This section describes the core functionalities ofclustimage.Many of the functionalities are written in a generic manner which allows to be used in various applications.
Core functionalities
The are 5 core functionalities ofclustimage that allows to preprocess the input images, robustly determines the optimal number of clusters, and then optimize the clusters if desired.
fit_transform()
extract_faces()
cluster()
find()
unique()
move_to_dir()
Fit and transform
Thefit_transform function allows to detect natural groups or clusters of images. It works using a multi-step proces of pre-processing, extracting the features, and evaluating the optimal number of clusters across the feature space.The optimal number of clusters are determined using well known methods such assilhouette, dbindex, and derivatives in combination with clustering methods, such asagglomerative, kmeans, dbscan and hdbscan.Based on the clustering results, the unique images are also gathered.
Examples can be found here:clustimage.clustimage.Clustimage.fit_transform()
The fit_transform contains 4 core functionalities that can also be used seperatly which provides more control:
import_data()
extract_feat()
embedding()
cluster()
import_data
The input for theclustimage.clustimage.Clustimage.import_data() can have multiple forms; path to directory, list of strings and and array-like input.The following steps are used for which the parameters needs to be set during initialization:
Images are imported with specific extention ([‘png’,’tiff’,’jpg’]),
Each input image can be grayscaled.
Resizing images in the same dimension such as (128,128). Note that if an array-like dataset [Samples x Features] is given as input, setting these dimensions are required to restore the image in case of plotting.
Independent of the input, a dict is returned in a consistent manner.
# Initializecl=Clustimage(method='pca')# Import dataX=cl.import_example(data='flowers')# Check whether in is dir, list of files or array-likeX=cl.import_data(X)print(cl.results.keys())# dict_keys(['img', 'feat', 'xycoord', 'pathnames', 'labels', 'filenames'])# Note that only the keys img, pathnames and filenames are filled.
extract_feat
Extracting of features is performed in theclustimage.clustimage.Clustimage.extract_feat() function.There are different options the extract features from the image as lised below. Note that these settings needs to be set during initialization.
‘pca’ : PCA feature extraction
‘hog’ : hog features extraced
‘pca-hog’ : PCA extracted features from the HOG desriptor
‘ahash’: Average hash
‘phash’: Perceptual hash
‘dhash’: Difference hash
‘whash-haar’: Haar wavelet hash
‘whash-db4’: Daubechies wavelet hash
‘colorhash’: HSV color hash
‘crop-resistant’: Crop-resistant hash
‘datetime’: datetime are extracted using EXIF metadata
‘latlon’: lat/lon coordinates are extracted using EXIF metadata
None : No feature extraction
# Initializecl=Clustimage(method='pca')# Import dataX=cl.import_example(data='flowers')# Check whether in is dir, list of files or array-likeX=cl.import_data(X)# Extract features using methodXfeat=cl.extract_feat(X)print(cl.results.keys())# dict_keys(['img', 'feat', 'xycoord', 'pathnames', 'labels', 'filenames'])# At this point, the key: 'feat' is filled.
embedding
The embedding is performed using tSNE in theclustimage.clustimage.Clustimage.embedding() function.The coordinates are used for vizualiation purposes only but if desired. However, when setting thecluster_space parameter to ‘low’ in thecluster function, the clustering will be performed in the low-dimensional tSNE space.
# Initializecl=Clustimage(method='pca')# Import dataX=cl.import_example(data='flowers')# Check whether in is dir, list of files or array-likeX=cl.import_data(X)# Extract features using methodXfeat=cl.extract_feat(X)# Embedding using tSNExycoord=cl.embedding(Xfeat)print(cl.results.keys())# dict_keys(['img', 'feat', 'xycoord', 'pathnames', 'labels', 'filenames'])# At this point, the key: 'xycoord' is filled.
cluster
Thecluster function is build on`clusteval`_, which is a python package that provides various evalution methods for unsupervised cluster validation.The optimal number of clusters are determined using well known methods such assilhouette, dbindex, and derivatives in combination with clustering methods, such asagglomerative, kmeans, dbscan and hdbscan.This function can be run after thefit_transform function to solely optimize the clustering results or try-out different evaluation approaches without repeately performing all the steps of preprocessing.Besides changing evaluation methods and metrics, it is also possible to cluster on the low-embedded feature space. This can be done setting the parametercluster_space='low'.
# Initializecl=Clustimage(method='pca')# Import dataX=cl.import_example(data='flowers')# Check whether in is dir, list of files or array-likeX=cl.import_data(X)# Extract features using methodXfeat=cl.extract_feat(X)# Embedding using tSNExycoord=cl.embedding(Xfeat)# Clusterlabels=cl.cluster(cluster='agglomerative',evaluate='silhouette',metric='euclidean',linkage='ward',min_clust=3,max_clust=25,cluster_space='high')print(cl.results.keys())# dict_keys(['img', 'feat', 'xycoord', 'pathnames', 'labels', 'filenames'])# At this point, the key: 'labels' is filled.
More examples can also be found here:clustimage.clustimage.Clustimage.cluster()
extract_faces
To cluster faces on images, we first need to detect, and extract the faces from the images.Theextract_faces function does this task.Faces and eyes are detected usinghaarcascade_frontalface_default.xml andhaarcascade_eye.xml inpython-opencv.
Examples can be found here:clustimage.clustimage.Clustimage.extract_faces()
find
Thefind functionclustimage.clustimage.Clustimage.find() allows to find images that are similar to that of the input image.Finding images can be performed in two manners:
Based on the k-nearest neighbour
Based on significance after probability density fitting
In both cases, the adjacency matrix is first computed using the distance metric (default Euclidean).In case of the k-nearest neighbour approach, the k nearest neighbours are determined.In case of significance, the adjacency matrix is used to to estimate the best fit for the loc/scale/arg parameters across various theoretical distribution.The tested disributions are[‘norm’, ‘expon’, ‘uniform’, ‘gamma’, ‘t’]. The fitted distribution is basically the similarity-distribution of samples.For each new (unseen) input image, the probability of similarity is computed across all images, and the images are returned that are P <=alpha in the lower bound of the distribution.If case bothk andalpha are specified, the union of detected samples is taken.Note that the metric can be changed in this function but this may lead to confusions as the results will not intuitively match with the scatter plots as these are determined using metric in the fit_transform() function.
Example to find similar images using 1D vector as input image.
fromclustimageimportClustimageimportmatplotlib.pyplotaspltimportpandasaspd# Init with default settingscl=Clustimage(method='pca')# load example with digitsX,y=cl.import_example(data='mnist')# Cluster digitsresults=cl.fit_transform(X)# Lets search for the following image:plt.figure();plt.imshow(X[0,:].reshape(cl.params['dim']),cmap='binary')# Find imagesresults_find=cl.find(X[0:3,:],k=None,alpha=0.05)# Show whatever is found. This looks pretty good.cl.plot_find()cl.scatter(zoom=3)# Extract the first input image namefilename=[*results_find.keys()][1]# Plot the probabilitiesplt.figure(figsize=(8,6))plt.plot(results_find[filename]['y_proba'],'.')plt.grid(True)plt.xlabel('samples')plt.ylabel('Pvalue')# Extract the cluster labels for the input imageresults_find[filename]['labels']# The majority of labels is for class 0print(pd.value_counts(results_find[filename]['labels']))# 0 171# 7 8# Name: labels, dtype: int64
|
|
|
|
** Example to find similar images based on the pathname as input.**
fromclustimageimportClustimage# Init with default settingscl=Clustimage(method='pca')# load example with flowerspathnames=cl.import_example(data='flowers')# Cluster flowersresults=cl.fit_transform(pathnames[1:])# Lets search for the following image:img=cl.imread(pathnames[10],colorscale=1)plt.figure();plt.imshow(img.reshape((128,128,3)));plt.axis('off')# Find imagesresults_find=cl.find(pathnames[10],k=None,alpha=0.05)# Show whatever is found. This looks pretty good.cl.plot_find()cl.scatter()
|
|
Examples can be found here:clustimage.clustimage.Clustimage.find()
unique
The unique images can be computed using the uniqueclustimage.clustimage.Clustimage.unique() and are detected by first computing the center of the cluster, and then taking the image closest to the center.Lets demonstrate this by example and the digits dataset.
fromclustimageimportClustimage# Init with default settingscl=Clustimage(method='pca')# load example with digitsX,y=cl.import_example(data='mnist')# Find natural groups of digitsresults=cl.fit_transform(X)# Show the unique detected imagescl.results_unique.keys()# Plot the digit that is located in the center of the clustercl.plot_unique(img_mean=False)# Average the image per cluster and plotcl.plot_unique()# Compute again with other metric desiredcl.unique()
|
|
Preprocessing
The preprocessing step is the functionclustimage.clustimage.Clustimage.imread(), and contains 3 functions to handle the import, scaling and resizing of images.This function requires the full path to the image for which the first step is reading the images and colour scaling it based on the input parametergrayscale.Ifgrayscale is set toTrue, thecv2.COLOR_GRAY2RGB setting frompython-opencv is used.
The pre-processing has 4 steps and are exectued in this order.
Import data.
Conversion to gray-scale (user defined)
Scaling color pixels between [0-255]
Resizing
# Import librariesfromclustimageimportClustimageimportmatplotlib.pyplotasplt# Initcl=Clustimage()# Load example datasetpathnames=cl.import_example(data='flowers')# Preprocessing of the first imageimg=cl.imread(pathnames[0],dim=(128,128))# Plotplt.figure()plt.imshow(img.reshape(128,128,3))plt.axis('off')
|
|
imscale
Theimscale functionclustimage.clustimage.Clustimage.imscale() is only applicable for 2D-arrays (images).Scaling data is an import pre-processing step to make sure all data is ranged between the minimum and maximum range.
The images are scaled between [0-255] by the following equation:
Ximg * (255 / max(Ximg) )
imresize
Theimresize functionclustimage.clustimage.imresize() resizes the images into 128x128 pixels (default) or to an user-defined size.The function depends on the functionality ofpython-opencv with the interpolation:interpolation=cv2.INTER_AREA.
Output
The results obtained from theclustimage.clustimage.Clustimage.fit_transform() orclustimage.clustimage.Clustimage.cluster() is a dictionary containing the following keys:
*img:Imagevectorofthepreprocessedimages.*feat:Extractedfeature.*xycoord:XandYcoordinatesfromtheembedding.*pathnames:Absolutepathlocationtotheimagefile.*filenames:Filenamesoftheimagefile.*labels:Clusterlabels.
For demonstration purposes I will load the flowers dataset and cluster the images.
# Import libraryfromclustimageimportClustimage# Initializecl=Clustimage(method='pca')# Import datapathnames=cl.import_example(data='flowers')# Cluster flowersresults=cl.fit_transform(pathnames)
# All results are stored in a dict:print(cl.results.keys())# Which is the same as:print(results.keys())# dict_keys(['img', 'feat', 'xycoord', 'pathnames', 'labels', 'filenames'])
Extract images that belong to a certain cluster and make some plots.
# Extracting images that belong to cluster label=0:label=0Iloc=cl.results['labels']==labelpathnames=cl.results['pathnames'][Iloc]# Extracting xy-coordinates for the scatterplot for cluster 0:importmatplotlib.pyplotaspltxycoord=cl.results['xycoord'][Iloc]plt.figure()plt.scatter(xycoord[:,0],xycoord[:,1])plt.title('Cluster%.0d'%label)# Plot the images for cluster 0:imgs=cl.results['img'][Iloc]# Make sure you get the right dimensiondim=cl.get_dim(cl.results['img'][Iloc][0,:])# Plotforimg,pathnameinzip(imgs,pathnames):plt.figure()plt.imshow(img.reshape(dim))plt.title(pathname)
Generic functions
clustimage contains various generic functionalities that are internally used but may be usefull too in other applications.
wget
Download files from the internet and store on disk.Examples can be found here:clustimage.clustimage.wget()
# Import libraryimportclustimageascl# Downloadimages=cl.wget('https://erdogant.github.io/datasets/flower_images.zip','c://temp//flower_images.zip')
unzip
Unzip files into a destination directory.Examples can be found here:clustimage.clustimage.unzip()
# Import libraryimportclustimageascl# Unzip to pathdirpath=cl.unzip('c://temp//flower_images.zip')
listdir
Recusively list the files in the directory.Examples can be found here:clustimage.clustimage.listdir()
# Import libraryimportclustimageascl# Unzip to pathdirpath='c://temp//flower_images'pathnames=cl.listdir(dirpath,ext=['png'])
set_logger
Change status of the logger.Examples can be found here:clustimage.clustimage.set_logger()
# Change to verbosity message of warnings and higherset_logger(verbose=30)
extract_hog
Histogram of Oriented Gradients (HOG), is a feature descriptor that is often used to extract features from image data.Examples:clustimage.clustimage.Clustimage.extract_hog() More detailed explanation can be found in theFeature Extraction -HOG section.
Merge/ Expand Clusters
The number of clusters are optimized using the clusteval library. However, when desired it is also possible to manually merge of expand the number of clusters.
Optimized Clusters
importnumpyasnpimportmatplotlib.pyplotaspltfromclustimageimportClustimage# Initializecl=Clustimage()# Import dataX=cl.import_example(data='flowers')# Fit transformcl.fit_transform(X)# Check number of clusterslen(np.unique(cl.results['labels']))# 9# Scattercl.scatter(dotsize=75)# Create dendrogramcl.dendrogram();
|
|
Force to K clusters
Let’s merge some of the clusters and set it to 5 clusters.
# Set to 5 clusterslabels=cl.cluster(min_clust=5,max_clust=5)# Check number of clusterslen(np.unique(cl.results['labels']))# 5# Scattercl.scatter(dotsize=75)# Create dendrogramcl.dendrogram();
|
|
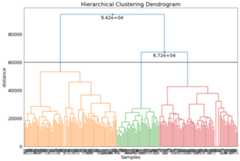
Set clusters by dendrogram threshold
Another manner to change the number of cluster is by specifying the height of the dendrogram (setting a threshold point or cut-off). The number of clusters is automatically derived from that point.
# Look at the dendrogram y-axis and specify the height to merge clustersdendro_results=cl.dendrogram(max_d=60000)# Check number of clusterslen(np.unique(cl.results['labels']))# 3# Scattercl.scatter(dotsize=75)
|
|
Selection on the cluster labels
# All results are stored in cl.resultscl.results.keys()['img','feat','xycoord','pathnames','labels','url','filenames','predict']# The cluster labels are stored in labelscl.results['labels']# Select cluster 0Iloc=cl.results['labels']==0# Select files for cluster 0cl.results['pathnames'][Iloc]# Select filenames for cluster 0cl.results['filenames'][Iloc]# Select xy-coordinates for cluster 0cl.results['xycoord'][Iloc]
Move files based on clusterlabels
Themove_to_dir function moves files physically files into subdirectories based on duplicates.First perform the desired undoubling approach and with the following steps it allows you to organize the images by physically moving them into subdirectories.
# Show the cluster labelsprint(cl.results['labels'])# Step 1. Use the plot function to determine what event the cluster of photos represents.# Do not plot cluster 0 (rest group), and only plot when a cluster contain 3 or more images.cl.plot(blacklist=[0],min_samples=3)# 2. Create a dict that specifies the cluster number with its folder names.# The first column is the cluster label and the second string is the destinated subfolder name.# Look at the clusters and specify the subdirectories. In my case, cluster 1 are holiday photos, cluster 2 are various photos and cluster 6 are screenshots.# All images that are not in the clusters, will remain untouched.target_labels={1:'holiday photos',2:'various',6:'screenshots',}# 3. Move the photos to the specified directories using the cluster labels.cl.move_to_dir(target_labels=target_labels)
Plotting
clustimage has various plotting functionalities.
plot: plots images in the clusters
plot_faces : plots the faces.
plot_find: Plot the input image together with the predicted images.
plot_unique: plot the unique images.
dendrogram: Plot Dendrogram.
plot_map: plots the images on map. Requires to use method=”exif”