- Notifications
You must be signed in to change notification settings - Fork3
Stand-alone software package of PureseqTM for transmembrane topology prediction from amino acid sequence only.
PureseqTM/PureseqTM_Package
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation
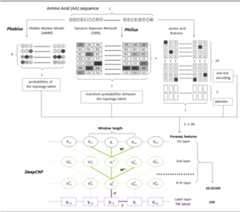
TransMembrane (TM) topology prediction from amino acid sequence only
Users may submit their sequences to the PureseqTM Server at:http://pureseqtm.predmp.com
Users may find the train, test data of PureseqTM at:https://github.com/PureseqTM/PureseqTM_Dataset
https://www.biorxiv.org/content/10.1101/627307v2.abstract
git clone https://github.com/PureseqTM/PureseqTM_Package.gitcd ./PureseqTM_Packagecd source_code makecd .././PureseqTM.sh -i example/4j7cK.fasta -m 0./PureseqTM.sh -i example/4j7cK.fasta./PureseqTM.sh -i example/4j7cK.fasta -l example/4j7cK.topIf the ground-truth label is provided, run below commands to evaluate the prediction accuracy:
grep -v "#" 4j7cK_PureTM/4j7cK.prob | awk '{print $NF" "$3}' > 4j7cK.pred_resoutil/TM2_Evaluation 4j7cK.pred_reso 0.5rm -f 4j7cK.pred_reso./PureseqTM.sh -i example/4j7cK.fasta -P 1Note that gnuplot is required to be installed in the local system.
./PureseqTM.sh -i example/1bhaA.tgtNote that to generate the evolutionary profile, users are suggested to use theTGT_Package to generate the TGT file:
./A3M_TGT_Gen.sh -i <input_fasta> -d uniprot20_2016_02 -h hhsuite2 -n 3 -E 10For more details on evolutionary profile, please gotohttps://github.com/realbigws/TGT_Package
As it takes about 2 hours for the whole Human proteome, below please find a toy example:
./PureseqTM_proteome.sh -i example/test_proteome.fastaTransfer PDBTM 9-state label to 0/1 TransMembrane (TM) label:
python util/pdbtm2binary.py example/1bhaA.pdbtmThere shall be 5 to 7 output files in XXX_PureTM by default, where XXX is the input name:
| File name | Description | Option |
|---|---|---|
| XXX.fasta_raw | original input sequence file in FASTA format. | |
| XXX.fasta | canonical sequence file without non-canonical characters. | |
| XXX.top | simple 2-state TransMembrane (TM) prediction in FASTA format. | |
| XXX.prob | detailed 2-state TransMembrane (TM) probability prediction. | |
| XXX.pred_mode | prediction mode (1 for sequence and 0 for profile). | |
| XXX.png | posterior probabilities plotted by GNUPLOT | if option -P 1 is set |
| XXX.gff | segment-level output | if option -P 1 is set |
Title: A Combined Transmembrane Topology and Signal Peptide Prediction MethodAuthors: Lukas Kall, Anders Krogh and Erik L. L. SonnhammerJournal: J. Mol. Biol. (2004) 338, 1027–1036Title: Transmembrane Topology and Signal Peptide Prediction Using Dynamic Bayesian NetworksAuthors: Sheila M. Reynolds, Lukas Kall, Michael E. Riffle, Jeff A. Bilmes, William Stafford NobleJournal: PLoS computational biology (2008) 4(11):e1000213Title: Protein secondary structure prediction using deep convolutional neural fieldsAuthors: Sheng Wang, Jian Peng, Jianzhu Ma, Jinbo XuJournal: Scientific reports (2016) 6:18962About
Stand-alone software package of PureseqTM for transmembrane topology prediction from amino acid sequence only.
Resources
Uh oh!
There was an error while loading.Please reload this page.