- Notifications
You must be signed in to change notification settings - Fork29
A repository for explaining feature attributions and feature interactions in deep neural networks.
License
suinleelab/path_explain
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation
A repository for explaining feature importances and feature interactions in deep neural networks using path attribution methods.
This repository contains tools to interpret and explain machine learning models usingIntegrated Gradients andExpected Gradients. In addition, it contains code to explaininteractions in deep networks using Integrated Hessians and Expected Hessians - methods that we introduced in our most recent paper:"Explaining Explanations: Axiomatic Feature Interactions for Deep Networks". If you use our work to explain your networks, please cite this paper.
@article{janizek2020explaining, author = {Joseph D. Janizek and Pascal Sturmfels and Su-In Lee}, title = {Explaining Explanations: Axiomatic Feature Interactions for Deep Networks}, journal = {Journal of Machine Learning Research}, year = {2021}, volume = {22}, number = {104}, pages = {1-54}, url = {http://jmlr.org/papers/v22/20-1223.html}}This repository contains two important directories: thepath_explain directory, which contains the packages used to interpret and explain machine learning models, and theexamples directory, which contains many examples using thepath_explain module to explain different models on different data types.
The easiest way to install this package is by using pip:
pip install path-explainAlternatively, you can clone this repository to re-run and explore the examples provided.
This package was written to support TensorFlow 2.0 (in eager execution mode) with Python 3. We have no current plans to support earlier versions of TensorFlow or Python.
Although we don't yet have formal API documentation, the underlying code does a pretty good job at explaining the API. See the code for generatingattributions andinteractions to better understand what the arguments to these functions mean.
For a simple, quick example to get started using this repository, see theexample_usage.ipynb notebook in the top-level directory of this repository. It gives an overview of the functionality provided by this repository. For more advanced examples, keep reading on.
Our repository can easily be adapted to explain attributions and interactions learned on tabular data.
# other import statements...frompath_explainimportPathExplainerTF,scatter_plot,summary_plot### Code to train a model would go herex_train,y_train,x_test,y_test=datset()model= ...model.fit(x_train,y_train, ...)###### Generating attributions using expected gradientsexplainer=PathExplainerTF(model)attributions=explainer.attributions(inputs=x_test,baseline=x_train,batch_size=100,num_samples=200,use_expectation=True,output_indices=0)###### Generating interactions using expected hessiansinteractions=explainer.interactions(inputs=x_test,baseline=x_train,batch_size=100,num_samples=200,use_expectation=True,output_indices=0)###
Once we've generated attributions and interactions, we can use the provided plotting modules to help visualize them. First we plot a summary of the top features and their attribution values:
### First we need a list of strings denoting the name of each featurefeature_names= ...###summary_plot(attributions=attributions,feature_values=x_test,feature_names=feature_names,plot_top_k=10)
Second, we plot an interaction our model has learned between maximum achieved heart rate and gender:
scatter_plot(attributions=attributions,feature_values=x_test,feature_index='max. achieved heart rate',interactions=interactions,color_by='is male',feature_names=feature_names,scale_y_ind=True)
The model used to generate the above interactions is a two layer neural network trained on theUCI Heart Disease Dataset. Interactions learned by this model were featured in our paper. To learn more about this particular model and the experimental setup, seethe notebook used to train and explain the model.
As discussed in our paper, we can use Integrated Hessians to get interactions in language models. We explain a transformer from theHuggingFace Transformers Repository.
fromtransformersimportDistilBertTokenizer,TFDistilBertForSequenceClassification, \DistilBertConfig,glue_convert_examples_to_features, \glue_processors# This is a custom explainer to explain huggingface modelsfrompath_explainimportEmbeddingExplainerTF,text_plot,matrix_interaction_plot,bar_interaction_plottokenizer=DistilBertTokenizer.from_pretrained('distilbert-base-uncased')config=DistilBertConfig.from_pretrained('distilbert-base-uncased',num_labels=num_labels)model=TFDistilBertForSequenceClassification.from_pretrained('distilbert-base-uncased',config=config)### Some custom code to fine-tune the model on a sentiment analysis task...max_length=128data,info=tensorflow_datasets.load('glue/sst-2',with_info=True)train_dataset=glue_convert_examples_to_features(data['train'],tokenizer,max_length, 'sst-2)valid_dataset=glue_convert_examples_to_features(data['validation'],tokenizer,max_length,'sst-2')...### we won't include the whole fine-tuning code. See the HuggingFace repository for more.### Here we define functions that represent two pieces of the model:### embedding and predictiondefembedding_model(batch_ids):batch_embedding=model.distilbert.embeddings(batch_ids)returnbatch_embeddingdefprediction_model(batch_embedding):# Note: this isn't exactly the right way to use the attention mask.# It should actually indicate which words are real words. This# makes the coding easier however, and the output is fairly similar,# so it suffices for this tutorial.attention_mask=tf.ones(batch_embedding.shape[:2])attention_mask=tf.cast(attention_mask,dtype=tf.float32)head_mask= [None]*model.distilbert.num_hidden_layerstransformer_output=model.distilbert.transformer([batch_embedding,attention_mask,head_mask],training=False)[0]pooled_output=transformer_output[:,0]pooled_output=model.pre_classifier(pooled_output)logits=model.classifier(pooled_output)returnlogits###### We need some data to explainforbatchinvalid_dataset.take(1):batch_input=batch[0]batch_ids=batch_input['input_ids']batch_embedding=embedding_model(batch_ids)baseline_ids=np.zeros((1,128),dtype=np.int64)baseline_embedding=embedding_model(baseline_ids)###### We are finally ready to explain our modelexplainer=EmbeddingExplainerTF(prediction_model)attributions=explainer.attributions(inputs=batch_embedding,baseline=baseline_embedding,batch_size=32,num_samples=256,use_expectation=False,output_indices=1)###### For interactions, the hessian is rather large so we use a very small batch sizeinteractions=explainer.interactions(inputs=batch_embedding,baseline=baseline_embedding,batch_size=1,num_samples=256,use_expectation=False,output_indices=1)###
We can plot the learned attributions and interactions as follows. First we plot the attributions:
### First we need to decode the tokens from the batch ids.batch_sentences= ...### Doing so will depend on how you tokenized your model!text_plot(batch_sentences[0],attributions[0],include_legend=True)
Then we plot the interactions:
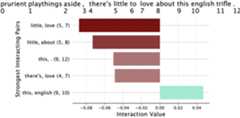
bar_interaction_plot(interactions[0],batch_sentences[0],top_k=5)
If you would rather plot the full matrix of attributions rather than the top interactions in a bar plot, our package also supports this. First we show the attributions:
text_plot(batch_sentences[1],attributions[1],include_legend=True)
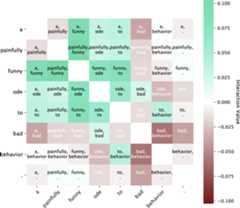
And then we show the full interaction matrix. Here we've zeroed out the diagonals so you can better see the off-diagonal terms.
matrix_interaction_plot(interaction_list[1],token_list[1])
This example - interpretingDistilBERT - was also featured in our paper. You can examine the setup morehere. For more examples, see theexamples directory in this repository.
About
A repository for explaining feature attributions and feature interactions in deep neural networks.
Topics
Resources
License
Uh oh!
There was an error while loading.Please reload this page.
Stars
Watchers
Forks
Releases
Packages0
Uh oh!
There was an error while loading.Please reload this page.
Contributors5
Uh oh!
There was an error while loading.Please reload this page.