- Notifications
You must be signed in to change notification settings - Fork7
R package and web app for spatial visualisation of admixture on a projected map
License
Tom-Jenkins/mapmixture
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation
mapmixture is an R package and Shiny app that enables users tovisualise admixture as pie charts on a projected map. It also allowsusers to visualise admixture as traditional structure barplots or facetbarplots.
mapmixture requiresR (>= 4.2) to beinstalled on your system. Clickhere to download thelatest version of R for Windows.
Install the latest stable release from CRAN:
install.packages("mapmixture")Install the latest development version from GitHub:
# install.packages("devtools")devtools::install_github("Tom-Jenkins/mapmixture")
mapmixture()# main functionstructure_plot()# plot traditional structure or facet barplotscatter_plot()# plot PCA or DAPC resultslaunch_mapmixture()# launch mapmixture Shiny app
Jenkins TL (2024).mapmixture: an Rpackage and web app for spatial visualisation of admixture andpopulation structure.Molecular Ecology Resources, 24: e13943. DOI:10.1111/1755-0998.13943.
Code
# Load packagelibrary(mapmixture)# Read in admixture file format 1file<- system.file("extdata","admixture1.csv",package="mapmixture")admixture1<- read.csv(file)# Read in coordinates filefile<- system.file("extdata","coordinates.csv",package="mapmixture")coordinates<- read.csv(file)# Run mapmixturemap1<- mapmixture(admixture1,coordinates,crs=3035)# map1
Code
# Load packageslibrary(mapmixture)library(rnaturalearthhires)# Install rnaturalearthhires package using:# install.packages("rnaturalearthhires", repos = "https://ropensci.r-universe.dev", type = "source")# Read in admixture file format 1file<- system.file("extdata","admixture1.csv",package="mapmixture")admixture1<- read.csv(file)# Read in coordinates filefile<- system.file("extdata","coordinates.csv",package="mapmixture")coordinates<- read.csv(file)# Run mapmixturemap2<- mapmixture(admixture_df=admixture1,coords_df=coordinates,cluster_cols= c("#f1a340","#998ec3"),cluster_names= c("Group A","Group B"),crs=3035,basemap=rnaturalearthhires::countries10[, c("geometry")],boundary= c(xmin=-15,xmax=16,ymin=40,ymax=62),pie_size=1,pie_border=0.3,pie_border_col="white",pie_opacity=1,land_colour="#d9d9d9",sea_colour="#deebf7",expand=TRUE,arrow=TRUE,arrow_size=1.5,arrow_position="bl",scalebar=TRUE,scalebar_size=1.5,scalebar_position="tl",plot_title="Admixture Map",plot_title_size=12,axis_title_size=10,axis_text_size=8)# map2
Code
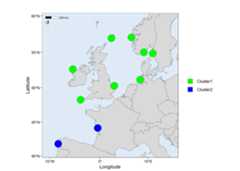
# Load packagelibrary(mapmixture)# Read in admixture file format 3file<- system.file("extdata","admixture3.csv",package="mapmixture")admixture3<- read.csv(file)# Read in coordinates filefile<- system.file("extdata","coordinates.csv",package="mapmixture")coordinates<- read.csv(file)# Run mapmixturemap3<- mapmixture(admixture3,coordinates,crs=3035)# map3
Code
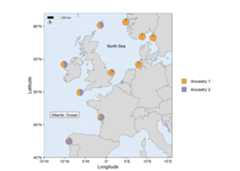
# Load packageslibrary(mapmixture)library(ggplot2)# Read in admixture file format 1file<- system.file("extdata","admixture1.csv",package="mapmixture")admixture1<- read.csv(file)# Read in coordinates filefile<- system.file("extdata","coordinates.csv",package="mapmixture")coordinates<- read.csv(file)# Run mapmixturemap4<- mapmixture(admixture_df=admixture1,coords_df=coordinates,cluster_cols= c("#f1a340","#998ec3"),cluster_names= c("Ancestry 1","Ancestry 2"),crs=4326,boundary= c(xmin=-15,xmax=16,ymin=40,ymax=62),pie_size=1,)+# Add additional label to the map annotate("label",x=-10,y=46.5,label="Atlantic Ocean",size=3, )+# Add additional text to the map annotate("text",x=2.5,y=57,label="North Sea",size=3, )+# Adjust ggplot theme options theme(axis.title= element_text(size=10),axis.text= element_text(size=8), )+# Adjust the size of the legend keys guides(fill= guide_legend(override.aes=list(size=5,alpha=1)))# map4
Code
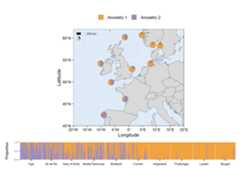
# Load packageslibrary(mapmixture)library(ggplot2)library(gridExtra)# Read in admixture file format 1file<- system.file("extdata","admixture1.csv",package="mapmixture")admixture1<- read.csv(file)# Read in coordinates filefile<- system.file("extdata","coordinates.csv",package="mapmixture")coordinates<- read.csv(file)# Run mapmixturemap5<- mapmixture(admixture_df=admixture1,coords_df=coordinates,cluster_cols= c("#f1a340","#998ec3"),cluster_names= c("Ancestry 1","Ancestry 2"),crs=4326,boundary= c(xmin=-20,xmax=20,ymin=40,ymax=62),pie_size=1.3,)+# Adjust theme options theme(legend.position="top",plot.margin= margin(l=10,r=10), )+# Adjust the size of the legend keys guides(fill= guide_legend(override.aes=list(size=5,alpha=1)))# Traditional structure barplotstructure_barplot<- structure_plot(admixture_df=admixture1,type="structure",cluster_cols= c("#f1a340","#998ec3"),site_dividers=TRUE,divider_width=0.4,site_order= c("Vigo","Ile de Re","Isles of Scilly","Mullet Peninsula","Shetland","Cromer","Helgoland","Flodevigen","Lysekil","Bergen" ),labels="site",flip_axis=FALSE,site_ticks_size=-0.05,site_labels_y=-0.35,site_labels_size=2.2)+# Adjust theme options theme(axis.title.y= element_text(size=8,hjust=1),axis.text.y= element_text(size=5), )# Arrange plots# grid.arrange(map5, structure_barplot, nrow = 2, heights = c(4,1))
Code
# Load packageslibrary(mapmixture)library(ggplot2)library(gridExtra)# Read in admixture file format 1file<- system.file("extdata","admixture1.csv",package="mapmixture")admixture1<- read.csv(file)# Read in coordinates filefile<- system.file("extdata","coordinates.csv",package="mapmixture")coordinates<- read.csv(file)# Run mapmixturemap6<- mapmixture(admixture_df=admixture1,coords_df=coordinates,cluster_cols= c("#f1a340","#998ec3"),cluster_names= c("Ancestry 1","Ancestry 2"),crs=4326,boundary= c(xmin=-20,xmax=20,ymin=40,ymax=62),pie_size=1.3,)+# Adjust theme options theme(legend.position="top",plot.margin= margin(l=10,r=10), )+# Adjust the size of the legend keys guides(fill= guide_legend(override.aes=list(size=5,alpha=1)))# Facet structure barplotfacet_barplot<- structure_plot(admixture1,type="facet",cluster_cols= c("#f1a340","#998ec3"),facet_col=2,ylabel="Admixture proportions",)+ theme(axis.title.y= element_text(size=10),axis.text.y= element_text(size=5),strip.text= element_text(size=6,vjust=1,margin= margin(t=1.5,r=0,b=1.5,l=0)), )# Arrange plots# grid.arrange(map6, facet_barplot, ncol = 2, widths = c(3,2))
The raster (TIFF) used in the example below was downloaded from NaturalEarthhere.You need to install theterrapackage to use this feature. Currently, thebasemap argument accepts aSpatRaster or asf object.
Code
# Load packageslibrary(mapmixture)library(terra)# Create SpatRaster objectearth<-terra::rast("../NE1_50M_SR_W/NE1_50M_SR_W.tif")# Read in admixture file format 1file<- system.file("extdata","admixture1.csv",package="mapmixture")admixture1<- read.csv(file)# Read in coordinates filefile<- system.file("extdata","coordinates.csv",package="mapmixture")coordinates<- read.csv(file)# Run mapmixturemap7<- mapmixture(admixture1,coordinates,crs=3035,basemap=earth)# map7
The vector data (shapefile) used in the example below was downloadedfrom the Natural England Open Data Geoportalhere.
Code
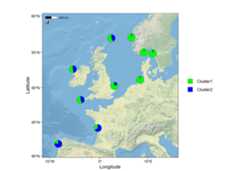
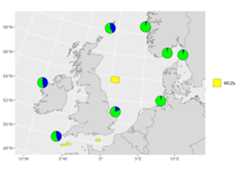
# Load packageslibrary(mapmixture)library(rnaturalearthhires)library(ggplot2)library(dplyr)library(sf)# Read in admixture file format 1file<- system.file("extdata","admixture1.csv",package="mapmixture")admixture1<- read.csv(file)# Read in coordinates filefile<- system.file("extdata","coordinates.csv",package="mapmixture")coordinates<- read.csv(file)# Parameterscrs<-3035boundary<- c(xmin=-11,xmax=13,ymin=50,ymax=60)|> transform_bbox(bbox= _,crs)# Read in world countries from Natural Earth and transform to CRSworld<-rnaturalearthhires::countries10[, c("geometry")]world<- st_transform(world,crs=crs)# Read in Marine Conservation Zones shapefile# Extract polygons for Western Channel, Offshore Brighton and Swallow Sand# Transform to CRSmczs<- st_read("../Marine_Conservation_Zones_England/Marine_Conservation_Zones___Natural_England_and_JNCC.shp",quiet=TRUE)|>dplyr::filter(.data= _,MCZ_NAME%in% c("Western Channel","Offshore Brighton","Swallow Sand"))|> st_transform(x= _,crs=crs)# Run mapmixture helper functions to prepare admixture and coordinates dataadmixture_df<- standardise_data(admixture1,type="admixture")|> transform_admix_data(data= _)coords_df<- standardise_data(coordinates,type="coordinates")admix_coords<- merge_coords_data(coords_df,admixture_df)|> transform_df_coords(df= _,crs=crs)# Plot map and add pie chartsmap8<- ggplot()+ geom_sf(data=world,colour="black",fill="#d9d9d9",size=0.1)+ geom_sf(data=mczs, aes(fill="MCZs"),linewidth=0.3)+ scale_fill_manual(values= c("yellow"))+ coord_sf(xlim= c(boundary[["xmin"]],boundary[["xmax"]]),ylim= c(boundary[["ymin"]],boundary[["ymax"]]) )+ add_pie_charts(admix_coords,admix_columns=4:ncol(admix_coords),lat_column="lat",lon_column="lon",pie_colours= c("green","blue"),border=0.3,opacity=1,pie_size=0.8 )+ theme(legend.title= element_blank(), )# map8
Code
# Load packageslibrary(mapmixture)library(ggplot2)library(adegenet)library(RColorBrewer)library(gridExtra)# Load example genotypesdata("dapcIllus")geno=dapcIllus$a# Change population labelspopNames(geno)= c("Pop1","Pop2","Pop3","Pop4","Pop5","Pop6")# Region namesregion_names<- rep(c("Region1","Region2"),each=300)# Define colour palettecols= brewer.pal(nPop(geno),"RdYlBu")# Perform PCApca1= dudi.pca(geno,scannf=FALSE,nf=3)# Percent of genetic variance explained by each axispercent= round(pca1$eig/sum(pca1$eig)*100,digits=1)# Scatter plot with centroids and segmentsscatter1<- scatter_plot(dataframe=pca1$li,group_ids=geno$pop,type="points",axes= c(1,2),percent=percent,colours=cols,point_size=2,point_type=21,centroid_size=2,stroke=0.1,plot_title="PCA coloured by group_ids")+ theme(legend.position="none",axis.title= element_text(size=8),axis.text= element_text(size=6),plot.title= element_text(size=10), )# Same as scatter1 but no segments and axis 1 and 3 are shownscatter2<- scatter_plot(dataframe=pca1$li,group_ids=geno$pop,type="points",axes= c(1,3),percent=percent,colours=cols,point_size=2,point_type=21,centroids=TRUE,centroid_size=2,segments=FALSE,stroke=0.1,plot_title="PCA no segments and axis 1 and 3 shown")+ theme(legend.position="none",axis.title= element_text(size=8),axis.text= element_text(size=6),plot.title= element_text(size=10), )# Same as scatter1 but coloured by regionscatter3<- scatter_plot(dataframe=pca1$li,group_ids=geno$pop,other_group=region_names,type="points",axes= c(1,2),percent=percent,colours=cols,point_size=2,point_type=21,centroid_size=2,stroke=0.1,plot_title="PCA coloured by other_group")+ theme(legend.position="none",axis.title= element_text(size=8),axis.text= element_text(size=6),plot.title= element_text(size=10), )# Scatter plot with labels instead of pointsscatter4<- scatter_plot(dataframe=pca1$li,group_ids=geno$pop,type="labels",labels= rownames(pca1$li),colours=cols,size=2,label.size=0.10,label.padding= unit(0.10,"lines"),plot_title="PCA using labels instead of points")+ theme(legend.position="none",axis.title= element_text(size=8),axis.text= element_text(size=6),plot.title= element_text(size=10), )# Arrange plots# grid.arrange(scatter1, scatter2, scatter3, scatter4)
# Load packagelibrary(mapmixture)# Launch Shiny applaunch_mapmixture()# Tested with the following package versions:# shiny v1.8.0 (important)# shinyFeedback v0.4.0# shinyjs v2.1.0# shinyWidgets 0.8.4# bslib 0.7.0# colourpicker 1.3.0# htmltools v0.5.8.1# waiter 0.2.5
https://tomjenkins.shinyapps.io/mapmixture/
# Load packagelibrary(mapmixture)# Admixture Format 1file<- system.file("extdata","admixture1.csv",package="mapmixture")admixture1<- read.csv(file)head(admixture1)#> Site Ind Cluster1 Cluster2#> 1 Bergen Ber01 0.9999 1e-04#> 2 Bergen Ber02 0.9999 1e-04#> 3 Bergen Ber03 0.9999 1e-04#> 4 Bergen Ber04 0.9999 1e-04#> 5 Bergen Ber05 0.9999 1e-04#> 6 Bergen Ber06 0.9999 1e-04# Admixture Format 2file<- system.file("extdata","admixture2.csv",package="mapmixture")admixture2<- read.csv(file)admixture2#> Site Ind Cluster1 Cluster2#> 1 Bergen Bergen 0.9675212 0.03247879#> 2 Cromer Cromer 0.8217114 0.17828857#> 3 Flodevigen Flodevigen 0.9843806 0.01561944#> 4 Helgoland Helgoland 0.9761543 0.02384571#> 5 Ile de Re Ile de Re 0.3529000 0.64710000#> 6 Isles of Scilly Isles of Scilly 0.5632444 0.43675556#> 7 Lysekil Lysekil 0.9661722 0.03382778#> 8 Mullet Peninsula Mullet Peninsula 0.5316833 0.46831667#> 9 Shetland Shetland 0.5838028 0.41619722#> 10 Vigo Vigo 0.2268444 0.77315556# Admixture Format 3file<- system.file("extdata","admixture3.csv",package="mapmixture")admixture3<- read.csv(file)admixture3#> Site Ind Cluster1 Cluster2#> 1 Bergen Bergen 1 0#> 2 Cromer Cromer 1 0#> 3 Flodevigen Flodevigen 1 0#> 4 Helgoland Helgoland 1 0#> 5 Ile de Re Ile de Re 0 1#> 6 Isles of Scilly Isles of Scilly 1 0#> 7 Lysekil Lysekil 1 0#> 8 Mullet Peninsula Mullet Peninsula 1 0#> 9 Shetland Shetland 1 0#> 10 Vigo Vigo 0 1# Coordinatesfile<- system.file("extdata","coordinates.csv",package="mapmixture")coordinates<- read.csv(file)coordinates#> Site Lat Lon#> 1 Bergen 60.65 4.77#> 2 Cromer 52.94 1.31#> 3 Flodevigen 58.42 8.76#> 4 Helgoland 54.18 7.90#> 5 Ile de Re 46.13 -1.25#> 6 Isles of Scilly 49.92 -6.33#> 7 Lysekil 58.26 11.37#> 8 Mullet Peninsula 54.19 -10.15#> 9 Shetland 60.17 -1.40#> 10 Vigo 42.49 -8.99
About
R package and web app for spatial visualisation of admixture on a projected map
Topics
Resources
License
Uh oh!
There was an error while loading.Please reload this page.
Stars
Watchers
Forks
Packages0
Uh oh!
There was an error while loading.Please reload this page.