- Notifications
You must be signed in to change notification settings - Fork4
A fast and flexible framework for data reduction in R
License
Unknown, MIT licenses found
Licenses found
USCbiostats/partition
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation
partition is a fast and flexible framework for agglomerativepartitioning. partition uses an approach called Direct-Measure-Reduce tocreate new variables that maintain the user-specified minimum level ofinformation. Each reduced variable is also interpretable: the originalvariables map to one and only one variable in the reduced data set.partition is flexible, as well: how variables are selected to reduce,how information loss is measured, and the way data is reduced can all becustomized.
You can install the partition from CRAN with:
install.packages("partition")Or you can install the development version of partition GitHub with:
# install.packages("remotes")remotes::install_github("USCbiostats/partition")
library(partition)set.seed(1234)df<- simulate_block_data(c(3,4,5),lower_corr=.4,upper_corr=.6,n=100)# don't accept reductions where information < .6prt<- partition(df,threshold=.6)prt#> Partitioner:#> Director: Minimum Distance (Pearson)#> Metric: Intraclass Correlation#> Reducer: Scaled Mean#>#> Reduced Variables:#> 1 reduced variables created from 2 observed variables#>#> Mappings:#> reduced_var_1 = {block2_x3, block2_x4}#>#> Minimum information:#> 0.602# return reduced datapartition_scores(prt)#> # A tibble: 100 × 11#> block1_x1 block1_x2 block1_x3 block2_x1 block2_x2 block3_x1 block3_x2#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>#> 1 -1.00 -0.344 1.35 -0.526 -1.25 1.13 0.357#> 2 0.518 -0.434 -0.361 -1.48 -1.53 -0.317 0.290#> 3 -1.77 -0.913 -0.722 0.122 0.224 -0.529 0.114#> 4 -1.49 -0.998 0.189 0.149 -0.994 -0.433 0.0120#> 5 0.616 0.0211 0.895 1.09 -1.25 0.440 -0.550#> 6 0.0765 0.522 1.20 -0.152 -0.419 -0.912 -0.362#> 7 1.74 0.0993 -0.654 -1.26 -0.502 -0.792 -1.03#> 8 1.05 2.19 0.913 0.254 0.328 -1.07 -0.976#> 9 -1.07 -0.292 -0.763 0.437 0.739 0.899 -0.342#> 10 -1.02 -0.959 -1.33 -1.57 -1.11 0.618 0.153#> # ℹ 90 more rows#> # ℹ 4 more variables: block3_x3 <dbl>, block3_x4 <dbl>, block3_x5 <dbl>,#> # reduced_var_1 <dbl># access mapping keysmapping_key(prt)#> # A tibble: 11 × 4#> variable mapping information indices#> <chr> <list> <dbl> <list>#> 1 block1_x1 <chr [1]> 1 <int [1]>#> 2 block1_x2 <chr [1]> 1 <int [1]>#> 3 block1_x3 <chr [1]> 1 <int [1]>#> 4 block2_x1 <chr [1]> 1 <int [1]>#> 5 block2_x2 <chr [1]> 1 <int [1]>#> 6 block3_x1 <chr [1]> 1 <int [1]>#> 7 block3_x2 <chr [1]> 1 <int [1]>#> 8 block3_x3 <chr [1]> 1 <int [1]>#> 9 block3_x4 <chr [1]> 1 <int [1]>#> 10 block3_x5 <chr [1]> 1 <int [1]>#> 11 reduced_var_1 <chr [2]> 0.602 <int [2]>unnest_mappings(prt)#> # A tibble: 12 × 4#> variable mapping information indices#> <chr> <chr> <dbl> <int>#> 1 block1_x1 block1_x1 1 1#> 2 block1_x2 block1_x2 1 2#> 3 block1_x3 block1_x3 1 3#> 4 block2_x1 block2_x1 1 4#> 5 block2_x2 block2_x2 1 5#> 6 block3_x1 block3_x1 1 8#> 7 block3_x2 block3_x2 1 9#> 8 block3_x3 block3_x3 1 10#> 9 block3_x4 block3_x4 1 11#> 10 block3_x5 block3_x5 1 12#> 11 reduced_var_1 block2_x3 0.602 6#> 12 reduced_var_1 block2_x4 0.602 7# use a lower threshold of information losspartition(df,threshold=.5,partitioner= part_kmeans())#> Partitioner:#> Director: <custom director>#> Metric: <custom metric>#> Reducer: <custom reducer>#>#> Reduced Variables:#> 2 reduced variables created from 7 observed variables#>#> Mappings:#> reduced_var_1 = {block3_x1, block3_x2, block3_x5}#> reduced_var_2 = {block2_x1, block2_x2, block2_x3, block2_x4}#>#> Minimum information:#> 0.508# use a custom partitionerpart_icc_rowmeans<- replace_partitioner(part_icc,reduce= as_reducer(rowMeans))partition(df,threshold=.6,partitioner=part_icc_rowmeans)#> Partitioner:#> Director: Minimum Distance (Pearson)#> Metric: Intraclass Correlation#> Reducer: <custom reducer>#>#> Reduced Variables:#> 1 reduced variables created from 2 observed variables#>#> Mappings:#> reduced_var_1 = {block2_x3, block2_x4}#>#> Minimum information:#> 0.602
partition also supports a number of ways to visualize partitions andpermutation tests; these functions all start withplot_*(). Thesefunctions all return ggplots and can thus be extended using ggplot2.
plot_stacked_area_clusters(df)+ggplot2::theme_minimal(14)
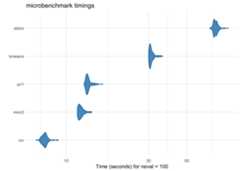
partition has been meticulously benchmarked and profiled to improveperformance, and key sections are written in C++ or use C++-basedpackages. Using a data frame with 1 million rows on a 2017 MacBook Prowith 16 GB RAM, here’s how each of the built-in partitioners perform:
large_df<- simulate_block_data(c(3,4,5),lower_corr=.4,upper_corr=.6,n=1e6)basic_benchmarks<-microbenchmark::microbenchmark(icc= partition(large_df,.3),kmeans= partition(large_df,.3,partitioner= part_kmeans()),minr2= partition(large_df,.3,partitioner= part_minr2()),pc1= partition(large_df,.3,partitioner= part_pc1()),stdmi= partition(large_df,.3,partitioner= part_stdmi()))
As the features (columns) in the data set become greater than the numberof observations (rows), the default ICC method scales more linearly thanK-Means-based methods. While K-Means is often faster at lowerdimensions, it becomes slower as the features outnumber theobservations. For example, using three data sets with increasing numbersof columns, K-Means starts as the fastest and gets increasingly slower,although in this case it is still comparable to ICC:
narrow_df<- simulate_block_data(3:5,lower_corr=.4,upper_corr=.6,n=100)wide_df<- simulate_block_data(rep(3:10,2),lower_corr=.4,upper_corr=.6,n=100)wider_df<- simulate_block_data(rep(3:20,4),lower_corr=.4,upper_corr=.6,n=100)icc_kmeans_benchmarks<-microbenchmark::microbenchmark(icc_narrow= partition(narrow_df,.3),icc_wide= partition(wide_df,.3),icc_wider= partition(wider_df,.3),kmeans_narrow= partition(narrow_df,.3,partitioner= part_kmeans()),kmeans_wide= partition(wide_df,.3,partitioner= part_kmeans()),kmeans_wider= partition(wider_df,.3,partitioner= part_kmeans()))
For more information, seeour paper inBioinformatics, whichdiscusses these issues in more depth (Millstein et al. 2020).
Please read theContributorGuidelinesprior to submitting a pull request to partition. Also note that thisproject is released with aContributor Code ofConduct.By participating in this project you agree to abide by its terms.
Millstein, Joshua, Francesca Battaglin, Malcolm Barrett, Shu Cao, WuZhang, Sebastian Stintzing, Volker Heinemann, and Heinz-Josef Lenz.2020. “Partition: A Surjective Mapping Approach for DimensionalityReduction.”Bioinformatics 36 (3): 676–81.https://doi.org/10.1093/bioinformatics/btz661.
About
A fast and flexible framework for data reduction in R
Topics
Resources
License
Unknown, MIT licenses found
Licenses found
Code of conduct
Contributing
Uh oh!
There was an error while loading.Please reload this page.
Stars
Watchers
Forks
Packages0
Uh oh!
There was an error while loading.Please reload this page.
Contributors4
Uh oh!
There was an error while loading.Please reload this page.