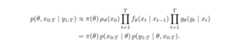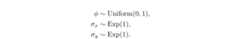- Notifications
You must be signed in to change notification settings - Fork0
Bayesian Methods for State Space Models
License
Unknown, MIT licenses found
Licenses found
BjarkeHautop/bayesSSM
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation
bayesSSM is an R package offering a set of tools for performing Bayesianinference in state-space models (SSMs). It implements the ParticleMarginal Metropolis-Hastings (PMMH) in the main functionpmmh forBayesian inference in SSMs.
While there are several alternative packages available for performingParticle MCMC, bayesSSM is designed to be simple and easy to use. It wasdeveloped alongside my Master’s thesis about Particle MCMC, since I wasimplementing everything from scratch anyway.
In some state-space models, the full joint density of the parameters andthe latent states
can be written out explicitly. In these cases, inference tools like Stan(which uses Hamiltonian Monte Carlo) are very efficient.
However, many real-world models define the latent dynamics only throughasimulator, not via an explicit transition density. This includes:
- Epidemic models with stochastic transmission dynamics
- Agent-based models
- Ecological or physical systems with complex latent dynamics
In these cases, the joint posterior density cannot be computed directly,and the marginal likelihood is intractable. Thus, standard MCMC methodslike Hamiltonian Monte Carlo (HMC) or Metropolis-Hastings (MH) cannot beapplied directly.
Particle Markov Chain Monte Carlo (PMCMC) methods, such as the ParticleMarginal Metropolis-Hastings (PMMH) implemented in this package, aredesigned to handle these situations. They use particle filters toapproximate the marginal likelihood and allow for efficient samplingfrom the joint posterior density.
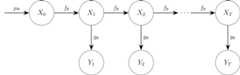
A state-space model (SSM) has the structure given in the followingdiagram, where we omitted potential time dependency in the transitionand observation densities for simplicity.
The core function,pmmh, implements Particle MarginalMetropolis-Hastings, which is an algorithm that first generates a set of
You can install the latest stable version of bayesSSM from CRAN with:
install.packages("bayesSSM")or the development version from GitHub with:
# install.packages("pak")pak::pak("BjarkeHautop/bayesSSM")
To illustrate how to use thepmmh function, we will simulate a simplestate-space model (SSM) and perform Bayesian inference on it. Note:While this example usespmmh, the model is simple enough that standardMCMC methods could also be applied. For a more complicated example wherestandard MCMC methods cannot be used, see the article Stochastic SIRModelhere.
We will simulate a state-space model with the following structure:
Let’s first simulate 20 data points from this model with
set.seed(1405)t_val<-20phi<-0.8sigma_x<-1sigma_y<-0.5init_state<- rnorm(1,mean=0,sd=1)x<-numeric(t_val)y<-numeric(t_val)x[1]<-phi*init_state+ sin(init_state)+ rnorm(1,mean=0,sd=sigma_x)y[1]<-x[1]+ rnorm(1,mean=0,sd=sigma_y)for (tin2:t_val) {x[t]<-phi*x[t-1]+ sin(x[t-1])+ rnorm(1,mean=0,sd=sigma_x)y[t]<-x[t]+ rnorm(1,mean=0,sd=sigma_y)}x<- c(init_state,x)
We define the priors for our model as follows:
We can usepmmh to perform Bayesian inference on this model. To usepmmh we need to define the functions for the SSM and the priors.
The functionsinit_fn,transition_fn should be functions thatsimulate the latent states.init_fn must contain the argumentnum_particles for initializing the particles, andtransition_fn mustcontain the argumentparticles, which is a vector of particles, andcan contain any other arguments for model-specific parameters.
The functionlog_likelihood_fn should be a function that calculatesthe log-likelihood of the observed data given the latent statevariables. It must contain the argumentsy for the data andparticles. Time dependency can be implemented by giving at argumentintransition_fn andlog_likelihood_fn.
init_fn<-function(num_particles) { rnorm(num_particles,mean=0,sd=1)}transition_fn<-function(particles,phi,sigma_x) {phi*particles+ sin(particles)+ rnorm(length(particles),mean=0,sd=sigma_x)}log_likelihood_fn<-function(y,particles,sigma_y) { dnorm(y,mean=particles,sd=sigma_y,log=TRUE)}
The priors for the parameters must be defined as log-prior functions.Every parameter frominit_fn,transition_fn, andlog_likelihood_fnmust have a corresponding log-prior function.
log_prior_phi<-function(phi) { dunif(phi,min=0,max=1,log=TRUE)}log_prior_sigma_x<-function(sigma) { dexp(sigma,rate=1,log=TRUE)}log_prior_sigma_y<-function(sigma) { dexp(sigma,rate=1,log=TRUE)}log_priors<-list(phi=log_prior_phi,sigma_x=log_prior_sigma_x,sigma_y=log_prior_sigma_y)
Now we can run the PMMH algorithm using thepmmh function. For thisREADME we use a lower number of samples and a smaller burn-in period,and also modify the pilot chains to only use 200 samples. This is doneto make the example run faster.
library(bayesSSM)result<- pmmh(y=y,m=500,# number of MCMC samplesinit_fn=init_fn,transition_fn=transition_fn,log_likelihood_fn=log_likelihood_fn,log_priors=log_priors,pilot_init_params=list( c(phi=0.4,sigma_x=0.4,sigma_y=0.4), c(phi=0.8,sigma_x=0.8,sigma_y=0.8) ),burn_in=50,num_chains=2,seed=1405,tune_control= default_tune_control(pilot_m=200,pilot_burn_in=10))#> Running chain 1...#> Running pilot chain for tuning...#> Using 50 particles for PMMH:#> Running Particle MCMC chain with tuned settings...#> Running chain 2...#> Running pilot chain for tuning...#> Using 50 particles for PMMH:#> Running Particle MCMC chain with tuned settings...#> PMMH Results Summary:#> Parameter Mean SD Median 2.5% 97.5% ESS Rhat#> phi 0.80 0.10 0.82 0.60 0.97 10 1.228#> sigma_x 0.58 0.50 0.46 0.00 1.71 2 1.376#> sigma_y 0.97 0.35 1.05 0.13 1.46 5 1.272#> Warning in pmmh(y = y, m = 500, init_fn = init_fn, transition_fn =#> transition_fn, : Some ESS values are below 400, indicating poor mixing.#> Consider running the chains for more iterations.#> Warning in pmmh(y = y, m = 500, init_fn = init_fn, transition_fn = transition_fn, :#> Some Rhat values are above 1.01, indicating that the chains have not converged.#> Consider running the chains for more iterations and/or increase burn_in.
We get convergence warnings as expected due to the small number ofiterations.
About
Bayesian Methods for State Space Models
Topics
Resources
License
Unknown, MIT licenses found
Licenses found
Uh oh!
There was an error while loading.Please reload this page.
Stars
Watchers
Forks
Packages0
Uh oh!
There was an error while loading.Please reload this page.