- Notifications
You must be signed in to change notification settings - Fork58
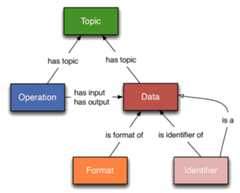
EDAM is an ontology of data analysis and data management in bio- and other sciences, and science-based applications. It comprises concepts related to analysis, modelling, optimisation, and data life cycle. Targeting usability by diverse users, EDAM's structure is relatively simple, divided into 4 sections: Topic, Operation, Data, and Format.
License
edamontology/edamontology
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation
DOI representing all stable versions, resolving to the latest:
DOI of the latest stable EDAM version 1.25:
Current status of the 'main' development file:
Twitter:@edamontology (follow).
EDAM is a comprehensive ontology of well-established, familiar concepts that are prevalent within computational biology, bioinformatics, andbioimage informatics. EDAM includes types of data and data identifiers, data formats, operations, and topics related to data analysis in life sciences. EDAM provides a set of concepts with preferred terms and synonyms, related terms, definitions, and other information - organised into a simple and intuitive hierarchy for convenient use (see figure).
EDAM is particularly suitable for semantic annotations and categorisation of diverse resources related to bioscientific data analysis:e.g. tools, workflows, or training materials. EDAM is also useful in data management, for recording provenance metadata of processed bioscientific data.
EDAM can be browsed online at theNCBO BioPortal, atOLS, and in theEDAM Browser.
The all-newestunstable version can be browsed and commented at theNCBO BioPortal andWebProtégé (free registration required).
The lateststable version is always downloable fromhttp://edamontology.org/EDAM.owl |tsv |csv. For older versions, seehttp://edamontology.org/page#Download or/releases.
Comprehensive documentation and guidelines are available viareadthedocs (maintainedhere).
A quick overview is at thehttps://edamontology.org home page.
If you refer to EDAM or its part in a scholarly publication, please cite:
Ison, J., Kalaš, M., Jonassen, I., Bolser, D., Uludag, M., McWilliam, H., Malone, J., Lopez, R., Pettifer, S. and Rice, P. (2013).EDAM: an ontology of bioinformatics operations, types of data and identifiers, topics and formats.Bioinformatics,29(10): 1325-1332. PMID:23479348Open access
Melissa Black, Lucie Lamothe, Hager Eldakroury, Mads Kierkegaard, Ankita Priya, Anne Machinda, Uttam Singh Khanduja, Drashti Patoliya, Rashika Rathi, Tawah Peggy Che Nico, Gloria Umutesi, Claudia Blankenburg, Anita Op, Precious Chieke, Omodolapo Babatunde, Steve Laurie, Steffen Neumann, Veit Schwämmle, Ivan Kuzmin, Chris Hunter, Jonathan Karr, Jon Ison, Alban Gaignard, Bryan Brancotte, Hervé Ménager, Matúš Kalaš (2022).EDAM: the bioscientific data analysis ontology (update 2021) [version 1; not peer reviewed].F1000Research,11(ISCB Comm J): 1. Poster.Open access
EDAM releases are citable with DOIs too, for cases when that is needed. represents all releases and resolves to the DOI of the last stable release.
Please note that this repository is participating in a study into sustainabilityof open source projects. Data will be gathered about this repository forapproximately the next 12 months, starting from June 2021.
Data collected will include number of contributors, number of PRs, time taken toclose/merge these PRs, and issues closed.
For more information, please visitour informational page or download ourparticipant information sheet.
About
EDAM is an ontology of data analysis and data management in bio- and other sciences, and science-based applications. It comprises concepts related to analysis, modelling, optimisation, and data life cycle. Targeting usability by diverse users, EDAM's structure is relatively simple, divided into 4 sections: Topic, Operation, Data, and Format.
Topics
Resources
License
Code of conduct
Contributing
Uh oh!
There was an error while loading.Please reload this page.
Stars
Watchers
Forks
Packages0
Uh oh!
There was an error while loading.Please reload this page.