- Notifications
You must be signed in to change notification settings - Fork1
mulea - an R package for enrichment analysis using various ontologies and empirical false discovery rate
ELTEbioinformatics/mulea
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation

mulea - an R Package for Enrichment Analysis Using Multiple Ontologies and Empirical False Discovery Rate
Themulea R package (Turek et al. 2024) is a comprehensive tool forfunctional enrichment analysis. It provides two different approaches:
For unranked sets of elements, such as significantly up- ordown-regulated genes,
muleaemploys the set-basedoverrepresentation analysis (ORA).Alternatively, if the data consists of ranked elements, forinstance, genes ordered byp-value or log fold-change calculatedby the differential expression analysis,
muleaoffers thegeneset enrichment (GSEA) approach.
For the overrepresentation analysis,mulea employs a progressiveempirical false discovery rate (eFDR) method, specifically designedfor interconnected biological data, to accurately identify significantterms within diverse ontologies.
mulea expands beyond traditional tools by incorporating awide rangeof ontologies, encompassing Gene Ontology, pathways, regulatoryelements, genomic locations, and protein domains for 27 model organisms,covering 22 ontology types from 16 databases and various identifiersresulting in 879 files available at theELTEbioinformatics/GMT_files_for_muleaGitHub repository and through themuleaDataExperimentData Bioconductor package.
Install the dependencyfgsea BioConductor package:
# Installing the BiocManager package if neededif (!require("BiocManager",quietly=TRUE)) install.packages("BiocManager")# Installing the fgsea package with the BiocManagerBiocManager::install("fgsea")
To installmulea fromCRAN:
install.packages("mulea")To install the development version ofmulea from GitHub:
# Installing the devtools package if neededif (!require("devtools",quietly=TRUE)) install.packages("devtools")# Installing the mulea package from GitHubdevtools::install_github("https://github.com/ELTEbioinformatics/mulea")
First, load themulea anddplyr libraries. Thedplyrlibrary is not essential but is used here to facilitate datamanipulation and inspection.
library(mulea)library(tidyverse)
This section demonstrates how to import the desired ontology, such astranscription factors and their target genes downloaded from the database, into a dataframe suitable for enrichment analysis. We present multiple methods forimporting the ontology. Ensure that the identifier type (e.g.,UniProt protein ID,Entrez ID, Gene Symbol,Ensembl gene ID)matches between the ontology and the elements you wish to investigate.
database, into a dataframe suitable for enrichment analysis. We present multiple methods forimporting the ontology. Ensure that the identifier type (e.g.,UniProt protein ID,Entrez ID, Gene Symbol,Ensembl gene ID)matches between the ontology and the elements you wish to investigate.
TheGMT (Gene MatrixTransposed)format contains collections of genes or proteins associated withspecific ontology terms in a tab-delimited text file. The GMT file canbe read into R as a data frame using theread_gmt function from themulea package. Each term is represented by a single row in both theGMT file and the data frame. Each row includes three types of elements:
Ontology identifier (“ontology_id”): This uniquely identifieseach term within the file or data frame.
Ontology name or description (“ontology_name”): This providesa user-friendly label or textual description for each term.
Associated gene or protein identifiers: These are listed in the“list_of_values” column, with identifiers separated by spaces, andbelong to each term.1
Alongside with themulea package we provide ontologies collected from16 publicly available databases, in a standardised GMT format for 27model organisms, from Bacteria to human. These files are available attheELTEbioinformatics/GMT_files_for_muleaGitHub repository.
To read a downloaded GMT file locally:
# Reading the mulea GMT file locallytf_ontology<- read_gmt("Transcription_factor_RegulonDB_Escherichia_coli_GeneSymbol.gmt")
Alternatively, one can read it directly from the GitHub repository:
# Reading the GMT file from the GitHub repositorytf_ontology<- read_gmt("https://raw.githubusercontent.com/ELTEbioinformatics/GMT_files_for_mulea/main/GMT_files/Escherichia_coli_83333/Transcription_factor_RegulonDB_Escherichia_coli_GeneSymbol.gmt")
mulea is compatible withGMTfiles provided with theEnricher R package (Kuleshov et al. 2016). Download and read such aGMT file (e. g. TRRUST_Transcription_Factors_2019.txt) locally.Note that this ontology is not suitable for analyzing the Escherichiacoli differential expression data described in the sectionTheDifferential Expression Dataset toAnalyse.
# Reading the Enrichr GMT file locallytf_enrichr_ontology<- read_gmt("TRRUST_Transcription_Factors_2019.txt")# The ontology_name is empty, therefore we need to fill it with the ontology_idtf_enrichr_ontology$ontology_name<-tf_enrichr_ontology$ontology_id
mulea is compatible with the MsigDB (Subramanian et al. 2005)GMTfiles. Download and read sucha GMT file (e. g. c3.tft.v2023.2.Hs.symbols.gmt) locally.Notethat this ontology is not suitable for analyzing the Escherichia colidifferential expression data described in the sectionThe DifferentialExpression Dataset toAnalyse.
# Reading the MsigDB GMT file locallytf_msigdb_ontology<- read_gmt("c3.tft.v2023.2.Hs.symbols.gmt")
Alternatively, you can retrieve the ontology using themuleaDataExperimentData Bioconductor package:
# Installing the ExperimentHub package from BioconductorBiocManager::install("ExperimentHub")# Calling the ExperimentHub library.library(ExperimentHub)# Downloading the metadata from ExperimentHub.eh<- ExperimentHub()# Creating the muleaData variable.muleaData<- query(eh,"muleaData")# Looking for the ExperimentalHub ID of the ontology.EHID<- mcols(muleaData) %>% as.data.frame() %>%dplyr::filter(title=="Transcription_factor_RegulonDB_Escherichia_coli_GeneSymbol.rds") %>% rownames()# Reading the ontology from the muleaData package.tf_ontology<-muleaData[[EHID]]# Change the headertf_ontology<-tf_ontology %>% rename(ontology_id="ontologyId",ontology_name="ontologyName",list_of_values="listOfValues")
Enrichment analysis results can sometimes be skewed by overly specificor broad entries.mulea allows you to customise the size of ontologyentries – the number of genes or proteins belonging to a term – ensuringyour analysis aligns with your desired scope.
Let’s exclude ontology entries with less than 3 or more than 400 genesymbols.
# Filtering the ontologytf_ontology_filtered<- filter_ontology(gmt=tf_ontology,min_nr_of_elements=3,max_nr_of_elements=400)
You can save the ontology as a GMT file using thewrite_gmtfunction.
# Saving the ontology to GMT filewrite_gmt(gmt=tf_ontology_filtered,file="Filtered.gmt")
Themulea package provides thelist_to_gmt function to convert alist of gene sets into an ontology data frame. The following exampledemonstrates how to use this function:
# Creating a list of gene setsontology_list<-list(gene_set1= c("gene1","gene2","gene3"),gene_set2= c("gene4","gene5","gene6"))# Converting the list to a ontology (GMT) objectnew_ontology_df<- list_to_gmt(ontology_list)
For further steps we will analyse a dataset from a microarray experiment(GSE55662)in the NCBI Gene Expression Omnibus . The study byMéhi et al. (2014) investigated antibiotic resistance evolution inEscherichia coli. Gene expression changes were compared betweenciprofloxacin antibiotic-treatedEscherichia coli bacteria andnon-treated controls.
. The study byMéhi et al. (2014) investigated antibiotic resistance evolution inEscherichia coli. Gene expression changes were compared betweenciprofloxacin antibiotic-treatedEscherichia coli bacteria andnon-treated controls.
The expression levels of these groups were compared with theGEO2R tool:
- Non-treated control samples (2 replicates):WT_noCPR_1,WT_noCPR_2
- Ciprofloxacin-treated samples (2 replicates):WT_CPR_1,WT_CPR_2
To see how the dataset were prepared go to theFormatting the Resultsof a Differential ExpressionAnalysissection.
Themulea package implements a set-based enrichment analysis approachusing thehypergeometric test, which is analogous to the one-tailedFisher’s exact test. This method identifies statistically significantoverrepresentation of elements from a target set (e.g., significantlyup- or downregulated genes) within a background set (e.g., all genesthat were investigated in the experiment). Therefore, a predefinedthreshold value, such as 0.05 for the correctedp-value or 2-foldchange, should be used in the preceding analysis.
The overrepresentation analysis is implemented in theora functionwhich requires three inputs:
Ontologydata frame: Fits the investigated taxa and theapplied gene or protein identifier type, such as GO, pathway,transcription factor regulation, microRNA regulation, geneexpression data, genomic location data, or protein domain content.
Target set: A vector of elements to investigate, containinggenes or proteins of interest, such as significantly overexpressedgenes in the experiment.
Background set: A vector of background elements representing thebroader context, often including all genes investigated in thestudy.
Let’s read the text files containing the identifiers (gene symbols) ofthe target and the background gene set directly from the GitHub website.To see how these files were prepared, refer to the section onFormatting the Results of a Differential ExpressionAnalysis.
# Taget settarget_set<- readLines("https://raw.githubusercontent.com/ELTEbioinformatics/mulea/master/inst/extdata/target_set.txt")# Background setbackground_set<- readLines("https://raw.githubusercontent.com/ELTEbioinformatics/mulea/master/inst/extdata/background_set.txt")
To perform the analysis, we will first establish a model using theorafunction. This model defines the parameters for the enrichment analysis.We then execute the test itself using therun_test function. It isimportant to note that for this example, we will employ 10,000permutations for theempirical false discovery rate (eFDR), which isthe recommended minimum, to ensure robust correction for multipletesting.
# Creating the ORA model using the GMT variableora_model<- ora(gmt=tf_ontology_filtered,# Test set variableelement_names=target_set,# Background set variablebackground_element_names=background_set,# p-value adjustment methodp_value_adjustment_method="eFDR",# Number of permutationsnumber_of_permutations=10000,# Number of processor threads to usenthreads=2,# Setting a random seed for reproducibilityrandom_seed=1)# Running the ORAora_results<- run_test(ora_model)
Theora_results data frame summarises the enrichment analysis, listingenriched ontology entries – in our case transcription factors –alongside their associatedp-values andeFDR values.
We can now determine the number of transcription factors classified as“enriched” based on these statistical measures (eFDR < 0.05).
ora_results %>%# Rows where the eFDR < 0.05 filter(eFDR<0.05) %>%# Number of such rows nrow()#> [1] 10
Inspect the significant results:
ora_results %>%# Arrange the rows by the eFDR values arrange(eFDR) %>%# Rows where the eFDR < 0.05 filter(eFDR<0.05)
| ontology_id | ontology_name | nr_common_with_tested_elements | nr_common_with_background_elements | p_value | eFDR |
|---|---|---|---|---|---|
| FNR | FNR | 26 | 259 | 0.0000003 | 0.0000000 |
| LexA | LexA | 14 | 53 | 0.0000000 | 0.0000000 |
| SoxS | SoxS | 7 | 37 | 0.0001615 | 0.0036667 |
| Rob | Rob | 5 | 21 | 0.0004717 | 0.0051200 |
| DnaA | DnaA | 4 | 13 | 0.0006281 | 0.0052000 |
| FadR | FadR | 5 | 20 | 0.0003692 | 0.0056000 |
| NsrR | NsrR | 8 | 64 | 0.0010478 | 0.0073714 |
| ArcA | ArcA | 12 | 148 | 0.0032001 | 0.0197500 |
| IHF | IHF | 14 | 205 | 0.0070758 | 0.0458600 |
| MarA | MarA | 5 | 37 | 0.0066068 | 0.0483111 |
To gain a comprehensive understanding of the enriched transcriptionfactors,mulea offers diverse visualisation tools, includinglollipop charts, bar plots, networks, and heatmaps. These visualisationseffectively reveal patterns and relationships among the enrichedfactors.
Initialising the visualisation with thereshape_results function:
# Reshapeing the ORA results for visualisationora_reshaped_results<- reshape_results(model=ora_model,model_results=ora_results,# Choosing which column to use for the# indication of significancep_value_type_colname="eFDR")
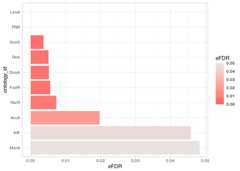
Visualising the Spread ofeFDR Values: Lollipop Plot
Lollipop charts provide a graphical representation of the distributionof enriched transcription factors. They-axis displays thetranscription factors, while thex-axis represents their correspondingeFDR values. The dots are coloured based on theireFDR values. Thisvisualisation helps us examine the spread ofeFDRs and identifyfactors exceeding the commonly used significance threshold of 0.05.
plot_lollipop(reshaped_results=ora_reshaped_results,# Column containing the names we wish to plotontology_id_colname="ontology_id",# Upper threshold for the value indicating the significancep_value_max_threshold=0.05,# Column that indicates the significance valuesp_value_type_colname="eFDR")
Visualising the Spread ofeFDR Values: Bar Plot
Bar charts offer a graphical representation similar to lollipop plots.They-axis displays the enriched ontology categories (e.g.,transcription factors), while thex-axis represents theircorrespondingeFDR values. The bars are coloured based on theireFDRvalues, aiding in examining the spread ofeFDRs and identifyingfactors exceeding the significance threshold of 0.05.
plot_barplot(reshaped_results=ora_reshaped_results,# Column containing the names we wish to plotontology_id_colname="ontology_id",# Upper threshold for the value indicating the significancep_value_max_threshold=0.05,# Column that indicates the significance valuesp_value_type_colname="eFDR")
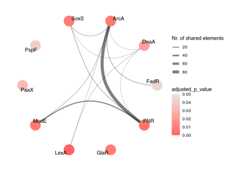
Visualising the Associations: Graph Plot
This function generates a network visualisation of the enriched ontologycategories (e.g., transcription factors). Each node represents aneriched ontology category, coloured based on itseFDR value. An edgeis drawn between two nodes if they share at least one common genebelonging to the target set, indicating co-regulation. The thickness ofthe edge reflects the number of shared genes.
plot_graph(reshaped_results=ora_reshaped_results,# Column containing the names we wish to plotontology_id_colname="ontology_id",# Upper threshold for the value indicating the significancep_value_max_threshold=0.05,# Column that indicates the significance valuesp_value_type_colname="eFDR")
Visualising the Associations: Heatmap
The heatmap displays the genes associated with the enriched ontologycategories (e.g., transcription factors). Each row represents acategory, coloured based on itseFDR value. Each column represents agene from the target set belonging to the enriched ontology category,indicating potential regulation by one or more enriched transcriptionfactors.
plot_heatmap(reshaped_results=ora_reshaped_results,# Column containing the names we wish to plotontology_id_colname="ontology_id",# Column that indicates the significance valuesp_value_type_colname="eFDR")
Comparing the significant results when applying the eFDR to the Benjamini-Hochberg and the Bonferroni corrections
Theora function allows you to choose between different methods forcalculating theFDR and adjusting thep-values:eFDR, and allmethod options from thestats::p.adjust documentation (holm,hochberg, hommel, bonferroni, BH, BY, and fdr). The following codesnippet demonstrates how to perform the analysis using theBenjamini-Hochberg andBonferroni corrections:
# Creating the ORA model using the Benjamini-Hochberg p-value correction methodBH_ora_model<- ora(gmt=tf_ontology_filtered,# Test set variableelement_names=target_set,# Background set variablebackground_element_names=background_set,# p-value adjustment methodp_value_adjustment_method="BH")# Running the ORABH_results<- run_test(BH_ora_model)# Creating the ORA model using the Bonferroni p-value correction methodBonferroni_ora_model<- ora(gmt=tf_ontology_filtered,# Test set variableelement_names=target_set,# Background set variablebackground_element_names=background_set,# p-value adjustment methodp_value_adjustment_method="bonferroni")# Running the ORABonferroni_results<- run_test(Bonferroni_ora_model)
To compare the significant results (using the conventional < 0.05threshold) of theeFDR,Benjamini-Hochberg, andBonferronicorrections, we can merge and filter the result tables:
# Merging the Benjamini-Hochberg and eFDR resultsmerged_results<-BH_results %>%# Renaming the column rename(BH_adjusted_p_value=adjusted_p_value) %>%# Selecting the necessary columns select(ontology_id,BH_adjusted_p_value) %>%# Joining with the eFDR results left_join(ora_results,.,by="ontology_id") %>%# Converting the data.frame to a tibble tibble()# Merging the Bonferroni results with the merged resultsmerged_results<-Bonferroni_results %>%# Renaming the column rename(Bonferroni_adjusted_p_value=adjusted_p_value) %>%# Selecting the necessary columns select(ontology_id,Bonferroni_adjusted_p_value) %>%# Joining with the eFDR results left_join(merged_results,.,by="ontology_id") %>%# Arranging by the p-value arrange(p_value)# filter the p-value < 0.05 resultsmerged_results_filtered<-merged_results %>% filter(p_value<0.05) %>%# remove the unnecessary columns select(-ontology_id,-nr_common_with_tested_elements,-nr_common_with_background_elements)
| ontology_name | p_value | eFDR | BH_adjusted_p_value | Bonferroni_adjusted_p_value |
|---|---|---|---|---|
| LexA | 0.0000000 | 0.0000000 | 0.0000001 | 0.0000001 |
| FNR | 0.0000003 | 0.0000000 | 0.0000208 | 0.0000416 |
| SoxS | 0.0001615 | 0.0036667 | 0.0082880 | 0.0248641 |
| FadR | 0.0003692 | 0.0056000 | 0.0142127 | 0.0568507 |
| Rob | 0.0004717 | 0.0051200 | 0.0145296 | 0.0726479 |
| DnaA | 0.0006281 | 0.0052000 | 0.0161218 | 0.0967306 |
| NsrR | 0.0010478 | 0.0073714 | 0.0230517 | 0.1613622 |
| ArcA | 0.0032001 | 0.0197500 | 0.0616014 | 0.4928114 |
| MarA | 0.0066068 | 0.0483111 | 0.1089670 | 1.0000000 |
| IHF | 0.0070758 | 0.0458600 | 0.1089670 | 1.0000000 |
| NarL | 0.0096065 | 0.0534000 | 0.1276532 | 1.0000000 |
| NikR | 0.0099470 | 0.0615833 | 0.1276532 | 1.0000000 |
| OxyR | 0.0174505 | 0.0786923 | 0.2067212 | 1.0000000 |
| ExuR | 0.0261046 | 0.1051867 | 0.2680073 | 1.0000000 |
| UxuR | 0.0261046 | 0.1051867 | 0.2680073 | 1.0000000 |
| NrdR | 0.0328500 | 0.1232750 | 0.3161817 | 1.0000000 |
| IscR | 0.0376038 | 0.1249412 | 0.3406459 | 1.0000000 |
| Nac | 0.0419701 | 0.1487556 | 0.3590774 | 1.0000000 |
| Fis | 0.0457307 | 0.1433053 | 0.3706596 | 1.0000000 |
A comparison of the significant results revealed that conventionalp-value corrections (Benjamini-Hochberg and Bonferroni) tend to beoverly conservative, leading to a reduction in the number of significanttranscription factors compared to theeFDR. As illustrated in thebelow figure, by applying theeFDR we were able to identify 10significant transcription factors, while with the Benjamini-Hochberg andBonferroni corrections only 7 and 3, respectively. This suggests thattheeFDR may be a more suitable approach for controlling falsepositives in this context.
To perform enrichment analysis using ranked lists, you need to providean ordered list of elements, such as genes or proteins. This ranking istypically based on the results of your prior analysis, using metricslikep-values,z-scores, fold-changes, or others. Crucially, theranked list should include all elements involved in your analysis. Forexample, in a differential expression study, it should encompass allgenes that were measured.
mulea utilises the Kolmogorov-Smirnov approach with a permutation test(developed by Subramanian et al. (2005)) to calculate gene setenrichment analyses. This functionality is implemented through theintegration of thefgseaBioconductor package (created by Korotkevich et al. (2021)).
GSEA requires input data about the genes analysed in our experiment.This data can be formatted in two ways:
Data frame: This format should include all genes investigatedand their respective log fold change values (or other values forordering the genes) obtained from the differential expressionanalysis.
Two vectors: Alternatively, you can provide two separatevectors. One vector should contain the gene symbols (or IDs), andthe other should hold the corresponding log fold change values (orother values for ordering the genes) for each gene.
Let’s read the TSV file containing the identifiers (gene symbols) andthe log fold change values of the investigated set directly from theGitHub website. For details on how this file was prepared, please referto theFormatting the Results of a Differential ExpressionAnalysissection.
# Reading the tsv containing the ordered setordered_set<- read_tsv("https://raw.githubusercontent.com/ELTEbioinformatics/mulea/master/inst/extdata/ordered_set.tsv")
To perform the analysis, we will first establish a model using thegsea function. This model defines the parameters for the enrichmentanalysis. Subsequently, we will execute the test itself using therun_test function. We will employ 10,000 permutations for the falsediscovery rate, to ensure robust correction for multiple testing.
# Creating the GSEA model using the GMT variablegsea_model<- gsea(gmt=tf_ontology_filtered,# Names of elements to testelement_names=ordered_set$Gene.symbol,# LogFC-s of elements to testelement_scores=ordered_set$logFC,# Consider elements having positive logFC values onlyelement_score_type="pos",# Number of permutationsnumber_of_permutations=10000)# Running the GSEAgsea_results<- run_test(gsea_model)
Thegsea_results data frame summarises the enrichment analysis,listing enriched ontology entries – in our case transcription factors –alongside their associatedp-values and adjustedp-value values.
We can now determine the number of transcription factors classified as“enriched” based on these statistical measures (adjustedp-value <0.05).
gsea_results %>%# rows where the adjusted_p_value < 0.05 filter(adjusted_p_value<0.05) %>%# the number of such rows nrow()#> [1] 10
Inspect the significant results:
gsea_results %>%# arrange the rows by the adjusted_p_value values arrange(adjusted_p_value) %>%# rows where the adjusted_p_value < 0.05 filter(adjusted_p_value<0.05)
| ontology_id | ontology_name | nr_common_with_tested_elements | p_value | adjusted_p_value |
|---|---|---|---|---|
| LexA | LexA | 53 | 0.0000000 | 0.0000047 |
| FNR | FNR | 259 | 0.0000660 | 0.0050484 |
| ArcA | ArcA | 148 | 0.0003076 | 0.0079598 |
| GlaR | GlaR | 3 | 0.0002188 | 0.0079598 |
| ModE | ModE | 45 | 0.0003122 | 0.0079598 |
| SoxS | SoxS | 37 | 0.0002848 | 0.0079598 |
| DnaA | DnaA | 13 | 0.0010217 | 0.0223306 |
| PaaX | PaaX | 14 | 0.0017028 | 0.0325652 |
| PspF | PspF | 7 | 0.0023494 | 0.0399397 |
| FadR | FadR | 20 | 0.0028304 | 0.0433046 |
Initializing the visualisation with thereshape_results function:
# Reshaping the GSEA results for visualisationgsea_reshaped_results<- reshape_results(model=gsea_model,model_results=gsea_results,# choosing which column to use for the# indication of significancep_value_type_colname="adjusted_p_value")
Visualising Relationships: Graph Plot
This function generates a network visualisation of the enriched ontologycategories (e.g., transcription factors). Each node represents acategory and is coloured based on its significance level. A connection(edge) is drawn between two nodes if they share at least one common genebelonging to theranked list, meaning that both transcriptionfactors regulate the expression of the same target gene. The thicknessof the edge reflects the number of shared genes.
plot_graph(reshaped_results=gsea_reshaped_results,# the column containing the names we wish to plotontology_id_colname="ontology_id",# upper threshold for the value indicating the significancep_value_max_threshold=0.05,# column that indicates the significance valuesp_value_type_colname="adjusted_p_value")
Other plot types such as lollipop plots, bar plots, and heatmaps canalso be used to investigate the GSEA results.
This section aims to elucidate the structure and essential components ofthe provided DE results table. It offers guidance to users oninterpreting the data effectively for subsequent analysis withmulea.
Let’s read the differential expression result file namedGSE55662.table_wt_non_vs_cipro.tsvlocated in theinst/extdata/folder directly from the GitHub website.
# Importing necessary libraries and reading the DE results tablegeo2r_result_tab<- read_tsv("https://raw.githubusercontent.com/ELTEbioinformatics/mulea/master/inst/extdata/GSE55662.table_wt_non_vs_cipro.tsv")
Let’s delve into thegeo2r_result_tab data frame by examining itsinitial rows:
# Printing the first few rows of the data framegeo2r_result_tab %>% head(3)
| ID | adj.P.Val | P.Value | t | B | logFC | Gene.symbol | Gene.title |
|---|---|---|---|---|---|---|---|
| 1765336_s_at | 0.0186 | 2.4e-06 | 21.5 | 4.95769 | 3.70 | gnsB | Qin prophage; multicopy suppressor of secG(Cs) and fabA6(Ts) |
| 1760422_s_at | 0.0186 | 3.8e-06 | 19.6 | 4.68510 | 3.14 | NA | NA |
| 1764904_s_at | 0.0186 | 5.7e-06 | 18.2 | 4.43751 | 2.54 | sulA///sulA///sulA///ECs1042 | SOS cell division inhibitor///SOS cell division inhibitor///SOS cell division inhibitor///SOS cell division inhibitor |
Preparing the data frame appropriately for enrichment analysis iscrucial. This involves specific steps tailored to the microarrayexperiment type. Here, we undertake the following transformations:
Gene Symbol Extraction: We isolate the primary gene symbol fromthe
Gene.symbolcolumn, eliminating any extraneous information.Handling Missing Values: Rows with missing gene symbols (
NA) areexcluded.Sorting by Fold Change: The data frame is sorted by log-foldchange (
logFC) in descending order, prioritizing genes with the mostsignificant expression alterations.
# Formatting the data framegeo2r_result_tab<-geo2r_result_tab %>%# Extracting the primary gene symbol and removing extraneous information mutate(Gene.symbol= str_remove(string=Gene.symbol,pattern="\\/.*")) %>%# Filtering out rows with NA gene symbols filter(!is.na(Gene.symbol)) %>%# Sorting by logFC arrange(desc(logFC))
Before proceeding with enrichment analysis, let’s examine the initialrows of the formattedgeo2r_result_tab data frame:
# Printing the first few rows of the formatted data framegeo2r_result_tab %>% head(3)
| ID | adj.P.Val | P.Value | t | B | logFC | Gene.symbol | Gene.title |
|---|---|---|---|---|---|---|---|
| 1765336_s_at | 0.0186 | 2.40e-06 | 21.5 | 4.95769 | 3.70 | gnsB | Qin prophage; multicopy suppressor of secG(Cs) and fabA6(Ts) |
| 1764904_s_at | 0.0186 | 5.70e-06 | 18.2 | 4.43751 | 2.54 | sulA | SOS cell division inhibitor///SOS cell division inhibitor///SOS cell division inhibitor///SOS cell division inhibitor |
| 1761763_s_at | 0.0186 | 1.54e-05 | 15.0 | 3.73568 | 2.16 | recN | recombination and repair protein///recombination and repair protein///recombination and repair protein///recombination and repair protein |
Following these formatting steps, the data frame is primed for furtheranalysis.
A vector containing the gene symbols of significantly overexpressed(adjustedp-value < 0.05) genes with greater than 2 fold-change(logFC > 1).
target_set<-geo2r_result_tab %>%# Filtering for adjusted p-value < 0.05 and logFC > 1 filter(adj.P.Val<0.05&logFC>1) %>%# Selecting the Gene.symbol column select(Gene.symbol) %>%# Converting the tibble to a vector pull() %>%# Removing duplicates unique()
The first 10 elements of the target set:
target_set %>% head(10)#> [1] "gnsB" "sulA" "recN" "c4435" "dinI" "c2757" "c1431"#> [8] "gabP" "recA" "ECs5456"
The number of genes in the target set:
target_set %>% length()#> [1] 241
A vector containing the gene symbols of all genes were included in thedifferential expression analysis.
background_set<-geo2r_result_tab %>%# Selecting the Gene.symbol column select(Gene.symbol) %>%# Converting the tibble to a vector pull() %>%# Removing duplicates unique()
The number of genes in the background set:
background_set %>% length()#> [1] 7381
Save the target and the background set vectors to text file:
# Save taget set to text filetarget_set %>% writeLines("target_set.txt")# Save background set to text filebackground_set %>% writeLines("inst/extdata/background_set.txt")
# If there are duplicated Gene.symbols keep the first one onlyordered_set<-geo2r_result_tab %>%# Grouping by Gene.symbol to be able to filter group_by(Gene.symbol) %>%# Keeping the first row for each Gene.symbol from rows with the same# Gene.symbol filter(row_number()==1) %>%# Ungrouping ungroup() %>%# Arranging by logFC in descending order arrange(desc(logFC)) %>% select(Gene.symbol,logFC)
The number of gene symbols in theordered_set vector:
ordered_set %>% nrow()#> [1] 7381
Save the ordered set data frame to tab delimited file:
# Save ordered set to text fileordered_set %>% write_tsv("ordered_set.tsv")
sessionInfo()#> R version 4.4.2 (2024-10-31)#> Platform: x86_64-pc-linux-gnu#> Running under: Ubuntu 22.04.5 LTS#>#> Matrix products: default#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0#>#> locale:#> [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C#> [3] LC_TIME=hu_HU.UTF-8 LC_COLLATE=en_US.UTF-8#> [5] LC_MONETARY=hu_HU.UTF-8 LC_MESSAGES=en_US.UTF-8#> [7] LC_PAPER=hu_HU.UTF-8 LC_NAME=C#> [9] LC_ADDRESS=C LC_TELEPHONE=C#> [11] LC_MEASUREMENT=hu_HU.UTF-8 LC_IDENTIFICATION=C#>#> time zone: Europe/Budapest#> tzcode source: system (glibc)#>#> attached base packages:#> [1] stats graphics grDevices utils datasets methods base#>#> other attached packages:#> [1] lubridate_1.9.3 forcats_1.0.0 stringr_1.5.1 dplyr_1.1.4#> [5] purrr_1.0.2 readr_2.1.5 tidyr_1.3.1 tibble_3.2.1#> [9] ggplot2_3.5.1 tidyverse_2.0.0 mulea_1.0.2#>#> loaded via a namespace (and not attached):#> [1] fastmatch_1.1-4 gtable_0.3.5 xfun_0.47#> [4] ggrepel_0.9.6 lattice_0.22-6 tzdb_0.4.0#> [7] vctrs_0.6.5 tools_4.4.2 generics_0.1.3#> [10] curl_5.2.2 parallel_4.4.2 fansi_1.0.6#> [13] highr_0.11 pkgconfig_2.0.3 Matrix_1.7-0#> [16] data.table_1.16.0 lifecycle_1.0.4 compiler_4.4.2#> [19] farver_2.1.2 munsell_0.5.1 ggforce_0.4.2#> [22] fgsea_1.30.0 graphlayouts_1.2.0 codetools_0.2-20#> [25] htmltools_0.5.8.1 yaml_2.3.10 crayon_1.5.3#> [28] pillar_1.9.0 MASS_7.3-61 BiocParallel_1.38.0#> [31] cachem_1.1.0 viridis_0.6.5 tidyselect_1.2.1#> [34] digest_0.6.37 stringi_1.8.4 labeling_0.4.3#> [37] cowplot_1.1.3 polyclip_1.10-7 fastmap_1.2.0#> [40] grid_4.4.2 colorspace_2.1-1 cli_3.6.3#> [43] magrittr_2.0.3 ggraph_2.2.1 tidygraph_1.3.1#> [46] utf8_1.2.4 withr_3.0.1 scales_1.3.0#> [49] bit64_4.0.5 timechange_0.3.0 rmarkdown_2.28#> [52] bit_4.0.5 igraph_2.0.3 gridExtra_2.3#> [55] hms_1.1.3 memoise_2.0.1 evaluate_0.24.0#> [58] knitr_1.48 viridisLite_0.4.2 rlang_1.1.4#> [61] Rcpp_1.0.13 glue_1.7.0 tweenr_2.0.3#> [64] vroom_1.6.5 rstudioapi_0.16.0 R6_2.5.1#> [67] plyr_1.8.9
To cite packagemulea in publications use:
Turek, Cezary, Márton Ölbei, Tamás Stirling, Gergely Fekete, ErvinTasnádi, Leila Gul, Balázs Bohár, Balázs Papp, Wiktor Jurkowski, andEszter Ari. 2024. “mulea: An R Package for Enrichment Analysis UsingMultiple Ontologies and Empirical False Discovery Rate.”BMCBioinformatics 25 (1): 334.https://doi.org/10.1186/s12859-024-05948-7.
Korotkevich, Gennady, Vladimir Sukhov, Nikolay Budin, Boris Shpak, MaximN. Artyomov, and Alexey Sergushichev. 2021. “Fast Gene Set EnrichmentAnalysis.”bioRxiv, February.https://doi.org/10.1101/060012.
Kuleshov, Maxim V., Matthew R. Jones, Andrew D. Rouillard, Nicolas F.Fernandez, Qiaonan Duan, Zichen Wang, Simon Koplev, et al. 2016.“Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016Update.”Nucleic Acids Research 44 (W1): W90–97.https://doi.org/10.1093/nar/gkw377.
Méhi, Orsolya, Balázs Bogos, Bálint Csörgő, Ferenc Pál, Ákos Nyerges,Balázs Papp, and Csaba Pál. 2014. “Perturbation of Iron HomeostasisPromotes the Evolution of Antibiotic Resistance.”Molecular Biology andEvolution 31 (10): 2793–2804.https://doi.org/10.1093/molbev/msu223.
Subramanian, Aravind, Pablo Tamayo, Vamsi K. Mootha, Sayan Mukherjee,Benjamin L. Ebert, Michael A. Gillette, Amanda Paulovich, et al. 2005.“Gene Set Enrichment Analysis: A Knowledge-Based Approach forInterpreting Genome-Wide Expression Profiles.”Proceedings of theNational Academy of Sciences 102 (43): 15545–50.https://doi.org/10.1073/pnas.0506580102.
Turek, Cezary, Márton Ölbei, Tamás Stirling, Gergely Fekete, ErvinTasnádi, Leila Gul, Balázs Bohár, Balázs Papp, Wiktor Jurkowski, andEszter Ari. 2024. “mulea: An R Package forEnrichment Analysis Using Multiple Ontologies and Empirical FalseDiscovery Rate.”BMC Bioinformatics 25 (1): 334.https://doi.org/10.1186/s12859-024-05948-7.
Footnotes
The format of the actually used ontology slightly deviates fromstandard GMT files. In
tf_ontology, both theontology_idandontology_namecolumns containgene symbols of the transcriptionfactors, unlike other ontologies such as GO, where these columnshold specific identifiers and corresponding names.↩
About
mulea - an R package for enrichment analysis using various ontologies and empirical false discovery rate
Topics
Resources
Uh oh!
There was an error while loading.Please reload this page.
Stars
Watchers
Forks
Packages0
Languages
- R89.8%
- C++7.6%
- TeX2.6%