numerical approximation ofthe Jacobian matrix. While symbolic differentiation of some models (e.g., basic linear models) is possible usingD() andderiv(), R's modelling language (the "formula" class) is sufficiently general to enable the construction of model formulae that contain terms that fall outside of R's symbolic differentiation rule table (e.g.,y ~ factor(x) ory ~ I(FUN(x)) for any arbitraryFUN()). By relying on numeric differentiation,margins() supportsany model that can be expressed in R formula syntax. Even Stata'smargins command is limited in its ability to handle variable transformations (e.g., includingx andlog(x) as predictors) and quadratic terms (e.g.,x^3); these scenarios are easily expressed in an R formula and easily handled, correctly, bymargins().
Replicating Stata's results is incredibly simple using just themargins() method to obtain average marginal effects:
library("margins")mod1<- lm(mpg~cyl*hp+wt,data=mtcars)(marg1<- margins(mod1))
## Average marginal effects## lm(formula = mpg ~ cyl * hp + wt, data = mtcars)## cyl hp wt## 0.03814 -0.04632 -3.12summary(marg1)## factor AME SE z p lower upper## cyl 0.0381 0.5999 0.0636 0.9493 -1.1376 1.2139## hp -0.0463 0.0145 -3.1909 0.0014 -0.0748 -0.0179## wt -3.1198 0.6613 -4.7176 0.0000 -4.4160 -1.8236With the exception of differences in rounding, the above results match identically what Stata'smargins command produces. A slightly more concise expression relies on the syntactic sugar provided bymargins_summary():
margins_summary(mod1)## Error in margins_summary(mod1): could not find function "margins_summary"If you are only interested in obtaining the marginal effects (without corresponding variances or the overhead of creating a "margins" object), you can callmarginal_effects(x) directly. Furthermore, thedydx() function enables the calculation of the marginal effect of a single named variable:
# all marginal effects, as a data.framehead(marginal_effects(mod1))
## dydx_cyl dydx_hp dydx_wt## 1 -0.6572244 -0.04987248 -3.119815## 2 -0.6572244 -0.04987248 -3.119815## 3 -0.9794364 -0.08777977 -3.119815## 4 -0.6572244 -0.04987248 -3.119815## 5 0.5747624 -0.01196519 -3.119815## 6 -0.7519926 -0.04987248 -3.119815# subset of all marginal effects, as a data.framehead(marginal_effects(mod1,variables= c("cyl","hp")))
## dydx_cyl dydx_hp## 1 -0.6572244 -0.04987248## 2 -0.6572244 -0.04987248## 3 -0.9794364 -0.08777977## 4 -0.6572244 -0.04987248## 5 0.5747624 -0.01196519## 6 -0.7519926 -0.04987248# marginal effect of one variablehead(dydx(mtcars,mod1,"cyl"))
## dydx_cyl## 1 -0.6572244## 2 -0.6572244## 3 -0.9794364## 4 -0.6572244## 5 0.5747624## 6 -0.7519926These functions may be useful for plotting, getting a quick impression of the results, or for using unit-specific marginal effects in further analyses.
The package also implement's one of the best features ofmargins, which is theat specification that allows for the estimation of average marginal effects for counterfactual datasets in which particular variables are held at fixed values:
# webuse margexlibrary("webuse")webuse::webuse("margex")# logistic outcome treatment##group age c.age#c.age treatment#c.agemod2<- glm(outcome~treatment*group+age+ I(age^2)*treatment,data=margex,family=binomial)# margins, dydx(*)summary(margins(mod2))
## factor AME SE z p lower upper## age 0.0096 0.0008 12.3763 0.0000 0.0081 0.0112## group -0.0479 0.0129 -3.7044 0.0002 -0.0733 -0.0226## treatment 0.0432 0.0147 2.9320 0.0034 0.0143 0.0720# margins, dydx(treatment) at(age=(20(10)60))summary(margins(mod2,at=list(age= c(20,30,40,50,60)),variables="treatment"))
## factor age AME SE z p lower upper## treatment 20.0000 -0.0009 0.0043 -0.2061 0.8367 -0.0093 0.0075## treatment 30.0000 0.0034 0.0107 0.3199 0.7490 -0.0176 0.0245## treatment 40.0000 0.0301 0.0170 1.7736 0.0761 -0.0032 0.0634## treatment 50.0000 0.0990 0.0217 4.5666 0.0000 0.0565 0.1415## treatment 60.0000 0.1896 0.0384 4.9341 0.0000 0.1143 0.2649This functionality removes the need to modify data before performing such calculations, which can be quite unwieldy when many specifications are desired.
If one desiressubgroup effects, simply pass a subset of data to thedata argument:
# effects for mensummary(margins(mod2,data= subset(margex,sex==0)))
## factor AME SE z p lower upper## age 0.0043 0.0007 5.7723 0.0000 0.0028 0.0057## group -0.0753 0.0105 -7.1745 0.0000 -0.0959 -0.0547## treatment 0.0381 0.0070 5.4618 0.0000 0.0244 0.0517# effects for wommensummary(margins(mod2,data= subset(margex,sex==1)))
## factor AME SE z p lower upper## age 0.0150 0.0013 11.5578 0.0000 0.0125 0.0176## group -0.0206 0.0236 -0.8742 0.3820 -0.0669 0.0256## treatment 0.0482 0.0231 2.0909 0.0365 0.0030 0.0934The package implements several useful additional features for summarizing model objects, including:
- A
plot()method for the new "margins" class that ports Stata'smarginsplotcommand. - A plotting function
cplot()to provide the commonly needed visual summaries of predictions or average marginal effects conditional on a covariate. - A
persp()method for "lm", "glm", and "loess" objects to provide three-dimensional representations of response surfaces or marginal effects over two covariates. - An
image()method for the same that produces flat, two-dimensional heatmap-style representations ofpersp()-type plots.
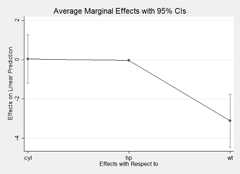
Using theplot() method yields an aesthetically similar result to Stata'smarginsplot:
library("webuse")webuse::webuse("nhanes2")mod3<- glm(highbp~sex*agegrp*bmi,data=nhanes2,family=binomial)summary(marg3<- margins(mod3))
## factor AME SE z p lower upper## agegrp 0.0846 0.0021 39.4392 0.0000 0.0804 0.0888## bmi 0.0261 0.0009 28.4995 0.0000 0.0243 0.0279## sex -0.0911 0.0085 -10.7063 0.0000 -0.1077 -0.0744plot(marg3)In addition to the estimation procedures andplot() generic,margins offers several plotting methods for model objects. First, there is a new genericcplot() that displays predictions or marginal effects (from an "lm" or "glm" model) of a variable conditional across values of third variable (or itself). For example, here is a graph of predicted probabilities from a logit model:
mod4<- glm(am~wt*drat,data=mtcars,family=binomial)cplot(mod4,x="wt",se.type="shade")
## xvals yvals upper lower## 1 1.513000 0.927274748 1.25767803 0.59687146## 2 1.675958 0.896156250 1.31282164 0.47949086## 3 1.838917 0.853821492 1.36083558 0.34680740## 4 2.001875 0.798115859 1.38729030 0.20894142## 5 2.164833 0.727945940 1.37431347 0.08157841## 6 2.327792 0.644257693 1.30643930 -0.01792391## 7 2.490750 0.550714595 1.17940279 -0.07797360## 8 2.653708 0.453441410 1.00638808 -0.09950526## 9 2.816667 0.359598025 0.81514131 -0.09594526## 10 2.979625 0.275390447 0.63577343 -0.08499254## 11 3.142583 0.204601856 0.48756886 -0.07836515## 12 3.305542 0.148285654 0.37415646 -0.07758515## 13 3.468500 0.105415989 0.28892829 -0.07809631## 14 3.631458 0.073865178 0.22356331 -0.07583296## 15 3.794417 0.051216829 0.17224934 -0.06981569## 16 3.957375 0.035248556 0.13162443 -0.06112732## 17 4.120333 0.024132208 0.09961556 -0.05135115## 18 4.283292 0.016461806 0.07467832 -0.04175471## 19 4.446250 0.011201450 0.05550126 -0.03309836## 20 4.609208 0.007609032 0.04093572 -0.02571766And fitted values with a factor independent variable:
cplot(lm(Sepal.Length~Species,data=iris))
## xvals yvals upper lower## 1 setosa 5.006 5.14869 4.86331## 2 versicolor 5.936 6.07869 5.79331## 3 virginica 6.588 6.73069 6.44531and a graph of the effect ofdrat across levels ofwt:
cplot(mod4,x="wt",dx="drat",what="effect",se.type="shade")
cplot() also returns a data frame of values, so that it can be used just for calculating quantities of interest before plotting them with another graphics package, such asggplot2:
library("ggplot2")dat<- cplot(mod4,x="wt",dx="drat",what="effect",draw=FALSE)head(dat)
## xvals yvals upper lower factor## 1.5130 0.3250 1.3927 -0.7427 drat## 1.6760 0.3262 1.1318 -0.4795 drat## 1.8389 0.3384 0.9214 -0.2447 drat## 2.0019 0.3623 0.7777 -0.0531 drat## 2.1648 0.3978 0.7110 0.0846 drat## 2.3278 0.4432 0.7074 0.1789 dratggplot(dat, aes(x=xvals))+ geom_ribbon(aes(ymin=lower,ymax=upper),fill="gray70")+ geom_line(aes(y=yvals))+ xlab("Vehicle Weight (1000s of lbs)")+ ylab("Average Marginal Effect of Rear Axle Ratio")+ ggtitle("Predicting Automatic/Manual Transmission from Vehicle Characteristics")+ theme_bw()
Second, the package implements methods for "lm" and "glm" class objects for thepersp() generic plotting function. This enables three-dimensional representations of predicted outcomes:
persp(mod1,xvar="cyl",yvar="hp")
and marginal effects:
persp(mod1,xvar="cyl",yvar="hp",what="effect",nx=10)
And if three-dimensional plots aren't your thing, there are also analogous methods for theimage() generic, to produce heatmap-style representations:
image(mod1,xvar="cyl",yvar="hp",main="Predicted Fuel Efficiency,\nby Cylinders and Horsepower")
The numerous package vignettes and help files contain extensive documentation and examples of all package functionality.
While there is still work to be done to improve performance,margins is reasonably speedy:
library("microbenchmark")microbenchmark(marginal_effects(mod1))
## Unit: milliseconds## expr min lq mean median uq max neval## marginal_effects(mod1) 5.708847 9.310043 11.38549 10.6044 13.46623 20.93396 100microbenchmark(margins(mod1))## Unit: milliseconds## expr min lq mean median uq max neval## margins(mod1) 35.37752 58.19732 70.77973 66.56501 82.34321 118.8913 100The most computationally expensive part ofmargins() is variance estimation. If you don't need variances, usemarginal_effects() directly or specifymargins(..., vce = "none").
The development version of this package can be installed directly from GitHub usingremotes:
if (!require("remotes")) { install.packages("remotes") library("remotes")}install_github("leeper/prediction")install_github("leeper/margins")# building vignettes takes a moment, so for a quicker install set:install_github("leeper/margins",build_vignettes=FALSE)
About
An R Port of Stata's 'margins' Command
Topics
Resources
License
Unknown, MIT licenses found
Licenses found
Uh oh!
There was an error while loading.Please reload this page.
Stars
Watchers
Forks
Packages0
Uh oh!
There was an error while loading.Please reload this page.
Contributors8
Uh oh!
There was an error while loading.Please reload this page.