- Notifications
You must be signed in to change notification settings - Fork111
Optimal binning: monotonic binning with constraints. Support batch & stream optimal binning. Scorecard modelling and counterfactual explanations.
License
guillermo-navas-palencia/optbinning
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation
OptBinning is a library written in Python implementing a rigorous and flexible mathematical programming formulation to solve the optimal binning problem for a binary, continuous and multiclass target type, incorporating constraints not previously addressed.
- Papers:
- Optimal binning: mathematical programming formulation.http://arxiv.org/abs/2001.08025
- Optimal counterfactual explanations for scorecard modelling.https://arxiv.org/abs/2104.08619
- Blog: Optimal binning for streaming data.http://gnpalencia.org/blog/2020/binning_data_streams/
To install the current release of OptBinning from PyPI:
pip install optbinningTo include batch and stream binning algorithms (this option is not required for most users):
pip install optbinning[distributed]To include support for theecos solver:
pip install optbinning[ecos]To install from source, download or clone the git repository
git clone https://github.com/guillermo-navas-palencia/optbinning.gitcd optbinningpython setup.py installOptBinning requires
- matplotlib
- numpy (>=1.16.1)
- ortools (>=9.4)
- pandas
- ropwr (>=1.0.0)
- scikit-learn (>=1.6.0)
- scipy (>=1.6.0)
OptBinning[distributed] requires additional packages
- pympler
- tdigest
Please visit the OptBinning documentation (current release)http://gnpalencia.org/optbinning/. If your are new to OptBinning, you can get started following thetutorials and checking the API references.
- Optimal binning tutorials
- Binning process tutorials
- Scorecard and counterfactual tutorials
- Optimal piecewise binning tutorials
- Batch and stream optimal binning tutorials
- Optimal binning under uncertainty
- Optimal binning 2D
Let's load a well-known dataset from the UCI repository and choose a variable to discretize and the binary target.
importpandasaspdfromsklearn.datasetsimportload_breast_cancerdata=load_breast_cancer()df=pd.DataFrame(data.data,columns=data.feature_names)variable="mean radius"x=df[variable].valuesy=data.target
Import and instantiate anOptimalBinning object class. We pass the variable name, its data type, and a solver, in this case, we choose the constraint programming solver. Fit the optimal binning object with arraysx andy.
fromoptbinningimportOptimalBinningoptb=OptimalBinning(name=variable,dtype="numerical",solver="cp")optb.fit(x,y)
Check status and retrieve optimal split points
>>>optb.status'OPTIMAL'>>>optb.splitsarray([11.42500019,12.32999992,13.09499979,13.70499992,15.04500008,16.92500019])
The optimal binning algorithms return a binning table; a binning table displays the binned data and several metrics for each bin. Call the methodbuild, which returns a pandas.DataFrame.
>>>optb.binning_table.build()
Bin Count Count (%) Non-event Event Event rate WoE IV JS0 [-inf, 11.43) 118 0.207381 3 115 0.974576 -3.12517 0.962483 0.0872051 [11.43, 12.33) 79 0.138840 3 76 0.962025 -2.71097 0.538763 0.0521982 [12.33, 13.09) 68 0.119508 7 61 0.897059 -1.64381 0.226599 0.0255133 [13.09, 13.70) 49 0.086116 10 39 0.795918 -0.839827 0.052131 0.0063314 [13.70, 15.05) 83 0.145870 28 55 0.662651 -0.153979 0.003385 0.0004235 [15.05, 16.93) 54 0.094903 44 10 0.185185 2.00275 0.359566 0.0386786 [16.93, inf) 118 0.207381 117 1 0.008475 5.28332 2.900997 0.1834367 Special 0 0.000000 0 0 0.000000 0 0.000000 0.0000008 Missing 0 0.000000 0 0 0.000000 0 0.000000 0.000000Totals 569 1.000000 212 357 0.627417 5.043925 0.393784You can use the methodplot to visualize the histogram and WoE or event rate curve. Note that the Bin ID corresponds to the binning table index.
>>>optb.binning_table.plot(metric="woe")

Optionally, you can show the binning plot with the actual bin widths.
>>>optb.binning_table.plot(metric="woe",style="actual",add_special=False,add_missing=False)

Now that we have checked the binned data, we can transform our original data into WoE or event rate values.
x_transform_woe=optb.transform(x,metric="woe")x_transform_event_rate=optb.transform(x,metric="event_rate")
Theanalysis method performs a statistical analysis of the binning table, computing the statistics Gini index, Information Value (IV), Jensen-Shannon divergence, and the quality score. Additionally, several statistical significance tests between consecutive bins of the contingency table are performed.
>>>optb.binning_table.analysis()
---------------------------------------------OptimalBinning: Binary Binning Table Analysis--------------------------------------------- General metrics Gini index 0.87541620 IV (Jeffrey) 5.04392547 JS (Jensen-Shannon) 0.39378376 Hellinger 0.47248971 Triangular 1.25592041 KS 0.72862164 HHI 0.15727342 HHI (normalized) 0.05193260 Cramer's V 0.80066760 Quality score 0.00000000 Monotonic trend descending Significance tests Bin A Bin B t-statistic p-value P[A > B] P[B > A] 0 1 0.252432 6.153679e-01 0.684380 3.156202e-01 1 2 2.432829 1.188183e-01 0.948125 5.187465e-02 2 3 2.345804 1.256207e-01 0.937874 6.212635e-02 3 4 2.669235 1.023052e-01 0.955269 4.473083e-02 4 5 29.910964 4.523477e-08 1.000000 9.814594e-12 5 6 19.324617 1.102754e-05 0.999999 1.216668e-06Print overview information about the options settings, problem statistics, and the solution of the computation.
>>>optb.information(print_level=2)
optbinning (Version 0.21.0)Copyright (c) 2019-2025 Guillermo Navas-Palencia, Apache License 2.0 Begin options name mean radius * U dtype numerical * d prebinning_method cart * d solver cp * d divergence iv * d max_n_prebins 20 * d min_prebin_size 0.05 * d min_n_bins no * d max_n_bins no * d min_bin_size no * d max_bin_size no * d min_bin_n_nonevent no * d max_bin_n_nonevent no * d min_bin_n_event no * d max_bin_n_event no * d monotonic_trend auto * d min_event_rate_diff 0 * d max_pvalue no * d max_pvalue_policy consecutive * d gamma 0 * d class_weight no * d cat_cutoff no * d user_splits no * d user_splits_fixed no * d special_codes no * d split_digits no * d mip_solver bop * d time_limit 100 * d verbose False * d End options Name : mean radius Status : OPTIMAL Pre-binning statistics Number of pre-bins 9 Number of refinements 1 Solver statistics Type cp Number of booleans 26 Number of branches 58 Number of conflicts 0 Objective value 5043922 Best objective bound 5043922 Timing Total time 0.04 sec Pre-processing 0.00 sec ( 0.33%) Pre-binning 0.00 sec ( 5.54%) Solver 0.04 sec ( 93.03%) model generation 0.03 sec ( 85.61%) optimizer 0.01 sec ( 14.39%) Post-processing 0.00 sec ( 0.30%)In this case, we choose two variables to discretized and the binary target.
importpandasaspdfromsklearn.datasetsimportload_breast_cancerdata=load_breast_cancer()df=pd.DataFrame(data.data,columns=data.feature_names)variable1="mean radius"variable2="worst concavity"x=df[variable1].valuesy=df[variable2].valuesz=data.target
Import and instantiate anOptimalBinning2D object class. We pass the variable names, and monotonic trends. Fit the optimal binning object with arraysx,y andz.
fromoptbinningimportOptimalBinning2Doptb=OptimalBinning2D(name_x=variable1,name_y=variable2,monotonic_trend_x="descending",monotonic_trend_y="descending",min_bin_size=0.05)optb.fit(x,y,z)
Show binning table:
>>>optb.binning_table.build()
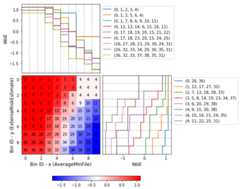
Bin x Bin y Count Count (%) Non-event Event Event rate WoE IV JS0 (-inf, 13.70) (-inf, 0.21) 219 0.384886 1 218 0.995434 -4.863346 2.946834 0.1994301 [13.70, inf) (-inf, 0.21) 48 0.084359 5 43 0.895833 -1.630613 0.157946 0.0178112 (-inf, 13.09) [0.21, 0.38) 48 0.084359 1 47 0.979167 -3.328998 0.422569 0.0370103 [13.09, 15.05) [0.21, 0.38) 46 0.080844 17 29 0.630435 -0.012933 0.000013 0.0000024 [15.05, inf) [0.21, 0.32) 32 0.056239 29 3 0.093750 2.789833 0.358184 0.0342715 [15.05, inf) [0.32, inf) 129 0.226714 128 1 0.007752 5.373180 3.229133 0.2012946 (-inf, 15.05) [0.38, inf) 47 0.082601 31 16 0.340426 1.182548 0.119920 0.0141737 Special Special 0 0.000000 0 0 0.000000 0.000000 0.000000 0.0000008 Missing Missing 0 0.000000 0 0 0.000000 0.000000 0.000000 0.000000Totals 569 1.000000 212 357 0.627417 7.234600 0.503991Similar to the optimal binning, you can generate a histogram 2D to visualize WoE and event rate.
>>>optb.binning_table.plot(metric="event_rate")

Let's load the California housing dataset.
importpandasaspdfromsklearn.datasetsimportfetch_california_housingfromsklearn.linear_modelimportHuberRegressorfromoptbinningimportBinningProcessfromoptbinningimportScorecarddata=fetch_california_housing()target="target"variable_names=data.feature_namesX=pd.DataFrame(data.data,columns=variable_names)y=data.target
Instantiate a binning process, an estimator, and a scorecard with scalingmethod and reverse mode.
binning_process=BinningProcess(variable_names)estimator=HuberRegressor(max_iter=200)scorecard=Scorecard(binning_process=binning_process,estimator=estimator,scaling_method="min_max",scaling_method_params={"min":0,"max":100},reverse_scorecard=True)scorecard.fit(X,y)
Print overview information about the options settings, problems statistics,and the number of selected variables after the binning process.
>>>scorecard.information(print_level=2)
optbinning (Version 0.21.0)Copyright (c) 2019-2025 Guillermo Navas-Palencia, Apache License 2.0 Begin options binning_process yes * U estimator yes * U scaling_method min_max * U scaling_method_params yes * U intercept_based False * d reverse_scorecard True * U rounding False * d verbose False * d End options Statistics Number of records 20640 Number of variables 8 Target type continuous Number of numerical 8 Number of categorical 0 Number of selected 8 Timing Total time 2.31 sec Binning process 1.83 sec ( 79.00%) Estimator 0.41 sec ( 17.52%) Build scorecard 0.08 sec ( 3.40%) rounding 0.00 sec ( 0.00%)>>>scorecard.table(style="summary")
Two scorecard styles are available:style="summary" shows the variable name, and their corresponding bins and assigned points;style="detailed" adds information from the corresponding binning table.
Variable Bin Points0 MedInc [-inf, 1.90) 9.8692241 MedInc [1.90, 2.16) 10.8969402 MedInc [2.16, 2.37) 11.4829973 MedInc [2.37, 2.66) 12.6078054 MedInc [2.66, 2.88) 13.609078.. ... ... ...2 Longitude [-118.33, -118.26) 10.4704013 Longitude [-118.26, -118.16) 9.0923914 Longitude [-118.16, inf) 10.2239365 Longitude Special 1.3768626 Longitude Missing 1.376862[94 rows x 3 columns]>>>scorecard.table(style="detailed")
Variable Bin id Bin Count Count (%) ... Zeros count WoE IV Coefficient Points0 MedInc 0 [-inf, 1.90) 2039 0.098789 ... 0 -0.969609 0.095786 0.990122 9.8692241 MedInc 1 [1.90, 2.16) 1109 0.053731 ... 0 -0.836618 0.044952 0.990122 10.8969402 MedInc 2 [2.16, 2.37) 1049 0.050824 ... 0 -0.760779 0.038666 0.990122 11.4829973 MedInc 3 [2.37, 2.66) 1551 0.075145 ... 0 -0.615224 0.046231 0.990122 12.6078054 MedInc 4 [2.66, 2.88) 1075 0.052083 ... 0 -0.485655 0.025295 0.990122 13.609078.. ... ... ... ... ... ... ... ... ... ... ...2 Longitude 2 [-118.33, -118.26) 1120 0.054264 ... 0 -0.011006 0.000597 0.566265 10.4704013 Longitude 3 [-118.26, -118.16) 1127 0.054603 ... 0 -0.322802 0.017626 0.566265 9.0923914 Longitude 4 [-118.16, inf) 6530 0.316376 ... 0 -0.066773 0.021125 0.566265 10.2239365 Longitude 5 Special 0 0.000000 ... 0 -2.068558 0.000000 0.566265 1.3768626 Longitude 6 Missing 0 0.000000 ... 0 -2.068558 0.000000 0.566265 1.376862[94 rows x 14 columns]Compute score and predicted target using the fitted estimator.
score=scorecard.score(X)y_pred=scorecard.predict(X)
First, we load the dataset and a scorecard previously developed.
importpandasaspdfromoptbinningimportScorecardfromoptbinning.scorecardimportCounterfactualfromsklearn.datasetsimportload_bostondata=load_boston()X=pd.DataFrame(data.data,columns=data.feature_names)scorecard=Scorecard.load("myscorecard.pkl")
We create a new Counterfactual instance that is fitted with the datasetused during the scorecard development. Then, we select a sample from which to generatecounterfactual explanations.
cf=Counterfactual(scorecard=scorecard)cf.fit(X)query=X.iloc[0, :].to_frame().T
The scorecard model predicts 26.8. However, we would like to find out what needs to bechanged to return a prediction greater or equal to 30.
>>>queryCRIMZNINDUSCHASNOXRMAGEDISRADTAXPTRATIOBLSTAT00.0063218.02.310.00.5386.57565.24.091.0296.015.3396.94.98>>>scorecard.predict(query)array([26.83423364])
We can generate a single counterfactual explanation:
>>>cf.generate(query=query,y=30,outcome_type="continuous",n_cf=1,max_changes=3,hard_constraints=["min_outcome"])>>>cf.status'OPTIMAL'>>>cf.display(show_only_changes=True,show_outcome=True)CRIMZNINDUSCHASNOXRMAGEDISRADTAXPTRATIOBLSTAToutcome0 [0.04,0.07)--- [0.45,0.50) [6.94,7.44)-------31.28763
Or simultaneously three counterfactuals, enforcing diversity on the feature values and selecting only a few actionable features.
>>>cf.generate(query=query,y=30,outcome_type="continuous",n_cf=3,max_changes=3,hard_constraints=["diversity_values","min_outcome"],actionable_features=["CRIM","NOX","RM","PTRATIO"])>>>cf.status'OPTIMAL'>>>cf.display(show_only_changes=True,show_outcome=True)CRIMZNINDUSCHASNOXRMAGEDISRADTAXPTRATIOBLSTAToutcome0 [0.03,0.04)--- [0.42,0.45) [6.94,7.44)-------31.7378440 [0.04,0.07)---- [7.44,inf)---- [17.85,18.55)--36.3700860---- [0.45,0.50) [6.68,6.94)---- [-inf,15.15)--30.095258
The following table shows how OptBinning compares toscorecardpy 0.1.9.1.1 on a selection of variables from the public dataset, Home Credit Default Risk - Kaggle’s competitionLink. This dataset contains 307511 samples.The experiments were run on Intel(R) Core(TM) i5-3317 CPU at 1.70GHz, using a single core, running Linux. For scorecardpy, we use default settings only increasing the maximum number of binsbin_num_limit=20. For OptBinning, we use default settings (max_n_prebins=20) only changing the maximum allowed p-value between consecutive bins,max_pvalue=0.05.
To compare softwares we use the shifted geometric mean, typically used in mathematical optimization benchmarks:http://plato.asu.edu/bench.html. Using the shifted (by 1 second) geometric mean we found thatOptBinning is17x faster than scorecardpy, with an average IV increment of12%. Besides the speed and IV gains, OptBinning includes many more constraints and monotonicity options.
| Variable | scorecardpy_time | scorecardpy_IV | optbinning_time | optbinning_IV |
|---|---|---|---|---|
| AMT_INCOME_TOTAL | 6.18 s | 0.010606 | 0.363 s | 0.011705 |
| NAME_CONTRACT_TYPE (C) | 3.72 s | 0.015039 | 0.148 s | 0.015039 |
| AMT_CREDIT | 7.10 s | 0.053593 | 0.634 s | 0.059311 |
| ORGANIZATION_TYPE (C) | 6.31 s | 0.063098 | 0.274 s | 0.071520 |
| AMT_ANNUITY | 6.51 s | 0.024295 | 0.648 s | 0.031179 |
| AMT_GOODS_PRICE | 6.95 s | 0.056923 | 0.401 s | 0.092032 |
| NAME_HOUSING_TYPE (C) | 3.57 s | 0.015055 | 0.140 s | 0.015055 |
| REGION_POPULATION_RELATIVE | 4.33 s | 0.026578 | 0.392 s | 0.035567 |
| DAYS_BIRTH | 5.18 s | 0.081270 | 0.564 s | 0.086539 |
| OWN_CAR_AGE | 4.85 s | 0.021429 | 0.055 s | 0.021890 |
| OCCUPATION_TYPE (C) | 4.24 s | 0.077606 | 0.201 s | 0.079540 |
| APARTMENTS_AVG | 5.61 s | 0.032247(*) | 0.184 s | 0.032415 |
| BASEMENTAREA_AVG | 5.14 s | 0.022320 | 0.119 s | 0.022639 |
| YEARS_BUILD_AVG | 4.49 s | 0.016033 | 0.055 s | 0.016932 |
| EXT_SOURCE_2 | 5.21 s | 0.298463 | 0.606 s | 0.321417 |
| EXT_SOURCE_3 | 5.08 s | 0.316352 | 0.303 s | 0.334975 |
| TOTAL | 84.47 s | 1.130907 | 5.087 s | 1.247756 |
(C): categorical variable.(*): max p-value between consecutive bins > 0.05.
The binning of variables with monotonicity trend peak or valley can benefit from the optionmonotonic_trend="auto_heuristic" at the expense of finding a suboptimal solution for some cases. The following table compares the optionsmonotonic_trend="auto" andmonotonic_trend="auto_heuristic",
| Variable | auto_time | auto_IV | heuristic_time | heuristic_IV |
|---|---|---|---|---|
| AMT_INCOME_TOTAL | 0.363 s | 0.011705 | 0.322 s | 0.011705 |
| AMT_CREDIT | 0.634 s | 0.059311 | 0.469 s | 0.058643 |
| AMT_ANNUITY | 0.648 s | 0.031179 | 0.505 s | 0.031179 |
| AMT_GOODS_PRICE | 0.401 s | 0.092032 | 0.299 s | 0.092032 |
| REGION_POPULATION_RELATIVE | 0.392 s | 0.035567 | 0.244 s | 0.035567 |
| TOTAL | 2.438 s | 0.229794 | 1.839 s | 0.229126 |
Observe that CPU time is reduced by 25% losing less than 1% in IV. The differences in CPU time are more noticeable as thenumber of bins increases, seehttp://gnpalencia.org/optbinning/tutorials/tutorial_binary_large_scale.html.
Found a bug? Want to contribute with a new feature, improve documentation, or add examples? We encourage you to create pull requests and/or open GitHub issues. Thanks!![]() 🎉 👍
🎉 👍
We would like to list companies using OptBinning. Please send a PR with your company name and @githubhandle if you may.
Currentlyofficially using OptBinning:
- Jeitto [@BrennerPablo &@ds-mauri &@GabrielSGoncalves]
- Bilendo [@FlorianKappert &@JakobBeyer]
- Aplazame
- Praelexis Credit
- ING
- DBRS Morningstar
- Loginom
- Risika
- Tamara
- BBVA AI Factory
- N26
- Home Credit International
- Farm Credit Canada
If you use OptBinning in your research/work, please cite the paper using the following BibTeX:
@article{Navas-Palencia2020OptBinning, title = {Optimal binning: mathematical programming formulation}, author = {Guillermo Navas-Palencia}, year = {2020}, eprint = {2001.08025}, archivePrefix = {arXiv}, primaryClass = {cs.LG}, volume = {abs/2001.08025}, url = {http://arxiv.org/abs/2001.08025},}@article{Navas-Palencia2021Counterfactual, title = {Optimal Counterfactual Explanations for Scorecard modelling}, author = {Guillermo Navas-Palencia}, year = {2021}, eprint = {2104.08619}, archivePrefix = {arXiv}, primaryClass = {cs.LG}, volume = {abs/2104.08619}, url = {http://arxiv.org/abs/2104.08619},}About
Optimal binning: monotonic binning with constraints. Support batch & stream optimal binning. Scorecard modelling and counterfactual explanations.
Topics
Resources
License
Uh oh!
There was an error while loading.Please reload this page.