- Notifications
You must be signed in to change notification settings - Fork1
Fast Change Point Detection in R
License
doccstat/fastcpd
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation
The fastcpd (fastchangepointdetection) is a fastimplmentation of change point detection methods in R/Python.
- R documentation:fastcpd.xingchi.li
- Python documentation:fastcpd.xingchi.li/python
# install.packages("devtools")devtools::install_github("doccstat/fastcpd")# or install from CRANinstall.packages("fastcpd")
# python -m ensurepip --upgradepip install.# or install from PyPIpip install fastcpd
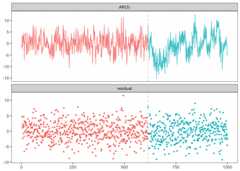
set.seed(1)n<-1000x<- rep(0,n+3)for (iin1:600) {x[i+3]<-0.6*x[i+2]-0.2*x[i+1]+0.1*x[i]+ rnorm(1,0,3)}for (iin601:1000) {x[i+3]<-0.3*x[i+2]+0.4*x[i+1]+0.2*x[i]+ rnorm(1,0,3)}result<-fastcpd::fastcpd.ar(x[3+ seq_len(n)],3,r.progress=FALSE)summary(result)#>#> Call:#> fastcpd::fastcpd.ar(data = x[3 + seq_len(n)], order = 3, r.progress = FALSE)#>#> Change points:#> 614#>#> Cost values:#> 2754.116 2038.945#>#> Parameters:#> segment 1 segment 2#> 1 0.57120256 0.2371809#> 2 -0.20985108 0.4031244#> 3 0.08221978 0.2290323plot(result)
importfastcpd.segmentationfromnumpyimportconcatenatefromnumpy.randomimportnormal,multivariate_normalcovariance_mat= [[100,0,0], [0,100,0], [0,0,100]]data=concatenate((multivariate_normal([0,0,0],covariance_mat,300),multivariate_normal([50,50,50],covariance_mat,400),multivariate_normal([2,2,2],covariance_mat,300)))fastcpd.segmentation.mean(data)importfastcpd.variance_estimationfastcpd.variance_estimation.mean(data)
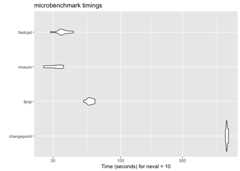
library(microbenchmark)set.seed(1)n<-5*10^6mean_data<- c(rnorm(n/2,0,1), rnorm(n/2,50,1))ggplot2::autoplot(microbenchmark(wbs=wbs::wbs(mean_data),not=not::not(mean_data,contrast="pcwsConstMean"),changepoint=changepoint::cpt.mean(mean_data,method="PELT"),jointseg=jointseg::jointSeg(mean_data,K=12),fpop=fpop::Fpop(mean_data,2* log(n)),mosum=mosum::mosum(c(mean_data),G=40),fastcpd=fastcpd::fastcpd.mean(mean_data,r.progress=FALSE,cp_only=TRUE,variance_estimation=1)))#> Warning in microbenchmark(wbs = wbs::wbs(mean_data), not = not::not(mean_data,#> : less accurate nanosecond times to avoid potential integer overflows
library(microbenchmark)set.seed(1)n<-10^8mean_data<- c(rnorm(n/2,0,1), rnorm(n/2,50,1))system.time(fastcpd::fastcpd.mean(mean_data,r.progress=FALSE,cp_only=TRUE,variance_estimation=1))#> user system elapsed#> 11.753 9.150 26.455system.time(mosum::mosum(c(mean_data),G=40))#> user system elapsed#> 5.518 11.516 38.368system.time(fpop::Fpop(mean_data,2* log(n)))#> user system elapsed#> 35.926 5.231 58.269system.time(changepoint::cpt.mean(mean_data,method="PELT"))#> user system elapsed#> 32.342 9.681 66.056ggplot2::autoplot(microbenchmark(changepoint=changepoint::cpt.mean(mean_data,method="PELT"),fpop=fpop::Fpop(mean_data,2* log(n)),mosum=mosum::mosum(c(mean_data),G=40),fastcpd=fastcpd::fastcpd.mean(mean_data,r.progress=FALSE,cp_only=TRUE,variance_estimation=1),times=10))
Some packages are not included in themicrobenchmark comparison due toeither memory constraints or long running time.
# Device: Mac mini (M1, 2020)# Memory: 8 GBsystem.time(CptNonPar::np.mojo(mean_data,G= floor(length(mean_data)/6)))#> Error: vector memory limit of 16.0 Gb reached, see mem.maxVSize()#> Timing stopped at: 0.061 0.026 0.092system.time(ecp::e.divisive(matrix(mean_data)))#> Error: vector memory limit of 16.0 Gb reached, see mem.maxVSize()#> Timing stopped at: 0.076 0.044 0.241system.time(strucchange::breakpoints(y~1,data=data.frame(y=mean_data)))#> Timing stopped at: 265.1 145.8 832.5system.time(breakfast::breakfast(mean_data))#> Timing stopped at: 45.9 89.21 562.3
- fastcpd: Fast Change Point Detection inR
- Sequential Gradient Descent and Quasi-Newton’s Method forChange-PointAnalysis
Should I install suggested packages?
The suggested packages are not required for the main functionality ofthe package. They are only required for the vignettes. If you want tolearn more about the package comparison and other vignettes, you couldeither check out vignettes onCRAN orpkgdown generateddocumentation.
I countered problems related to gfortran on Mac OSX or Linux!
The package should be able to install on Mac and any Linux distributionwithout any problems if all the dependencies are installed. However, ifyou encountered problems related to gfortran, it might be becauseRcppArmadillo is not installed previously. TryMac OSX stackoverflowsolution orLinux stackoversolution if you have troubleinstallingRcppArmadillo.
We welcome contributions from everyone. Please follow the instructionsbelow to make contributions.
Fork the repo.
Create a new branch from
mainbranch.Make changes and commit them.
- Please follow theGoogle’s R styleguide fornaming variables and functions.
- If you are adding a new family of models with new cost functionswith corresponding gradient and Hessian, please add them to
src/fastcpd_class_cost.ccwith proper example and tests invignettes/gallery.Rmdandtests/testthat/test-gallery.R. - Add the family name to
src/fastcpd_constants.h. - [Recommended] Add a new wrapper function in
R/fastcpd_wrappers.Rfor the new family of models and move theexamples to the new wrapper function as roxygen examples. - Add the new wrapper function to the corresponding section in
_pkgdown.yml.
Push the changes to your fork.
Create a pull request.
Make sure the pull request does not create new warnings or errors in
devtools::check().
Trouble installing Python package.
Python headers are required to install the Python package. If you areusing Ubuntu, you can install the headers with:
sudo apt install python3-dev
Encountered a bug or unintended behavior?
- File a ticket atGitHubIssues.
- Contact the authors specified inDESCRIPTION.
Special thanks toclODE.
About
Fast Change Point Detection in R
Topics
Resources
License
Uh oh!
There was an error while loading.Please reload this page.
Stars
Watchers
Forks
Uh oh!
There was an error while loading.Please reload this page.