Uh oh!
There was an error while loading.Please reload this page.
- Notifications
You must be signed in to change notification settings - Fork189
Building Convolutional Neural Networks From Scratch using NumPy
ahmedfgad/NumPyCNN
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation
NumPyCNN is a Python implementation for convolutional neural networks (CNNs) from scratch using NumPy.
IMPORTANTIf you are coming for the code of the tutorial titled Building Convolutional Neural Network using NumPy from Scratch, then it has been moved to theTutorialProject directory on 20 May 2020.
The project has a single module namedcnn.py which implements all classes and functions needed to build the CNN.
It is very important to note that the project only implements theforward pass of training CNNs and there isno learning algorithm used. Just the learning rate is used to make some changes to the weights after each epoch which is better than leaving the weights unchanged.
The project can be used for classification problems where only 1 class per sample is allowed.
The project will be extended totrain CNN using the genetic algorithm with the help of a library namedPyGAD. Check the library's documentation atRead The Docs:https://pygad.readthedocs.io
- Credit/Debit Card:https://donate.stripe.com/eVa5kO866elKgM0144
- Open Collective:opencollective.com/pygad
- PayPal: Use either this link:paypal.me/ahmedfgad or the e-mail addressahmed.f.gad@gmail.com
- Interac e-Transfer: Use e-mail addressahmed.f.gad@gmail.com
To installPyGAD, simply use pip to download and install the library fromPyPI (Python Package Index). The library is at PyPI at this pagehttps://pypi.org/project/pygad.
Install PyGAD with the following command:
pipinstallpygad
To get started with PyGAD, please read the documentation atRead The Docshttps://pygad.readthedocs.io.
The source code of the PyGAD' modules is found in the following GitHub projects:
- pygad: (https://github.com/ahmedfgad/GeneticAlgorithmPython)
- pygad.nn:https://github.com/ahmedfgad/NumPyANN
- pygad.gann:https://github.com/ahmedfgad/NeuralGenetic
- pygad.cnn:https://github.com/ahmedfgad/NumPyCNN
- pygad.gacnn:https://github.com/ahmedfgad/CNNGenetic
- pygad.kerasga:https://github.com/ahmedfgad/KerasGA
- pygad.torchga:https://github.com/ahmedfgad/TorchGA
The documentation of PyGAD is available atRead The Docshttps://pygad.readthedocs.io.
The documentation of the PyGAD library is available atRead The Docs at this link:https://pygad.readthedocs.io. It discusses the modules supported by PyGAD, all its classes, methods, attribute, and functions. For each module, a number of examples are given.
If there is an issue using PyGAD, feel free to post at issue in thisGitHub repositoryhttps://github.com/ahmedfgad/GeneticAlgorithmPython or by sending an e-mail toahmed.f.gad@gmail.com.
If you built a project that uses PyGAD, then please drop an e-mail toahmed.f.gad@gmail.com with the following information so that your project is included in the documentation.
- Project title
- Brief description
- Preferably, a link that directs the readers to your project
Please check theContact Us section for more contact details.
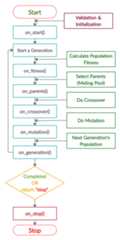
The next figure lists the different stages in the lifecycle of an instance of thepygad.GA class. Note that PyGAD stops when either all generations are completed or when the function passed to theon_generation parameter returns the stringstop.
The next code implements all the callback functions to trace the execution of the genetic algorithm. Each callback function prints its name.
importpygadimportnumpyfunction_inputs= [4,-2,3.5,5,-11,-4.7]desired_output=44deffitness_func(ga_instance,solution,solution_idx):output=numpy.sum(solution*function_inputs)fitness=1.0/ (numpy.abs(output-desired_output)+0.000001)returnfitnessfitness_function=fitness_funcdefon_start(ga_instance):print("on_start()")defon_fitness(ga_instance,population_fitness):print("on_fitness()")defon_parents(ga_instance,selected_parents):print("on_parents()")defon_crossover(ga_instance,offspring_crossover):print("on_crossover()")defon_mutation(ga_instance,offspring_mutation):print("on_mutation()")defon_generation(ga_instance):print("on_generation()")defon_stop(ga_instance,last_population_fitness):print("on_stop()")ga_instance=pygad.GA(num_generations=3,num_parents_mating=5,fitness_func=fitness_function,sol_per_pop=10,num_genes=len(function_inputs),on_start=on_start,on_fitness=on_fitness,on_parents=on_parents,on_crossover=on_crossover,on_mutation=on_mutation,on_generation=on_generation,on_stop=on_stop)ga_instance.run()
Based on the used 3 generations as assigned to thenum_generations argument, here is the output.
on_start()on_fitness()on_parents()on_crossover()on_mutation()on_generation()on_fitness()on_parents()on_crossover()on_mutation()on_generation()on_fitness()on_parents()on_crossover()on_mutation()on_generation()on_stop()Check thePyGAD's documentation for information about the implementation of this example.
importnumpyimportpygad.cnntrain_inputs=numpy.load("dataset_inputs.npy")train_outputs=numpy.load("dataset_outputs.npy")sample_shape=train_inputs.shape[1:]num_classes=4input_layer=pygad.cnn.Input2D(input_shape=sample_shape)conv_layer1=pygad.cnn.Conv2D(num_filters=2,kernel_size=3,previous_layer=input_layer,activation_function=None)relu_layer1=pygad.cnn.Sigmoid(previous_layer=conv_layer1)average_pooling_layer=pygad.cnn.AveragePooling2D(pool_size=2,previous_layer=relu_layer1,stride=2)conv_layer2=pygad.cnn.Conv2D(num_filters=3,kernel_size=3,previous_layer=average_pooling_layer,activation_function=None)relu_layer2=pygad.cnn.ReLU(previous_layer=conv_layer2)max_pooling_layer=pygad.cnn.MaxPooling2D(pool_size=2,previous_layer=relu_layer2,stride=2)conv_layer3=pygad.cnn.Conv2D(num_filters=1,kernel_size=3,previous_layer=max_pooling_layer,activation_function=None)relu_layer3=pygad.cnn.ReLU(previous_layer=conv_layer3)pooling_layer=pygad.cnn.AveragePooling2D(pool_size=2,previous_layer=relu_layer3,stride=2)flatten_layer=pygad.cnn.Flatten(previous_layer=pooling_layer)dense_layer1=pygad.cnn.Dense(num_neurons=100,previous_layer=flatten_layer,activation_function="relu")dense_layer2=pygad.cnn.Dense(num_neurons=num_classes,previous_layer=dense_layer1,activation_function="softmax")model=pygad.cnn.Model(last_layer=dense_layer2,epochs=1,learning_rate=0.01)model.summary()model.train(train_inputs=train_inputs,train_outputs=train_outputs)predictions=model.predict(data_inputs=train_inputs)print(predictions)num_wrong=numpy.where(predictions!=train_outputs)[0]num_correct=train_outputs.size-num_wrong.sizeaccuracy=100* (num_correct/train_outputs.size)print("Number of correct classifications : {num_correct}.".format(num_correct=num_correct))print("Number of wrong classifications : {num_wrong}.".format(num_wrong=num_wrong.size))print("Classification accuracy : {accuracy}.".format(accuracy=accuracy))
There are different resources that can be used to get started with the building CNN and its Python implementation.
To start with coding the genetic algorithm, you can check the tutorial titledGenetic Algorithm Implementation in Python available at these links:
This tutorial is prepared based on a previous version of the project but it still a good resource to start with coding the genetic algorithm.
Get started with the genetic algorithm by reading the tutorial titledIntroduction to Optimization with Genetic Algorithm which is available at these links:
Read about building neural networks in Python through the tutorial titledArtificial Neural Network Implementation using NumPy and Classification of the Fruits360 Image Dataset available at these links:
Read about training neural networks using the genetic algorithm through the tutorial titledArtificial Neural Networks Optimization using Genetic Algorithm with Python available at these links:
To start with coding the genetic algorithm, you can check the tutorial titledBuilding Convolutional Neural Network using NumPy from Scratch available at these links:
This tutorial) is prepared based on a previous version of the project but it still a good resource to start with coding CNNs.
Get started with the genetic algorithm by reading the tutorial titledDerivation of Convolutional Neural Network from Fully Connected Network Step-By-Step which is available at these links:
You can also check my book cited asAhmed Fawzy Gad 'Practical Computer Vision Applications Using Deep Learning with CNNs'. Dec. 2018, Apress, 978-1-4842-4167-7 which discusses neural networks, convolutional neural networks, deep learning, genetic algorithm, and more.
Find the book at these links:
If you used PyGAD, please consider adding a citation to the following paper about PyGAD:
@misc{gad2021pygad, title={PyGAD: An Intuitive Genetic Algorithm Python Library}, author={Ahmed Fawzy Gad}, year={2021}, eprint={2106.06158}, archivePrefix={arXiv}, primaryClass={cs.NE}}About
Building Convolutional Neural Networks From Scratch using NumPy
Topics
Resources
Uh oh!
There was an error while loading.Please reload this page.
Stars
Watchers
Forks
Releases
Sponsor this project
Uh oh!
There was an error while loading.Please reload this page.
Packages0
Uh oh!
There was an error while loading.Please reload this page.