OverviewData source: UniProt
| Official Gene Symbol | Other Aliases |
|---|---|
| MAN2B1 | LAMAN, MANB |
| Sequence Length (AA) | Molecular Weight (Da) |
|---|---|
| 1011 | 113744 |
| Protein Name |
|---|
| Lysosomal alpha-mannosidase |
| Sources | |
|---|---|
| UniProt PhosphoSitePlus ® GeneCards | Human Protein Atlas |
Protein Sequence hover to view complete sequence
| 10 | 20 | 30 | 40 | 50 |
| MGAYARASGV | CARGCLDSAG | PWTMSRALRP | PLPPLCFFLL | LLAAAGARAG |
| 60 | 70 | 80 | 90 | 100 |
| GYETCPTVQP | NMLNVHLLPH | THDDVGWLKT | VDQYFYGIKN | DIQHAGVQYI |
| 110 | 120 | 130 | 140 | 150 |
| LDSVISALLA | DPTRRFIYVE | IAFFSRWWHQ | QTNATQEVVR | DLVRQGRLEF |
| 160 | 170 | 180 | 190 | 200 |
| ANGGWVMNDE | AATHYGAIVD | QMTLGLRFLE | DTFGNDGRPR | VAWHIDPFGH |
| 210 | 220 | 230 | 240 | 250 |
| SREQASLFAQ | MGFDGFFFGR | LDYQDKWVRM | QKLEMEQVWR | ASTSLKPPTA |
| 260 | 270 | 280 | 290 | 300 |
| DLFTGVLPNG | YNPPRNLCWD | VLCVDQPLVE | DPRSPEYNAK | ELVDYFLNVA |
| 310 | 320 | 330 | 340 | 350 |
| TAQGRYYRTN | HTVMTMGSDF | QYENANMWFK | NLDKLIRLVN | AQQAKGSSVH |
| 360 | 370 | 380 | 390 | 400 |
| VLYSTPACYL | WELNKANLTW | SVKHDDFFPY | ADGPHQFWTG | YFSSRPALKR |
| 410 | 420 | 430 | 440 | 450 |
| YERLSYNFLQ | VCNQLEALVG | LAANVGPYGS | GDSAPLNEAM | AVLQHHDAVS |
| 460 | 470 | 480 | 490 | 500 |
| GTSRQHVAND | YARQLAAGWG | PCEVLLSNAL | ARLRGFKDHF | TFCQQLNISI |
| 510 | 520 | 530 | 540 | 550 |
| CPLSQTAARF | QVIVYNPLGR | KVNWMVRLPV | SEGVFVVKDP | NGRTVPSDVV |
| 560 | 570 | 580 | 590 | 600 |
| IFPSSDSQAH | PPELLFSASL | PALGFSTYSV | AQVPRWKPQA | RAPQPIPRRS |
| 610 | 620 | 630 | 640 | 650 |
| WSPALTIENE | HIRATFDPDT | GLLMEIMNMN | QQLLLPVRQT | FFWYNASIGD |
| 660 | 670 | 680 | 690 | 700 |
| NESDQASGAY | IFRPNQQKPL | PVSRWAQIHL | VKTPLVQEVH | QNFSAWCSQV |
| 710 | 720 | 730 | 740 | 750 |
| VRLYPGQRHL | ELEWSVGPIP | VGDTWGKEVI | SRFDTPLETK | GRFYTDSNGR |
| 760 | 770 | 780 | 790 | 800 |
| EILERRRDYR | PTWKLNQTEP | VAGNYYPVNT | RIYITDGNMQ | LTVLTDRSQG |
| 810 | 820 | 830 | 840 | 850 |
| GSSLRDGSLE | LMVHRRLLKD | DGRGVSEPLM | ENGSGAWVRG | RHLVLLDTAQ |
| 860 | 870 | 880 | 890 | 900 |
| AAAAGHRLLA | EQEVLAPQVV | LAPGGGAAYN | LGAPPRTQFS | GLRRDLPPSV |
| 910 | 920 | 930 | 940 | 950 |
| HLLTLASWGP | EMVLLRLEHQ | FAVGEDSGRN | LSAPVTLNLR | DLFSTFTITR |
| 960 | 970 | 980 | 990 | 1000 |
| LQETTLVANQ | LREAASRLKW | TTNTGPTPHQ | TPYQLDPANI | TLEPMEIRTF |
| 1010 | 1011 | |||
| LASVQWKEVD | G |
Data source: UniProt
Protein MapCollapse protein map
Position of Targeted Peptide Analytes Relative to SNPs, Isoforms, and PTMs
Uniprot Database EntryPhosphoSitePlus ®
loading

Assay Details for CPTAC-2225Collapse assay details
Data source: Panorama
- Official Gene Symbol
- MAN2B1
- Peptide Modified Sequence
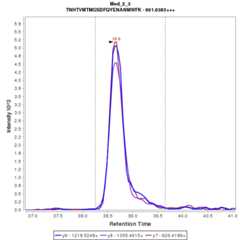
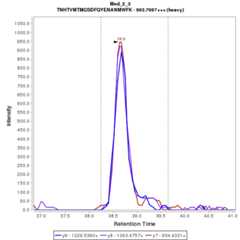
- TN[+1.0]HTVM[+16.0]TM[+16.0]GSDFQYENANM[+16.0]WFK
- Modification Type
- Deamidated (N), Oxidation (M), Oxidation (M), Oxidation (M)
- Protein - Site of Modification
- 310, 314, 316, 327
- Peptide - Site of Modification
- 2, 6, 8, 19
- Peptide Start
- 309
- Peptide End
- 330
- CPTAC ID
- CPTAC-2225
- Peptide Molecular Mass
- 2,700.0931
- Species
- Homosapiens (Human)
- Assay Type
- Direct MRM
- Matrix
- Human ovarian tissue digest
- Submitting Laboratory
- Johns Hopkins University
- Submitting Lab PI
- Daniel Chan, Hui Zhang, Zhen Zhang
Assay ParametersCollapse assay parameters
Data source: Panorama
- Instrument
- LCMS-8040
- Internal Standard
- Stable isotope-labeled peptides
- Peptide Standard Purity
- Crude (~60%)
- Peptide Standard Label Type
- 13C and 15N at C-terminus K
- LC
- Nexera X2 UHPLC
- Column Packing
- C18, 3 µm, 300 Å
- Column Dimensions
- 1.0 mm I.D. x 15 cm
- Flow Rate
- 50 µL/min
Assay MultiplexingExpand assay panel
Johns Hopkins University-G
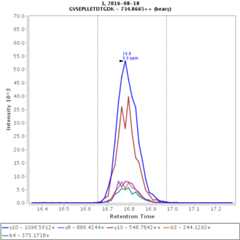
Chromatograms
Data source: Panorama
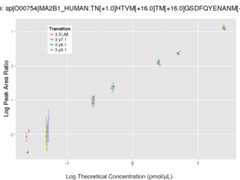
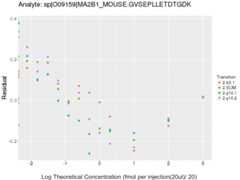
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| Average intra-assay CV (within day CV) | Average inter-assay CV (between day CV) | Total CV | n= | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| y7 (1+) | 26.3 | 8.2 | 10.3 | 24.9 | 9.3 | 11 | 36.2 | 12.4 | 15.1 | 15 | 15 | 15 |
| y8 (1+) | 22 | 11.6 | 7.3 | 19.8 | 9.5 | 6.3 | 29.6 | 15 | 9.6 | 15 | 15 | 15 |
| y9 (1+) | 19.4 | 8.1 | 12.2 | 22.2 | 7.5 | 8.4 | 29.5 | 11 | 14.8 | 15 | 15 | 15 |
| sum | 15.5 | 6.2 | 7.4 | 14.4 | 5.2 | 6.1 | 21.2 | 8.1 | 9.6 | 15 | 15 | 15 |
Additional Resources and Comments
Comments
Assay Details for non-CPTAC-3591Collapse assay details
Data source: Panorama
- Official Gene Symbol
- Man2b1
- Peptide Sequence
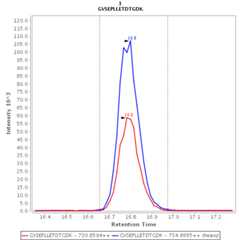
- GVSEPLLETDTGDK
- Modification Type
- unmodified
- Protein - Site of Modification
- N/A
- Peptide - Site of Modification
- N/A
- Peptide Start
- 825
- Peptide End
- 838
- CPTAC ID
- non-CPTAC-3591
- Peptide Molecular Mass
- 1,459.7042
- Species
- Musmusculus (Mouse)
- Assay Type
- Direct MRM
- Matrix
- Plasma
- Submitting Laboratory
- UVic-Genome BC Proteomics Centre
- Submitting Lab PI
- Christoph Borchers
Publication
View Details(opens in a new window)
Molecular phenotyping of laboratory mouse strains using 500 multiple reaction monitoring mass spectrometry plasma assays. Michaud SA, Sinclair NJ, Petrošová H, Palmer AL, Pistawka AJ, Zhang S, Hardie DB, Mohammed Y Eshghi A1, Richard VR, Sickmann A, Borchers CH. Commun Biol. 2018 Jun 27;1:78. doi: 10.1038/s42003-018-0087-6. eCollection 2018.
Assay ParametersCollapse assay parameters
Data source: Panorama
- Instrument
- Agilent 6490/6495 QQQ
- Internal Standard
- synthetic peptide
- Peptide Standard Purity
- >95%
- Peptide Standard Label Type
- 13C and 15N at C-terminus K
- LC
- 1290 LC (Agilent)
- Column Packing
- Zorbax Eclipse Plus C18, 1.8 µm
- Column Dimensions
- 2.1 x 150 mm
- Flow Rate
- 400 µL/min
Assay MultiplexingExpand assay panel
Johns Hopkins University-G
Chromatograms
Data source: Panorama
Response Curves
Data source: Panorama
Retrieving Data

Repeatability
Data source: Panorama
| Average intra-assay CV (within day CV) | Average inter-assay CV (between day CV) | Total CV | n= | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fragment ion / Transition | Low | Med | High | Low | Med | High | Low | Med | High | Low | Med | High |
| b4 (1+) | 19 | 5.7 | 5 | 17.5 | 8.1 | 9.9 | 25.8 | 9.9 | 11.1 | 15 | 15 | 15 |
| b3 (1+) | 20.1 | 5.3 | 3.7 | 24.5 | 8.1 | 13 | 31.7 | 9.7 | 13.5 | 15 | 15 | 15 |
| y10 (2+) | 10.7 | 6 | 4.7 | 12.7 | 8.2 | 6.7 | 16.6 | 10.2 | 8.2 | 15 | 15 | 15 |
| y8 (1+) | 9.1 | 6.1 | 7.4 | 8.2 | 8.7 | 7.5 | 12.2 | 10.6 | 10.5 | 15 | 15 | 15 |
| y10 (1+) | 5.3 | 5 | 5.2 | 7.2 | 9.2 | 8.2 | 8.9 | 10.5 | 9.7 | 15 | 15 | 15 |
| sum | 4.3 | 1.5 | 2.6 | 6.5 | 6.6 | 6.9 | 7.8 | 6.8 | 7.4 | 15 | 15 | 15 |