- Notifications
You must be signed in to change notification settings - Fork34
Column scatter / beeswarm-style plots in ggplot2
License
eclarke/ggbeeswarm
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation
Beeswarm plots (aka column scatter plots or violin scatter plots) are away of plotting points that would ordinarily overlap so that they fallnext to each other instead. In addition to reducing overplotting, ithelps visualize the density of the data at each point (similar to aviolin plot), while still showing each data point individually.
ggbeeswarm provides two different methods to create beeswarm-styleplots usingggplot2. It does this by adding twonew ggplot geom objects:
geom_quasirandom: Uses avan der Corputsequence orTukey texturing (Tukey and Tukey “Strips displaying empiricaldistributions: I. textured dot strips”) to space the dots to avoidoverplotting. This usessherrillmix/vipor.geom_beeswarm: Uses thebeeswarmlibrary to do point-size based offset.
Features:
- Can handle categorical variables on the y-axis (thanks @smsaladi,@koncina)
- Automatically dodges if a grouping variable is categorical and
dodge.widthis specified (thanks @josesho)
See the examples below.
This package is on CRAN so install should be a simple:
install.packages('ggbeeswarm')If you want the development version from GitHub, you can do:
devtools::install_github("eclarke/ggbeeswarm")
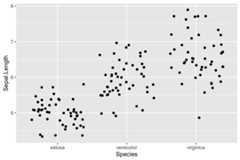
Here is a comparison betweengeom_jitter andgeom_quasirandom on theiris dataset:
set.seed(12345)library(ggplot2)library(ggbeeswarm)#compare to jitterggplot(iris,aes(Species,Sepal.Length))+ geom_jitter()
ggplot(iris,aes(Species,Sepal.Length))+ geom_quasirandom()
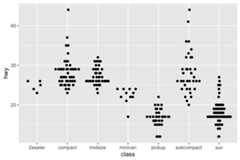
Usinggeom_quasirandom:
#default geom_quasirandomggplot(mpg,aes(class,hwy))+ geom_quasirandom()
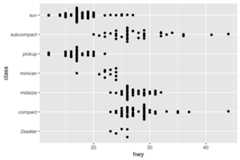
# With categorical y-axisggplot(mpg,aes(hwy,class))+ geom_quasirandom(groupOnX=FALSE)
# Some groups may have only a few points. Use `varwidth=TRUE` to adjust width dynamically.ggplot(mpg,aes(class,hwy))+ geom_quasirandom(varwidth=TRUE)
# Automatic dodgingsub_mpg<-mpg[mpg$class%in% c("midsize","pickup","suv"),]ggplot(sub_mpg, aes(class,displ,color=factor(cyl)))+ geom_quasirandom(dodge.width=1)
geom_quasirandom can also use several other methods to distributepoints. For example:
ggplot(iris, aes(Species,Sepal.Length))+ geom_quasirandom(method="tukey")+ ggtitle("Tukey texture")
ggplot(iris, aes(Species,Sepal.Length))+ geom_quasirandom(method="tukeyDense")+ ggtitle("Tukey + density")
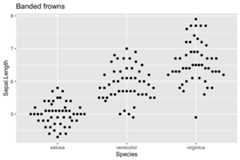
ggplot(iris, aes(Species,Sepal.Length))+ geom_quasirandom(method="frowney")+ ggtitle("Banded frowns")
ggplot(iris, aes(Species,Sepal.Length))+ geom_quasirandom(method="smiley")+ ggtitle("Banded smiles")
ggplot(iris, aes(Species,Sepal.Length))+ geom_quasirandom(method="pseudorandom")+ ggtitle("Jittered density")
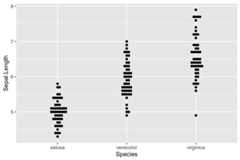
ggplot(iris, aes(Species,Sepal.Length))+ geom_beeswarm()+ ggtitle("Beeswarm")
Usinggeom_beeswarm:
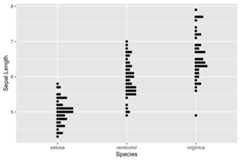
ggplot(iris,aes(Species,Sepal.Length))+ geom_beeswarm()
ggplot(iris,aes(Species,Sepal.Length))+ geom_beeswarm(side=1L)
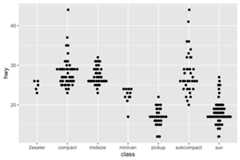
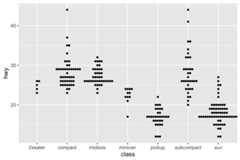
ggplot(mpg,aes(class,hwy))+ geom_beeswarm(size=.5)
# With categorical y-axisggplot(mpg,aes(hwy,class))+ geom_beeswarm(size=.5)
# Also watch out for points escaping from the plot with geom_beeswarmggplot(mpg,aes(hwy,class))+ geom_beeswarm(size=.5)+ scale_y_discrete(expand=expansion(add=c(0.5,1)))
ggplot(mpg,aes(class,hwy))+ geom_beeswarm(size=1.1)
# With automatic dodgingggplot(sub_mpg, aes(class,displ,color=factor(cyl)))+ geom_beeswarm(dodge.width=0.5)
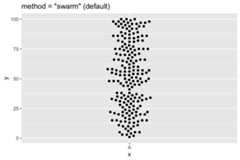
df<-data.frame(x="A",y= sample(1:100,200,replace=TRUE))ggplot(df, aes(x=x,y=y))+ geom_beeswarm(cex=2.5,method="swarm")+ ggtitle('method = "swarm" (default)')
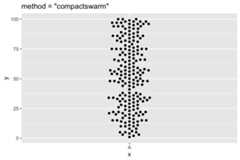
ggplot(df, aes(x=x,y=y))+ geom_beeswarm(cex=2.5,method="compactswarm")+ ggtitle('method = "compactswarm"')
ggplot(df, aes(x=x,y=y))+ geom_beeswarm(cex=2.5,method="hex")+ ggtitle('method = "hex"')
ggplot(df, aes(x=x,y=y))+ geom_beeswarm(cex=2.5,method="square")+ ggtitle('method = "square"')
ggplot(df, aes(x=x,y=y))+ geom_beeswarm(cex=2.5,method="center")+ ggtitle('method = "center"')
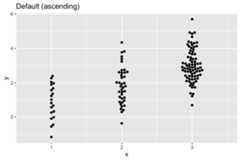
#With different beeswarm point distribution prioritydat<-data.frame(x=rep(1:3,c(20,40,80)))dat$y<-rnorm(nrow(dat),dat$x)ggplot(dat,aes(x,y))+ geom_beeswarm(cex=2)+ ggtitle('Default (ascending)')+ scale_x_continuous(expand=expansion(add=c(0.5,.5)))
ggplot(dat,aes(x,y))+ geom_beeswarm(cex=2,priority='descending')+ ggtitle('Descending')+ scale_x_continuous(expand=expansion(add=c(0.5,.5)))
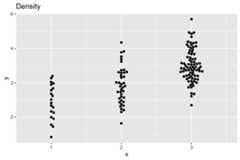
ggplot(dat,aes(x,y))+ geom_beeswarm(cex=2,priority='density')+ ggtitle('Density')+ scale_x_continuous(expand=expansion(add=c(0.5,.5)))
ggplot(dat,aes(x,y))+ geom_beeswarm(cex=2,priority='random')+ ggtitle('Random')+ scale_x_continuous(expand=expansion(add=c(0.5,.5)))
set.seed(1995)df2<-data.frame(y= rnorm(1000),id= sample(c("G1","G2","G3"),size=1000,replace=TRUE))p<- ggplot(df2, aes(x=id,y=y,colour=id))# use corral.width to control corral widthp+ geom_beeswarm(cex=2.5,corral="none",corral.width=0.9)+ ggtitle('corral = "none" (default)')
p+ geom_beeswarm(cex=2.5,corral="gutter",corral.width=0.9)+ ggtitle('corral = "gutter"')
p+ geom_beeswarm(cex=2.5,corral="wrap",corral.width=0.9)+ ggtitle('corral = "wrap"')
p+ geom_beeswarm(cex=2.5,corral="random",corral.width=0.9)+ ggtitle('corral = "random"')
p+ geom_beeswarm(cex=2.5,corral="omit",corral.width=0.9)+ ggtitle('corral = "omit"')
## Warning: Removed 303 rows containing missing values (geom_point).Authors: Erik Clarke, Scott Sherrill-Mix, and Charlotte Dawson
About
Column scatter / beeswarm-style plots in ggplot2
Resources
License
Uh oh!
There was an error while loading.Please reload this page.
Stars
Watchers
Forks
Packages0
Uh oh!
There was an error while loading.Please reload this page.
Contributors11
Uh oh!
There was an error while loading.Please reload this page.