Uh oh!
There was an error while loading.Please reload this page.
- Notifications
You must be signed in to change notification settings - Fork23
Robust, Fast, and Parallel Global Sensitivity Analysis (GSA) in Julia
License
SciML/GlobalSensitivity.jl
Folders and files
| Name | Name | Last commit message | Last commit date | |
|---|---|---|---|---|
Repository files navigation
GlobalSensitivity.jl package contains implementation of some the most popular GSA methods. Currently it supports Delta Moment-Independent, DGSM, EASI, eFAST, Morris, Mutual Information, Fractional Factorial, RBD-FAST, RSA, Sobol and Regression based sensitivity methods.
For information on using the package,see the stable documentation. Use thein-development documentation for the version ofthe documentation, which contains the unreleased features.
The GlobalSensitivity.jl package can be installed with julia's package manager as shown below:
using PkgPkg.add("GlobalSensitivity")
The general interface for performing global sensitivity analysis using this package is:
res=gsa(f, method, param_range; samples, batch=false)
Sobol method on theIshigami function.
Serial execution
functionishi(X) A=7 B=0.1sin(X[1])+ A*sin(X[2])^2+ B* X[3]^4*sin(X[1])endn=600000lb=-ones(4)* πub=ones(4)* πsampler=SobolSample()A, B= QuasiMonteCarlo.generate_design_matrices(n, lb, ub, sampler)res1=gsa(ishi,Sobol(order= [0,1,2]), A, B)
Using batching interface
functionishi_batch(X) A=7 B=0.1@.sin(X[1, :])+ A*sin(X[2, :])^2+ B* X[3, :]^4*sin(X[1, :])endres2=gsa(ishi_batch,Sobol(), A, B, batch=true)
using GlobalSensitivity, QuasiMonteCarlo, OrdinaryDiffEq, Statistics, CairoMakiefunctionf(du, u, p, t) du[1]= p[1]* u[1]- p[2]* u[1]* u[2]#prey du[2]=-p[3]* u[2]+ p[4]* u[1]* u[2]#predatorendu0= [1.0;1.0]tspan= (0.0,10.0)p= [1.5,1.0,3.0,1.0]prob=ODEProblem(f, u0, tspan, p)t=collect(range(0, stop=10, length=200))f1=function (p) prob1=remake(prob; p= p) sol=solve(prob1,Tsit5(); saveat= t)return [mean(sol[1, :]),maximum(sol[2, :])]endbounds= [[1,5], [1,5], [1,5], [1,5]]reg_sens=gsa(f1,RegressionGSA(true), bounds)fig=Figure(resolution= (600,400))ax,hm= CairoMakie.heatmap(fig[1,1], reg_sens.partial_correlation, figure= (resolution= (300,200),), axis= (xticksvisible=false, yticksvisible=false, yticklabelsvisible=false, xticklabelsvisible=false, title="Partial correlation"))Colorbar(fig[1,2], hm)ax,hm= CairoMakie.heatmap(fig[2,1], reg_sens.standard_regression, figure= (resolution= (300,200),), axis= (xticksvisible=false, yticksvisible=false, yticklabelsvisible=false, xticklabelsvisible=false, title="Standard regression"))Colorbar(fig[2,2], hm)fig
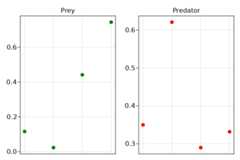
using StableRNGs_rng=StableRNG(1234)morris_sens=gsa(f1,Morris(), bounds, rng= _rng)fig=Figure(resolution= (300,200))scatter(fig[1,1], [1,2,3,4], morris_sens.means_star[1, :], color=:green, axis= (xticksvisible=false, xticklabelsvisible=false, title="Prey (Morris)"))scatter(fig[1,2], [1,2,3,4], morris_sens.means_star[2, :], color=:red, axis= (xticksvisible=false, xticklabelsvisible=false, title="Predator (Morris)"))fig
If you use this software in your work, please cite:
@article{dixit2022globalsensitivity,title={GlobalSensitivity. jl: Performant and Parallel Global Sensitivity Analysis with Julia},author={Dixit, Vaibhav Kumar and Rackauckas, Christopher},journal={Journal of Open Source Software},volume={7},number={76},pages={4561},year={2022}}
About
Robust, Fast, and Parallel Global Sensitivity Analysis (GSA) in Julia
Topics
Resources
License
Code of conduct
Contributing
Security policy
Uh oh!
There was an error while loading.Please reload this page.
Stars
Watchers
Forks
Sponsor this project
Uh oh!
There was an error while loading.Please reload this page.
Packages0
Uh oh!
There was an error while loading.Please reload this page.