- Notifications
You must be signed in to change notification settings - Fork0
CimpleG, an R package to find (small) CpG signatures.
License
CostaLab/CimpleG
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation
CimpleG, an R package to find (small) CpG signatures.
# Install from CRAN:install.packages("CimpleG")# Install dev version from github:devtools::install_github("costalab/CimpleG")
library("CimpleG")data(train_data)data(train_targets)data(test_data)data(test_targets)# check the train_targets table to see# what other columns can be used as targets# colnames(train_targets)# mini example with just 4 target signaturesset.seed(42)cimpleg_result<- CimpleG(train_data=train_data,train_targets=train_targets,test_data=test_data,test_targets=test_targets,method="CimpleG",has_annotation=TRUE,target_columns= c("neurons","glia","blood_cells","fibroblasts" ))cimpleg_result$results
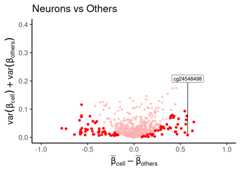
# check generated signaturescimpleg_result$signatures#> neurons glia blood_cells fibroblasts#> "cg24548498" "cg14501977" "cg04785083" "cg03369247"
# Get it directly from the results objectcimpleg_result$annotation#> # A tibble: 4 × 8#> IlmnID CHR_hg38 Start_hg38 End_hg38 UCSC_RefGene_Name UCSC_RefGene_Group#> <chr> <chr> <dbl> <dbl> <chr> <chr>#> 1 cg24548498 chr2 181684680 181684682 <NA> <NA>#> 2 cg14501977 chr12 123948446 123948448 CCDC92 5'UTR#> 3 cg04785083 chr1 8971202 8971204 CA6 Body#> 4 cg03369247 chr8 20174518 20174520 SLC18A1;SLC18A1;S… Body;Body;Body;Bo…#> # ℹ 2 more variables: UCSC_CpG_Islands_Name <chr>,#> # Relation_to_UCSC_CpG_Island <chr># or idependently through the "get_cpg_annotation" functionsignature_annotation<- get_cpg_annotation(cimpleg_result$signatures)# check signature annotationsignature_annotation#> # A tibble: 4 × 8#> IlmnID CHR_hg38 Start_hg38 End_hg38 UCSC_RefGene_Name UCSC_RefGene_Group#> <chr> <chr> <dbl> <dbl> <chr> <chr>#> 1 cg24548498 chr2 181684680 181684682 <NA> <NA>#> 2 cg14501977 chr12 123948446 123948448 CCDC92 5'UTR#> 3 cg04785083 chr1 8971202 8971204 CA6 Body#> 4 cg03369247 chr8 20174518 20174520 SLC18A1;SLC18A1;S… Body;Body;Body;Bo…#> # ℹ 2 more variables: UCSC_CpG_Islands_Name <chr>,#> # Relation_to_UCSC_CpG_Island <chr>
# adjust target names to match signature names# check generated signaturesplt<- signature_plot(cimpleg_result,train_data,train_targets,sample_id_column="gsm",true_label_column="cell_type")print(plt$plot)
We have two different functions to produce these plots, one with asimpler interface (and arguably cleaner look) than the other. I mightunify these interfaces in the future.
plt<- dmsv_plot(dat=train_data,target_vector=train_targets$neurons==1)print(plt)
plt<- diffmeans_sumvariance_plot(data=train_data,target_vector=train_targets$neurons==1)print(plt)
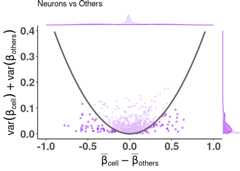
df_dmeansvar<- compute_diffmeans_sumvar(data=train_data,target_vector=train_targets$neurons==1)parab_param<-.7df_dmeansvar$is_selected<- select_features(x=df_dmeansvar$diff_means,y=df_dmeansvar$sum_variance,a=parab_param)
With the simpler interface
plt<- dmsv_plot(dat=df_dmeansvar,label_var1="Neurons",highlight_var="is_selected",display_var="is_selected",point_color="purple")print(plt)
With the more complex interface
plt<- diffmeans_sumvariance_plot(data=df_dmeansvar,label_var1="Neurons",color_all_points="purple",threshold_func=function(x,a) (a*x)^2,is_feature_selected_col="is_selected",func_factor=parab_param)print(plt)
# labeling best signature found by CimpleGdf_dmeansvar$best_neuron_sig<- (df_dmeansvar$id%in%cimpleg_result$signatures["neurons"])plt<- dmsv_plot(dat=df_dmeansvar,label_var1="Neurons",highlight_var="is_selected",display_var="best_neuron_sig",point_color="red")print(plt)
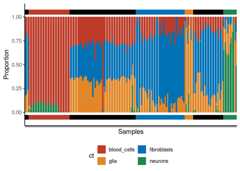
deconv_result<- run_deconvolution(cpg_obj=cimpleg_result,new_data=test_data)plt<- deconvolution_barplot(deconvoluted_data=deconv_result,meta_data=test_targets,sample_id="gsm",true_label="cell_type")print(plt$plot)
In this example, we’ll create two additional models made with CimpleG.One using only hypermethylated signatures, and the other using 3 CpGsper signature instead of just one. Then we will benchmark them againsteachother. This is similar to the approach that we use in the paperexcept there we use real data.
set.seed(42)cimpleg_hyper<- CimpleG(train_data=train_data,train_targets=train_targets,test_data=test_data,test_targets=test_targets,method="CimpleG",pred_type="hyper",target_columns= c("neurons","glia","blood_cells","fibroblasts" ))#> Training for target 'neurons' with 'CimpleG' has finished.: 0.251 sec elapsed#> Training for target 'glia' with 'CimpleG' has finished.: 0.253 sec elapsed#> Training for target 'blood_cells' with 'CimpleG' has finished.: 0.29 sec elapsed#> Training for target 'fibroblasts' with 'CimpleG' has finished.: 0.268 sec elapseddeconv_hyper<- run_deconvolution(cpg_obj=cimpleg_hyper,new_data=test_data)set.seed(42)cimpleg_3sigs<- CimpleG(train_data=train_data,train_targets=train_targets,test_data=test_data,test_targets=test_targets,method="CimpleG",n_sigs=3,target_columns= c("neurons","glia","blood_cells","fibroblasts" ))#> Training for target 'neurons' with 'CimpleG' has finished.: 0.315 sec elapsed#> Training for target 'glia' with 'CimpleG' has finished.: 0.296 sec elapsed#> Training for target 'blood_cells' with 'CimpleG' has finished.: 0.349 sec elapsed#> Training for target 'fibroblasts' with 'CimpleG' has finished.: 0.307 sec elapseddeconv_3sigs<- run_deconvolution(cpg_obj=cimpleg_3sigs,new_data=test_data)
deconv_3sigs$prop_3sigs<-deconv_3sigs$proportiondeconv_hyper$prop_hyper<-deconv_hyper$proportiondeconv_result$prop_cimpleg<-deconv_result$proportiondummy_deconvolution_data<-deconv_result|>dplyr::mutate(true_vals=proportion+ runif(nrow(deconv_result),min=-0.1,max=0.1))|>dplyr::select(cell_type,sample_id,prop_cimpleg,true_vals)|>dplyr::left_join(deconv_hyper|>dplyr::select(-proportion),by= c("sample_id","cell_type"))|>dplyr::left_join(deconv_3sigs|>dplyr::select(-proportion),by= c("sample_id","cell_type"))|>dplyr::mutate_if(is.numeric,function(x) { ifelse(x<0,0,x) })|>dplyr::mutate_if(is.numeric,function(x) { ifelse(x>1,1,x) })|>tibble::as_tibble()
scatter_plts<-CimpleG:::deconv_pred_obs_plot(deconv_df=dummy_deconvolution_data,true_values_col="true_vals",predicted_cols= c("prop_cimpleg","prop_hyper","prop_3sigs"),sample_id_col="sample_id",group_col="cell_type")scatter_panel<-scatter_plts|>patchwork::wrap_plots(ncol=1)print(scatter_panel)
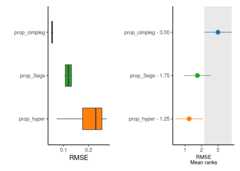
now, more interestingly, we can see in detail and rank one of the measures used to evaluate the deconvolution results
rank_plts<-CimpleG:::deconv_ranking_plot(deconv_df=dummy_deconvolution_data,true_values_col="true_vals",predicted_cols= c("prop_cimpleg","prop_hyper","prop_3sigs"),sample_id_col="sample_id",group_col="cell_type",metrics="rmse")rank_panel<-list(rank_plts$perf_boxplt[[1]],rank_plts$nemenyi_plt[[1]])|>patchwork::wrap_plots()print(rank_panel)
About
CimpleG, an R package to find (small) CpG signatures.
Topics
Resources
License
Uh oh!
There was an error while loading.Please reload this page.
Stars
Watchers
Forks
Packages0
Uh oh!
There was an error while loading.Please reload this page.