Heart Failure
The following example is a binary classification domain adaptation issue based on the heart failure UCI dataset:
https://archive.ics.uci.edu/ml/datasets/Heart+failure+clinical+records
The goal is to predict the survival of patients with heart failure (binary). Two domains are defined by splitting the dataset along the “sex” binary variable.
[15]:
importosimportadaptimportnumpyasnpimportpandasaspdimporttensorflowastfimportmatplotlib.pyplotaspltimportnumpyasnpfromsklearn.preprocessingimportOneHotEncoder,StandardScalerfromsklearn.decompositionimportPCAfromsklearn.manifoldimportTSNEfromsklearn.metricsimportaccuracy_scorefromsklearn.datasetsimportmake_moonsfromtensorflow.kerasimportModel,Sequentialfromtensorflow.keras.optimizersimportAdam,SGD,RMSprop,Adagradfromtensorflow.keras.layersimportDense,Input,Dropout,Conv2D,MaxPooling2D,Flatten,Reshape,GaussianNoise,BatchNormalizationfromtensorflow.keras.constraintsimportMinMaxNormfromtensorflow.keras.regularizersimportl2fromadapt.feature_basedimportDANN,ADDA,DeepCORAL,CORAL,MCD,MDD,WDGRL,CDAN
Setup
[16]:
df=pd.read_csv("https://archive.ics.uci.edu/ml/machine-learning-databases/00519/heart_failure_clinical_records_dataset.csv")df.head(3)
[16]:
| age | anaemia | creatinine_phosphokinase | diabetes | ejection_fraction | high_blood_pressure | platelets | serum_creatinine | serum_sodium | sex | smoking | time | DEATH_EVENT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 75.0 | 0 | 582 | 0 | 20 | 1 | 265000.00 | 1.9 | 130 | 1 | 0 | 4 | 1 |
| 1 | 55.0 | 0 | 7861 | 0 | 38 | 0 | 263358.03 | 1.1 | 136 | 1 | 0 | 6 | 1 |
| 2 | 65.0 | 0 | 146 | 0 | 20 | 0 | 162000.00 | 1.3 | 129 | 1 | 1 | 7 | 1 |
[17]:
Xs=df.loc[df["sex"]==0].drop(["DEATH_EVENT"],axis=1).drop(["sex","creatinine_phosphokinase"],axis=1).valuesys=df.loc[df["sex"]==0,"DEATH_EVENT"].valuesXt=df.loc[df["sex"]==1].drop(["DEATH_EVENT"],axis=1).drop(["sex","creatinine_phosphokinase"],axis=1).valuesyt=df.loc[df["sex"]==1,"DEATH_EVENT"].valuesstd_sc=StandardScaler().fit(np.concatenate((Xs,Xt)))Xs=std_sc.transform(Xs)Xt=std_sc.transform(Xt)
[18]:
pca=PCA(2).fit(np.concatenate((Xs,Xt)))Xs_pca=pca.transform(Xs)Xt_pca=pca.transform(Xt)x_min,y_min=np.min([Xs_pca.min(0),Xt_pca.min(0)],0)x_max,y_max=np.max([Xs_pca.max(0),Xt_pca.max(0)],0)x_grid,y_grid=np.meshgrid(np.linspace(x_min-0.1,x_max+0.1,100),np.linspace(y_min-0.1,y_max+0.1,100))X_grid=pca.inverse_transform(np.stack([x_grid.ravel(),y_grid.ravel()],-1))fig,ax1=plt.subplots(1,1,figsize=(6,5))ax1.set_title("Input space")ax1.scatter(Xs_pca[ys==0,0],Xs_pca[ys==0,1],label="source",edgecolors='k',c="red")ax1.scatter(Xs_pca[ys==1,0],Xs_pca[ys==1,1],label="source",edgecolors='k',c="blue")ax1.scatter(Xt_pca[:,0],Xt_pca[:,1],label="target",edgecolors='k',c="black",alpha=0.3)ax1.legend()ax1.set_yticklabels([])ax1.set_xticklabels([])ax1.tick_params(direction='in')plt.show()
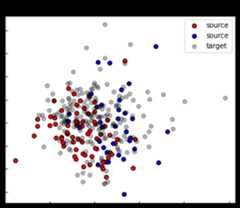
[19]:
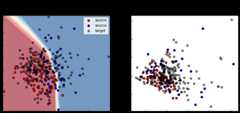
defshow_results(model,is_src_only=False):ys_pred=model.predict(Xs)yt_pred=model.predict(Xt)acc_s=accuracy_score(ys,ys_pred>0.5)acc_t=accuracy_score(yt,yt_pred>0.5)yp_grid=model.predict(X_grid).reshape(100,100)Xs_enc=model.transform(Xs)Xt_enc=model.transform(Xt)pca_enc=PCA(2).fit(np.concatenate((Xs_enc,Xt_enc)))Xs_enc_pca=pca_enc.transform(Xs_enc)Xt_enc_pca=pca_enc.transform(Xt_enc)cm=plt.cm.RdBufig,(ax1,ax2)=plt.subplots(1,2,figsize=(12,5))ax1.set_title("PCA input space")ax1.contourf(x_grid,y_grid,yp_grid,cmap=cm,alpha=0.6)ax1.scatter(Xs_pca[ys==0,0],Xs_pca[ys==0,1],label="source",edgecolors='k',c="red")ax1.scatter(Xs_pca[ys==1,0],Xs_pca[ys==1,1],label="source",edgecolors='k',c="blue")ax1.scatter(Xt_pca[:,0],Xt_pca[:,1],label="target",edgecolors='k',c="black",alpha=0.5)ax1.legend()ax1.set_yticklabels([])ax1.set_xticklabels([])ax1.tick_params(direction='in')ax2.set_title("PCA encoded space")ax2.scatter(Xs_enc_pca[ys==0,0],Xs_enc_pca[ys==0,1],label="source",edgecolors='k',c="red")ax2.scatter(Xs_enc_pca[ys==1,0],Xs_enc_pca[ys==1,1],label="source",edgecolors='k',c="blue")ax2.scatter(Xt_enc_pca[:,0],Xt_enc_pca[:,1],label="target",edgecolors='k',c="black",alpha=0.5)ax2.set_yticklabels([])ax2.set_xticklabels([])ax2.tick_params(direction='in')ifis_src_only:fig.suptitle("%s - Source Acc :%.3f - Target Acc :%.3f"%("Source Only",acc_s,acc_t))else:fig.suptitle("%s - Source Acc :%.3f - Target Acc :%.3f"%(model.__class__.__name__,acc_s,acc_t))plt.show()
Network
[20]:
defget_task(activation="sigmoid",units=1):model=Sequential()model.add(Flatten())model.add(Dense(10,activation="relu"))model.add(Dense(10,activation="relu"))model.add(Dense(units,activation=activation))returnmodel
Source Only
For source only, we use a DANN instance with lambda set to zero. Thus, the gradient of the discriminator is not back-propagated through the encoder.
[21]:
src_only=DANN(task=get_task(),loss="bce",optimizer=Adam(0.001,beta_1=0.5),copy=True,lambda_=0.,metrics=["acc"],random_state=0)
[22]:
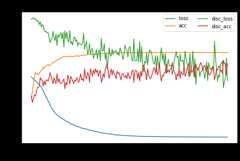
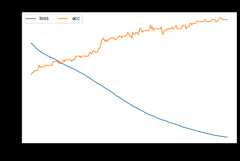
src_only.fit(Xs,ys,Xt,epochs=200,batch_size=34,verbose=0);pd.DataFrame(src_only.history_).plot(figsize=(8,5))plt.title("Training history",fontsize=14);plt.xlabel("Epochs");plt.ylabel("Scores")plt.legend(ncol=2)plt.show()
[23]:
show_results(src_only,True)
DANN
[24]:
dann=DANN(task=get_task(),loss="bce",optimizer=Adam(0.001,beta_1=0.5),copy=True,lambda_=1.,metrics=["acc"],random_state=0)
[25]:
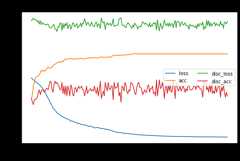
dann.fit(Xs,ys,Xt,epochs=200,batch_size=34,verbose=0);pd.DataFrame(dann.history_).plot(figsize=(8,5))plt.title("Training history",fontsize=14);plt.xlabel("Epochs");plt.ylabel("Scores")plt.legend(ncol=2)plt.show()
[26]:
show_results(dann,True)

ADDA
[ ]:
adda=ADDA(task=get_task(),loss="bce",optimizer=Adam(0.001,beta_1=0.5),copy=True,metrics=["acc"],random_state=0)
[ ]:
adda.fit(Xs,ys,Xt,epochs=200,batch_size=34,verbose=0);pd.DataFrame(adda.history_).plot(figsize=(8,5))plt.title("Training history",fontsize=14);plt.xlabel("Epochs");plt.ylabel("Scores")plt.legend(ncol=2)plt.show()
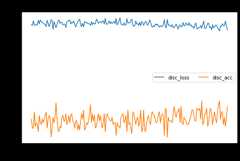
[ ]:
show_results(adda)

DeepCORAL
[ ]:
dcoral=DeepCORAL(encoder=src_only.encoder_,task=src_only.task_,lambda_=1000.,loss="bce",optimizer=Adam(0.001,beta_1=0.5),copy=True,metrics=["acc"],random_state=0)
[ ]:
dcoral.fit(Xs,ys,Xt,epochs=200,batch_size=34,verbose=0);pd.DataFrame(dcoral.history_).plot(figsize=(8,5))plt.title("Training history",fontsize=14);plt.xlabel("Epochs");plt.ylabel("Scores")plt.legend(ncol=2)plt.show()
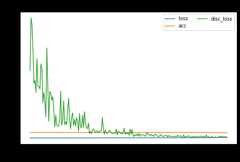
[ ]:
show_results(dcoral)
CORAL
[ ]:
coral=CORAL(estimator=get_task(),lambda_=0.,loss="bce",optimizer=Adam(0.001,beta_1=0.5),copy=True,metrics=["acc"],random_state=0)
[ ]:
coral.fit(Xs,ys,Xt,epochs=200,batch_size=34,verbose=0);pd.DataFrame(coral.estimator_.history.history).plot(figsize=(8,5))plt.title("Training history",fontsize=14);plt.xlabel("Epochs");plt.ylabel("Scores")plt.legend(ncol=2)plt.show()
Fit transform...Previous covariance difference: 0.106929New covariance difference: 0.000000Fit Estimator...
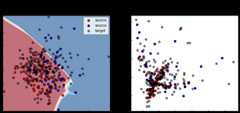
[ ]:
show_results(coral)

MCD
[ ]:
mcd=MCD(task=get_task(),loss="bce",optimizer=Adam(0.001,beta_1=0.5),copy=True,metrics=["acc"],random_state=0)
[ ]:
mcd.fit(Xs,ys,Xt,epochs=200,batch_size=34,verbose=0);pd.DataFrame(mcd.history_).plot(figsize=(8,5))plt.title("Training history",fontsize=14);plt.xlabel("Epochs");plt.ylabel("Scores")plt.legend(ncol=2)plt.show()
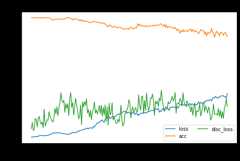
[ ]:
show_results(mcd)

MDD
[ ]:
mdd=MDD(task=get_task(),loss="bce",optimizer=Adam(0.001,beta_1=0.5),copy=True,metrics=["acc"],random_state=0)
[ ]:
mdd.fit(Xs,ys,Xt,epochs=200,batch_size=34,verbose=0);pd.DataFrame(mdd.history_).plot(figsize=(8,5))plt.title("Training history",fontsize=14);plt.xlabel("Epochs");plt.ylabel("Scores")plt.legend(ncol=2)plt.show()
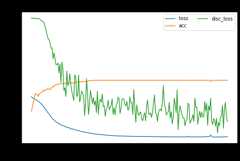
[ ]:
show_results(mdd)

WDGRL
[ ]:
wdgrl=WDGRL(task=get_task(None),gamma=0.,loss="bce",optimizer=Adam(0.001,beta_1=0.5),copy=True,metrics=["acc"],random_state=0)
[ ]:
wdgrl.fit(Xs,ys,Xt,epochs=200,batch_size=34,verbose=0);pd.DataFrame(wdgrl.history_).plot(figsize=(8,5))plt.title("Training history",fontsize=14);plt.xlabel("Epochs");plt.ylabel("Scores")plt.legend(ncol=2)plt.show()
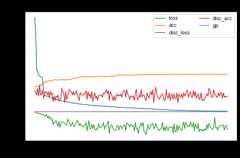
[ ]:
show_results(wdgrl)

CDAN
[ ]:
cdan=CDAN(task=get_task(),entropy=True,loss="bce",optimizer=Adam(0.001,beta_1=0.5),copy=True,random_state=0)
[ ]:
cdan.fit(Xs,ys,Xt,epochs=200,batch_size=34,verbose=0);pd.DataFrame(cdan.history_).plot(figsize=(8,5))plt.title("Training history",fontsize=14);plt.xlabel("Epochs");plt.ylabel("Scores")plt.legend(ncol=2)plt.show()

[ ]:
show_results(cdan)

[ ]: