profiling.sampling — Statistical profiler¶
Added in version 3.15.
Source code:Lib/profiling/sampling/

Theprofiling.sampling module, namedTachyon, provides statisticalprofiling of Python programs through periodic stack sampling. Tachyon canrun scripts directly or attach to any running Python process without requiringcode changes or restarts. Because sampling occurs externally to the targetprocess, overhead is virtually zero, making Tachyon suitable for bothdevelopment and production environments.
What is statistical profiling?¶
Statistical profiling builds a picture of program behavior by periodicallycapturing snapshots of the call stack. Rather than instrumenting every functioncall and return as deterministic profilers do, Tachyon reads the call stack atregular intervals to record what code is currently running.
This approach rests on a simple principle: functions that consume significantCPU time will appear frequently in the collected samples. By gathering thousandsof samples over a profiling session, Tachyon constructs an accurate statisticalestimate of where time is spent. The more samples collected, themore precise this estimate becomes.
How time is estimated¶
The time values shown in Tachyon’s output areestimates derived from samplecounts, not direct measurements. Tachyon counts how many times each functionappears in the collected samples, then multiplies by the sampling interval toestimate time.
For example, with a 100 microsecond sampling interval over a 10-second profile,Tachyon collects approximately 100,000 samples. If a function appears in 5,000samples (5% of total), Tachyon estimates it consumed 5% of the 10-secondduration, or about 500 milliseconds. This is a statistical estimate, not aprecise measurement.
The accuracy of these estimates depends on sample count. With 100,000 samples,a function showing 5% has a margin of error of roughly ±0.5%. With only 1,000samples, the same 5% measurement could actually represent anywhere from 3% to7% of real time.
This is why longer profiling durations and shorter sampling intervals producemore reliable results—they collect more samples. For most performanceanalysis, the default settings provide sufficient accuracy to identifybottlenecks and guide optimization efforts.
Because sampling is statistical, results will vary slightly between runs. Afunction showing 12% in one run might show 11% or 13% in the next. This isnormal and expected. Focus on the overall pattern rather than exact percentages,and don’t worry about small variations between runs.
When to use a different approach¶
Statistical sampling is not ideal for every situation.
For very short scripts that complete in under one second, the profiler may notcollect enough samples for reliable results. Useprofiling.tracinginstead, or run the script in a loop to extend profiling time.
When you need exact call counts, sampling cannot provide them. Samplingestimates frequency from snapshots, so if you need to know precisely how manytimes a function was called, useprofiling.tracing.
When comparing two implementations where the difference might be only 1-2%,sampling noise can obscure real differences. Usetimeit formicro-benchmarks orprofiling.tracing for precise measurements.
The key difference fromprofiling.tracing is how measurement happens.A tracing profiler instruments your code, recording every function call andreturn. This provides exact call counts and precise timing but adds overheadto every function call. A sampling profiler, by contrast, observes the programfrom outside at fixed intervals without modifying its execution. Think of thedifference like this: tracing is like having someone follow you and write downevery step you take, while sampling is like taking photographs every secondand inferring your path from those snapshots.
This external observation model is what makes sampling profiling practical forproduction use. The profiled program runs at full speed because there is noinstrumentation code running inside it, and the target process is never stoppedor paused during sampling—Tachyon reads the call stack directly from theprocess’s memory while it continues to run. You can attach to a live server,collect data, and detach without the application ever knowing it was observed.The trade-off is that very short-lived functions may be missed if they happento complete between samples.
Statistical profiling excels at answering the question, “Where is my programspending time?” It reveals hotspots and bottlenecks in production code wheredeterministic profiling overhead would be unacceptable. For exact call countsand complete call graphs, useprofiling.tracing instead.
Quick examples¶
Profile a script and see the results immediately:
python -m profiling.sampling run script.pyProfile a module with arguments:
python -m profiling.sampling run -m mypackage.module arg1 arg2Generate an interactive flame graph:
python -m profiling.sampling run --flamegraph -o profile.html script.pyAttach to a running process by PID:
python -m profiling.sampling attach 12345Use live mode for real-time monitoring (pressq to quit):
python -m profiling.sampling run --live script.pyProfile for 60 seconds with a faster sampling rate:
python -m profiling.sampling run -d 60 -i 50 script.pyGenerate a line-by-line heatmap:
python -m profiling.sampling run --heatmap script.pyEnable opcode-level profiling to see which bytecode instructions are executing:
python -m profiling.sampling run --opcodes --flamegraph script.pyCommands¶
Tachyon operates through two subcommands that determine how to obtain thetarget process.
Therun command¶
Therun command launches a Python script or module and profiles it fromstartup:
python -m profiling.sampling run script.pypython -m profiling.sampling run -m mypackage.module
When profiling a script, the profiler starts the target in a subprocess, waitsfor it to initialize, then begins collecting samples. The-m flagindicates that the target should be run as a module (equivalent topython-m). Arguments after the target are passed through to theprofiled program:
python -m profiling.sampling run script.py --config settings.yamlTheattach command¶
Theattach command connects to an already-running Python process by itsprocess ID:
python -m profiling.sampling attach 12345This command is particularly valuable for investigating performance issues inproduction systems. The target process requires no modification and need notbe restarted. The profiler attaches, collects samples for the specifiedduration, then detaches and produces output.
python -m profiling.sampling attach --live 12345python -m profiling.sampling attach --flamegraph -d 30 -o profile.html 12345
On most systems, attaching to another process requires appropriate permissions.SeePlatform requirements for platform-specific requirements.
Profiling in production¶
The sampling profiler is designed for production use. It imposes no measurableoverhead on the target process because it reads memory externally rather thaninstrumenting code. The target application continues running at full speed andis unaware it is being profiled.
When profiling production systems, keep these guidelines in mind:
Start with shorter durations (10-30 seconds) to get quick results, then extendif you need more statistical accuracy. The default 10-second duration is usuallysufficient to identify major hotspots.
If possible, profile during representative load rather than peak traffic.Profiles collected during normal operation are easier to interpret than thosecollected during unusual spikes.
The profiler itself consumes some CPU on the machine where it runs (not on thetarget process). On the same machine, this is typically negligible. Whenprofiling remote processes, network latency does not affect the target.
Results from production may differ from development due to different datasizes, concurrent load, or caching effects. This is expected and is oftenexactly what you want to capture.
Platform requirements¶
The profiler reads the target process’s memory to capture stack traces. Thisrequires elevated permissions on most operating systems.
Linux
On Linux, the profiler usesptrace orprocess_vm_readv to read thetarget process’s memory. This typically requires one of:
Running as root
Having the
CAP_SYS_PTRACEcapabilityAdjusting the Yama ptrace scope:
/proc/sys/kernel/yama/ptrace_scope
The default ptrace_scope of 1 restricts ptrace to parent processes only. Toallow attaching to any process owned by the same user, set it to 0:
echo 0 | sudo tee /proc/sys/kernel/yama/ptrace_scopemacOS
On macOS, the profiler usestask_for_pid() to access the target process.This requires one of:
Running as root
The profiler binary having the
com.apple.security.cs.debuggerentitlementSystem Integrity Protection (SIP) being disabled (not recommended)
Windows
On Windows, the profiler requires administrative privileges or theSeDebugPrivilege privilege to read another process’s memory.
Version compatibility¶
The profiler and target process must run the same Python minor version (forexample, both Python 3.15). Attaching from Python 3.14 to a Python 3.15 processis not supported.
Additional restrictions apply to pre-release Python versions: if either theprofiler or target is running a pre-release (alpha, beta, or release candidate),both must run the exact same version.
On free-threaded Python builds, the profiler cannot attach from a free-threadedbuild to a standard build, or vice versa.
Sampling configuration¶
Before exploring the various output formats and visualization options, it isimportant to understand how to configure the sampling process itself. Theprofiler offers several options that control how frequently samples arecollected, how long profiling runs, which threads are observed, and whatadditional context is captured in each sample.
The default configuration works well for most use cases:
Option | Default |
|---|---|
Default for | 100 µs between samples (~10,000 samples/sec) |
Default for | 10 seconds |
Default for | Main thread only |
Default for | No |
Default for |
|
Default for | Wall-clock mode (all samples recorded) |
Default for | Disabled |
Default for | Disabled |
Sampling interval and duration¶
The two most fundamental parameters are the sampling interval and duration.Together, these determine how many samples will be collected during a profilingsession.
The--interval option (-i) sets the time between samples inmicroseconds. The default is 100 microseconds, which produces approximately10,000 samples per second:
python -m profiling.sampling run -i 50 script.pyLower intervals capture more samples and provide finer-grained data at thecost of slightly higher profiler CPU usage. Higher intervals reduce profileroverhead but may miss short-lived functions. For most applications, thedefault interval provides a good balance between accuracy and overhead.
The--duration option (-d) sets how long to profile in seconds. Thedefault is 10 seconds:
python -m profiling.sampling run -d 60 script.pyLonger durations collect more samples and produce more statistically reliableresults, especially for code paths that execute infrequently. When profilinga program that runs for a fixed time, you may want to set the duration tomatch or exceed the expected runtime.
Thread selection¶
Python programs often use multiple threads, whether explicitly through thethreading module or implicitly through libraries that manage threadpools.
By default, the profiler samples only the main thread. The--all-threadsoption (-a) enables sampling of all threads in the process:
python -m profiling.sampling run -a script.pyMulti-thread profiling reveals how work is distributed across threads and canidentify threads that are blocked or starved. Each thread’s samples arecombined in the output, with the ability to filter by thread in some formats.This option is particularly useful when investigating concurrency issues orwhen work is distributed across a thread pool.
Special frames¶
The profiler can inject artificial frames into the captured stacks to provideadditional context about what the interpreter is doing at the moment eachsample is taken. These synthetic frames help distinguish different types ofexecution that would otherwise be invisible.
The--native option adds<native> frames to indicate when Python hascalled into C code (extension modules, built-in functions, or the interpreteritself):
python -m profiling.sampling run --native script.pyThese frames help distinguish time spent in Python code versus time spent innative libraries. Without this option, native code execution appears as timein the Python function that made the call. This is useful when optimizingcode that makes heavy use of C extensions like NumPy or database drivers.
By default, the profiler includes<GC> frames when garbage collection isactive. The--no-gc option suppresses these frames:
python -m profiling.sampling run --no-gc script.pyGC frames help identify programs where garbage collection consumes significanttime, which may indicate memory allocation patterns worth optimizing. If yousee substantial time in<GC> frames, consider investigating objectallocation rates or using object pooling.
Opcode-aware profiling¶
The--opcodes option enables instruction-level profiling that captureswhich Python bytecode instructions are executing at each sample:
python -m profiling.sampling run --opcodes --flamegraph script.pyThis feature provides visibility into Python’s bytecode execution, includingadaptive specialization optimizations. When a generic instruction likeLOAD_ATTR is specialized at runtime into a more efficient variant likeLOAD_ATTR_INSTANCE_VALUE, the profiler shows both the specialized nameand the base instruction.
Opcode information appears in several output formats:
Flame graphs: Hovering over a frame displays a tooltip with a bytecodeinstruction breakdown, showing which opcodes consumed time in that function
Heatmap: Expandable bytecode panels per source line show instructionbreakdown with specialization percentages
Live mode: An opcode panel shows instruction-level statistics for theselected function, accessible via keyboard navigation
Gecko format: Opcode transitions are emitted as interval markers in theFirefox Profiler timeline
This level of detail is particularly useful for:
Understanding the performance impact of Python’s adaptive specialization
Identifying hot bytecode instructions that might benefit from optimization
Analyzing the effectiveness of different code patterns at the instruction level
Debugging performance issues that occur at the bytecode level
The--opcodes option is compatible with--live,--flamegraph,--heatmap, and--gecko formats. It requires additional memory to storeopcode information and may slightly reduce sampling performance, but providesunprecedented visibility into Python’s execution model.
Real-time statistics¶
The--realtime-stats option displays sampling rate statistics duringprofiling:
python -m profiling.sampling run --realtime-stats script.pyThis shows the actual achieved sampling rate, which may be lower than requestedif the profiler cannot keep up. The statistics help verify that profiling isworking correctly and that sufficient samples are being collected. SeeSampling efficiency for details on interpreting these metrics.
Subprocess profiling¶
The--subprocesses option enables automatic profiling of subprocessesspawned by the target:
python -m profiling.sampling run --subprocesses script.pypython -m profiling.sampling attach --subprocesses 12345
When enabled, the profiler monitors the target process for child processcreation. When a new Python child process is detected, a separate profilerinstance is automatically spawned to profile it. This is useful forapplications that usemultiprocessing,subprocess,concurrent.futures withProcessPoolExecutor,or other process spawning mechanisms.
fromconcurrent.futuresimportProcessPoolExecutorimportmathdefcompute_factorial(n):total=0foriinrange(50):total+=math.factorial(n)returntotalif__name__=="__main__":numbers=[5000+i*100foriinrange(50)]withProcessPoolExecutor(max_workers=4)asexecutor:results=list(executor.map(compute_factorial,numbers))print(f"Computed{len(results)} factorials")
python -m profiling.sampling run --subprocesses --flamegraph worker_pool.pyThis produces separate flame graphs for the main process and each workerprocess:flamegraph_<main_pid>.html,flamegraph_<worker1_pid>.html,and so on.
Each subprocess receives its own output file. The filename is derived fromthe specified output path (or the default) with the subprocess’s process IDappended:
If you specify
-oprofile.html, subprocesses produceprofile_12345.html,profile_12346.html, and so onWith default output, subprocesses produce files like
flamegraph_12345.htmlor directories likeheatmap_12345For pstats format (which defaults to stdout), subprocesses produce files like
profile_12345.pstats
The subprocess profilers inherit most sampling options from the parent (interval,duration, thread selection, native frames, GC frames, async-aware mode, andoutput format). All Python descendant processes are profiled recursively,including grandchildren and further descendants.
Subprocess detection works by periodically scanning for new descendants ofthe target process and checking whether each new process is a Python processby probing the process memory for Python runtime structures. Non-Pythonsubprocesses (such as shell commands or external tools) are ignored.
There is a limit of 100 concurrent subprocess profilers to prevent resourceexhaustion in programs that spawn many processes. If this limit is reached,additional subprocesses are not profiled and a warning is printed.
The--subprocesses option is incompatible with--live modebecause live mode uses an interactive terminal interface that cannotaccommodate multiple concurrent profiler displays.
Sampling efficiency¶
Sampling efficiency metrics help assess the quality of the collected data.These metrics appear in the profiler’s terminal output and in the flame graphsidebar.
Sampling efficiency is the percentage of sample attempts that succeeded.Each sample attempt reads the target process’s call stack from memory. Anattempt can fail if the process is in an inconsistent state at the moment ofreading, such as during a context switch or while the interpreter is updatingits internal structures. A low efficiency may indicate that the profiler couldnot keep up with the requested sampling rate, often due to system load or anoverly aggressive interval setting.
Missed samples is the percentage of expected samples that were notcollected. Based on the configured interval and duration, the profiler expectsto collect a certain number of samples. Some samples may be missed if theprofiler falls behind schedule, for example when the system is under heavyload. A small percentage of missed samples is normal and does not significantlyaffect the statistical accuracy of the profile.
Both metrics are informational. Even with some failed attempts or missedsamples, the profile remains statistically valid as long as enough sampleswere collected. The profiler reports the actual number of samples captured,which you can use to judge whether the data is sufficient for your analysis.
Profiling modes¶
The sampling profiler supports four modes that control which samples arerecorded. The mode determines what the profile measures: total elapsed time,CPU execution time, time spent holding the global interpreter lock, orexception handling.
Wall-clock mode¶
Wall-clock mode (--mode=wall) captures all samples regardless of what thethread is doing. This is the default mode and provides a complete picture ofwhere time passes during program execution:
python -m profiling.sampling run --mode=wall script.pyIn wall-clock mode, samples are recorded whether the thread is activelyexecuting Python code, waiting for I/O, blocked on a lock, or sleeping.This makes wall-clock profiling ideal for understanding the overall timedistribution in your program, including time spent waiting.
If your program spends significant time in I/O operations, network calls, orsleep, wall-clock mode will show these waits as time attributed to the callingfunction. This is often exactly what you want when optimizing end-to-endlatency.
CPU mode¶
CPU mode (--mode=cpu) records samples only when the thread is actuallyexecuting on a CPU core:
python -m profiling.sampling run --mode=cpu script.pySamples taken while the thread is sleeping, blocked on I/O, or waiting fora lock are discarded. The resulting profile shows where CPU cycles are consumed,filtering out idle time.
CPU mode is useful when you want to focus on computational hotspots withoutbeing distracted by I/O waits. If your program alternates between computationand network calls, CPU mode reveals which computational sections are mostexpensive.
Comparing wall-clock and CPU profiles¶
Running both wall-clock and CPU mode profiles can reveal whether a function’stime is spent computing or waiting.
If a function appears prominently in both profiles, it is a true computationalhotspot—actively using the CPU. Optimization should focus on algorithmicimprovements or more efficient code.
If a function is high in wall-clock mode but low or absent in CPU mode, it isI/O-bound or waiting. The function spends most of its time waiting for network,disk, locks, or sleep. CPU optimization won’t help here; consider async I/O,connection pooling, or reducing wait time instead.
importtimedefdo_sleep():time.sleep(2)defdo_compute():sum(i**2foriinrange(1000000))if__name__=="__main__":do_sleep()do_compute()
python -m profiling.sampling run --mode=wall script.py # do_sleep ~98%, do_compute ~1%python -m profiling.sampling run --mode=cpu script.py # do_sleep absent, do_compute dominates
GIL mode¶
GIL mode (--mode=gil) records samples only when the thread holds Python’sglobal interpreter lock:
python -m profiling.sampling run --mode=gil script.pyThe GIL is held only while executing Python bytecode. When Python calls intoC extensions, performs I/O operations, or executes native code, the GIL istypically released. This means GIL mode effectively measures time spentrunning Python code specifically, filtering out time in native libraries.
In multi-threaded programs, GIL mode reveals which code is preventing otherthreads from running Python bytecode. Since only one thread can hold the GILat a time, functions that appear frequently in GIL mode profiles aremonopolizing the interpreter.
GIL mode helps answer questions like “which functions are monopolizing theGIL?” and “why are my other threads starving?” It can also be useful insingle-threaded programs to distinguish Python execution time from time spentin C extensions or I/O.
importhashlibdefhash_work():# C extension - releases GIL during computationfor_inrange(200):hashlib.sha256(b"data"*250000).hexdigest()defpython_work():# Pure Python - holds GIL during computationfor_inrange(3):sum(i**2foriinrange(1000000))if__name__=="__main__":hash_work()python_work()
python -m profiling.sampling run --mode=cpu script.py # hash_work ~42%, python_work ~38%python -m profiling.sampling run --mode=gil script.py # hash_work ~5%, python_work ~60%
Exception mode¶
Exception mode (--mode=exception) records samples only when a thread hasan active exception:
python -m profiling.sampling run --mode=exception script.pySamples are recorded in two situations: when an exception is being propagatedup the call stack (afterraise but before being caught), or when code isexecuting inside anexcept block where exception information is stillpresent in the thread state.
The following example illustrates which code regions are captured:
defexample():try:raiseValueError("error")# Captured: exception being raisedexceptValueError:process_error()# Captured: inside except blockfinally:cleanup()# NOT captured: exception already handleddefexample_propagating():try:try:raiseValueError("error")finally:cleanup()# Captured: exception propagating throughexceptValueError:passdefexample_no_exception():try:do_work()finally:cleanup()# NOT captured: no exception involved
Note thatfinally blocks are only captured when an exception is activelypropagating through them. Once anexcept block finishes executing, Pythonclears the exception information before running any subsequentfinallyblock. Similarly,finally blocks that run during normal execution (when noexception was raised) are not captured because no exception state is present.
This mode is useful for understanding where your program spends time handlingerrors. Exception handling can be a significant source of overhead in codethat uses exceptions for flow control (such asStopIteration in iterators)or in applications that process many error conditions (such as network servershandling connection failures).
Exception mode helps answer questions like “how much time is spent handlingexceptions?” and “which exception handlers are the most expensive?” It canreveal hidden performance costs in code that catches and processes manyexceptions, even when those exceptions are handled gracefully. For example,if a parsing library uses exceptions internally to signal format errors, thismode will capture time spent in those handlers even if the calling code neversees the exceptions.
Output formats¶
The profiler produces output in several formats, each suited to differentanalysis workflows. The format is selected with a command-line flag, andoutput goes to stdout, a file, or a directory depending on the format.
pstats format¶
The pstats format (--pstats) produces a text table similar to whatdeterministic profilers generate. This is the default output format:
python -m profiling.sampling run script.pypython -m profiling.sampling run --pstats script.py

The pstats format displays profiling results in a color-coded table showingfunction hotspots, sample counts, and timing estimates.¶
Output appears on stdout by default:
Profile Stats (Mode: wall): nsamples sample% tottime (ms) cumul% cumtime (ms) filename:lineno(function) 234/892 11.7% 234.00 44.6% 892.00 server.py:145(handle_request) 156/156 7.8% 156.00 7.8% 156.00 <built-in>:0(socket.recv) 98/421 4.9% 98.00 21.1% 421.00 parser.py:67(parse_message)
The columns show sampling counts and estimated times:
nsamples: Displayed as
direct/cumulative(for example,10/50).Direct samples are when the function was at the top of the stack, activelyexecuting. Cumulative samples are when the function appeared anywhere on thestack, including when it was waiting for functions it called. If a functionshows10/50, it was directly executing in 10 samples and was on the callstack in 50 samples total.sample% andcumul%: Percentages of total samples for direct andcumulative counts respectively.
tottime andcumtime: Estimated wall-clock time based on sample countsand the profiling duration. Time units are selected automatically based onthe magnitude: seconds for large values, milliseconds for moderate values,or microseconds for small values.
The output includes a legend explaining each column and a summary ofinteresting functions that highlights:
Hot spots: Functions with high direct/cumulative sample ratio (ratioclose to 1.0). These functions spend most of their time executing their owncode rather than waiting for callees. High ratios indicate where CPU timeis actually consumed.
Indirect calls: Functions with large differences between cumulative anddirect samples. These are orchestration functions that delegate work toother functions. They appear frequently on the stack but rarely at the top.
Call magnification: Functions where cumulative samples far exceed directsamples (high cumulative/direct multiplier). These are frequently-nestedfunctions that appear deep in many call chains.
Use--no-summary to suppress both the legend and summary sections.
To save pstats output to a file instead of stdout:
python -m profiling.sampling run -o profile.txt script.pyThe pstats format supports several options for controlling the display.The--sort option determines the column used for ordering results:
python -m profiling.sampling run --sort=tottime script.pypython -m profiling.sampling run --sort=cumtime script.pypython -m profiling.sampling run --sort=nsamples script.py
The--limit option restricts output to the top N entries:
python -m profiling.sampling run --limit=30 script.pyThe--no-summary option suppresses the header summary that precedes thestatistics table.
Collapsed stacks format¶
Collapsed stacks format (--collapsed) produces one line per unique callstack, with a count of how many times that stack was sampled:
python -m profiling.sampling run --collapsed script.pyThe output looks like:
main;process_data;parse_json;decode_utf8 42main;process_data;parse_json 156main;handle_request;send_response 89
Each line contains semicolon-separated function names representing the callstack from bottom to top, followed by a space and the sample count. Thisformat is designed for compatibility with external flame graph tools,particularly Brendan Gregg’sflamegraph.pl script.
To generate a flame graph from collapsed stacks:
python -m profiling.sampling run --collapsed script.py > stacks.txtflamegraph.pl stacks.txt > profile.svg
The resulting SVG can be viewed in any web browser and provides an interactivevisualization where you can click to zoom into specific call paths.
Flame graph format¶
Flame graph format (--flamegraph) produces a self-contained HTML file withan interactive flame graph visualization:
python -m profiling.sampling run --flamegraph script.pypython -m profiling.sampling run --flamegraph -o profile.html script.py
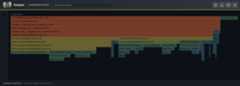
The flame graph visualization shows call stacks as nested rectangles, withwidth proportional to time spent. The sidebar displays runtime statistics,GIL metrics, and hotspot functions.¶
If no output file is specified, the profiler generates a filename based onthe process ID (for example,flamegraph.12345.html).
The generated HTML file requires no external dependencies and can be openeddirectly in a web browser. The visualization displays call stacks as nestedrectangles, with width proportional to time spent. Hovering over a rectangleshows details about that function including source code context, and clickingzooms into that portion of the call tree.
The flame graph interface includes:
A sidebar showing profile summary, thread statistics, sampling efficiencymetrics (seeSampling efficiency), and top hotspot functions
Search functionality supporting both function name matching and
file.py:42line patternsPer-thread filtering via dropdown
Dark/light theme toggle (preference saved across sessions)
SVG export for saving the current view
The thread statistics section shows runtime behavior metrics:
GIL Held: percentage of samples where a thread held the global interpreterlock (actively running Python code)
GIL Released: percentage of samples where no thread held the GIL
Waiting GIL: percentage of samples where a thread was waiting to acquirethe GIL
GC: percentage of samples during garbage collection
These statistics help identify GIL contention and understand how time isdistributed between Python execution, native code, and waiting.
Flame graphs are particularly effective for identifying deep call stacks andunderstanding the hierarchical structure of time consumption. Wide rectanglesat the top indicate functions that consume significant time either directlyor through their callees.
Gecko format¶
Gecko format (--gecko) produces JSON output compatible with the FirefoxProfiler:
python -m profiling.sampling run --gecko script.pypython -m profiling.sampling run --gecko -o profile.json script.py
TheFirefox Profiler is a sophisticatedweb-based tool originally built for profiling Firefox itself. It providesfeatures beyond basic flame graphs, including a timeline view, call treeexploration, and marker visualization. See theFirefox Profiler documentation fordetailed usage instructions.
To use the output, open the Firefox Profiler in your browser and load theJSON file. The profiler runs entirely client-side, so your profiling datanever leaves your machine.
Gecko format automatically collects additional metadata about GIL state andCPU activity, enabling analysis features specific to Python’s threading model.The profiler emits interval markers that appear as colored bands in theFirefox Profiler timeline:
GIL markers: show when threads hold or release the global interpreter lock
CPU markers: show when threads are executing on CPU versus idle
Code type markers: distinguish Python code from native (C extension) code
GC markers: indicate garbage collection activity
For this reason, the--mode option is not available with Gecko format;all relevant data is captured automatically.
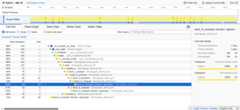
The Call Tree view shows the complete call hierarchy with sample countsand percentages. The sidebar displays detailed statistics for theselected function including running time and sample distribution.¶
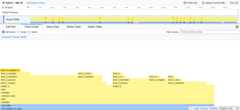
The Flame Graph visualization shows call stacks as nested rectangles.Functions names are visible in the call hierarchy.¶

The Marker Chart displays interval markers including CPU state, GILstatus, and opcodes. With--opcodes enabled, bytecode instructionslikeBINARY_OP_ADD_FLOAT,CALL_PY_EXACT_ARGS, andCALL_LIST_APPEND appear as markers showing execution over time.¶
Heatmap format¶
Heatmap format (--heatmap) generates an interactive HTML visualizationshowing sample counts at the source line level:
python -m profiling.sampling run --heatmap script.pypython -m profiling.sampling run --heatmap -o my_heatmap script.py

The heatmap overlays sample counts directly on your source code. Lines arecolor-coded from cool (few samples) to hot (many samples). Navigationbuttons (▲▼) let you jump between callers and callees.¶
Unlike other formats that produce a single file, heatmap output creates adirectory containing HTML files for each profiled source file. If no outputpath is specified, the directory is namedheatmap_PID.
The heatmap visualization displays your source code with a color gradientindicating how many samples were collected at each line. Hot lines (manysamples) appear in warm colors, while cold lines (few or no samples) appearin cool colors. This view helps pinpoint exactly which lines of code areresponsible for time consumption.
The heatmap interface provides several interactive features:
Coloring modes: toggle between “Self Time” (direct execution) and“Total Time” (cumulative, including time in called functions)
Cold code filtering: show all lines or only lines with samples
Call graph navigation: each line shows navigation buttons (▲ for callers,▼ for callees) that let you trace execution paths through your code. Whenmultiple functions called or were called from a line, a menu appears showingall options with their sample counts.
Scroll minimap: a vertical overview showing the heat distribution acrossthe entire file
Hierarchical index: files organized by type (stdlib, site-packages,project) with aggregate sample counts per folder
Dark/light theme: toggle with preference saved across sessions
Line linking: click line numbers to create shareable URLs
When opcode-level profiling is enabled with--opcodes, each hot linecan be expanded to show which bytecode instructions consumed time:
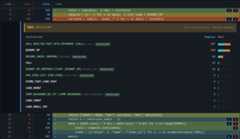
Expanding a hot line reveals the bytecode instructions executed, includingspecialized variants. The panel shows sample counts per instruction and theoverall specialization percentage for the line.¶
Heatmaps are especially useful when you know which file contains a performanceissue but need to identify the specific lines. Many developers prefer thisformat because it maps directly to their source code, making it easy to readand navigate. For smaller scripts and focused analysis, heatmaps provide anintuitive view that shows exactly where time is spent without requiringinterpretation of hierarchical visualizations.
Live mode¶
Live mode (--live) provides a terminal-based real-time view of profilingdata, similar to thetop command for system processes:
python -m profiling.sampling run --live script.pypython -m profiling.sampling attach --live 12345

Live mode displays real-time profiling statistics, showing combineddata from multiple threads in a multi-threaded application.¶
The display updates continuously as new samples arrive, showing the currenthottest functions. This mode requires thecurses module, which isavailable on Unix-like systems but not on Windows. The terminal must be atleast 60 columns wide and 12 lines tall; larger terminals display more columns.
The header displays the top 3 hottest functions, sampling efficiency metrics,and thread status statistics (GIL held percentage, CPU usage, GC time). Themain table shows function statistics with the currently sorted column indicatedby an arrow (▼).
When--opcodes is enabled, an additional opcode panel appears below themain table, showing instruction-level statistics for the currently selectedfunction. This panel displays which bytecode instructions are executing mostfrequently, including specialized variants and their base opcodes.

Live mode with--opcodes enabled shows an opcode panel with a bytecodeinstruction breakdown for the selected function.¶
Keyboard commands¶
Within live mode, keyboard commands control the display:
- q
Quit the profiler and return to the shell.
- s /S
Cycle through sort orders forward/backward (sample count, percentage,total time, cumulative percentage, cumulative time).
- p
Pause or resume display updates. Sampling continues in the backgroundwhile the display is paused, so you can freeze the view to examine resultswithout stopping data collection.
- r
Reset all statistics and start fresh. This is disabled after profilingfinishes to prevent accidental data loss.
- /
Enter filter mode to search for functions by name. The filter usescase-insensitive substring matching against the filename and function name.Type a pattern and press Enter to apply, or Escape to cancel. Glob patternsand regular expressions are not supported.
- c
Clear the current filter and show all functions again.
- t
Toggle between viewing all threads combined or per-thread statistics.In per-thread mode, a thread counter (for example,
1/4) appears showingyour position among the available threads.- ←→ or↑↓
In per-thread view, navigate between threads. Navigation wraps aroundfrom the last thread to the first and vice versa.
- + /-
Increase or decrease the display refresh rate. The range is 0.05 seconds(20 Hz, very responsive) to 1.0 second (1 Hz, lower overhead). Faster refreshrates use more CPU. The default is 0.1 seconds (10 Hz).
- x
Toggle trend indicators that show whether functions are becoming hotteror cooler over time. When enabled, increasing metrics appear in green anddecreasing metrics appear in red, comparing each update to the previous one.
- h or?
Show the help screen with all available commands.
- j /k (orUp /Down)
Navigate through opcode entries in the opcode panel (when
--opcodesisenabled). These keys scroll through the instruction-level statistics for thecurrently selected function.
When profiling finishes (duration expires or target process exits), the displayshows a “PROFILING COMPLETE” banner and freezes the final results. You canstill navigate, sort, and filter the results before pressingq to exit.
Live mode is incompatible with output format options (--collapsed,--flamegraph, and so on) because it uses an interactive terminalinterface rather than producing file output.
Async-aware profiling¶
For programs usingasyncio, the profiler offers async-aware mode(--async-aware) that reconstructs call stacks based on the task structurerather than the raw Python frames:
python -m profiling.sampling run --async-aware async_script.pyStandard profiling of async code can be confusing because the physical callstack often shows event loop internals rather than the logical flow of yourcoroutines. Async-aware mode addresses this by tracking which task is runningand presenting stacks that reflect theawait chain.
importasyncioasyncdeffetch(url):awaitasyncio.sleep(0.1)returnurlasyncdefmain():for_inrange(50):awaitasyncio.gather(fetch("a"),fetch("b"),fetch("c"))if__name__=="__main__":asyncio.run(main())
python -m profiling.sampling run --async-aware --flamegraph -o out.html script.pyNote
Async-aware profiling requires the target process to have theasynciomodule loaded. If you profile a script before it imports asyncio, async-awaremode will not be able to capture task information.
Async modes¶
The--async-mode option controls which tasks appear in the profile:
python -m profiling.sampling run --async-aware --async-mode=running async_script.pypython -m profiling.sampling run --async-aware --async-mode=all async_script.py
With--async-mode=running (the default), only the task currently executingon the CPU is profiled. This shows where your program is actively spending timeand is the typical choice for performance analysis.
With--async-mode=all, tasks that are suspended (awaiting I/O, locks, orother tasks) are also included. This mode is useful for understanding what yourprogram is waiting on, but produces larger profiles since every suspended taskappears in each sample.
Task markers and stack reconstruction¶
In async-aware profiles, you will see<task> frames that mark boundariesbetween asyncio tasks. These are synthetic frames inserted by the profiler toshow the task structure. The task name appears as the function name in theseframes.
When a task awaits another task, the profiler reconstructs the logical callchain by following theawait relationships. Only “leaf” tasks (tasks thatno other task is currently awaiting) generate their own stack entries. Tasksbeing awaited by other tasks appear as part of their awaiter’s stack instead.
If a task has multiple awaiters (a diamond pattern in the task graph), theprofiler deterministically selects one parent and annotates the task markerwith the number of parents, for exampleMyTask(2parents). This indicatesthat alternate execution paths exist but are not shown in this particular stack.
Option restrictions¶
Async-aware mode uses a different stack reconstruction mechanism and isincompatible with:--native,--no-gc,--all-threads, and--mode=cpu or--mode=gil.
Command-line interface¶
The complete command-line interface for reference.
Global options¶
- run¶
Run and profile a Python script or module.
- attach¶
Attach to and profile a running process by PID.
Sampling options¶
- -i<microseconds>,--interval<microseconds>¶
Sampling interval in microseconds. Default: 100.
- -d<seconds>,--duration<seconds>¶
Profiling duration in seconds. Default: 10.
- -a,--all-threads¶
Sample all threads, not just the main thread.
- --realtime-stats¶
Display sampling statistics during profiling.
- --native¶
Include
<native>frames for non-Python code.
- --no-gc¶
Exclude
<GC>frames for garbage collection.
- --async-aware¶
Enable async-aware profiling for asyncio programs.
- --opcodes¶
Gather bytecode opcode information for instruction-level profiling. Showswhich bytecode instructions are executing, including specializations.Compatible with
--live,--flamegraph,--heatmap, and--geckoformats only.
- --subprocesses¶
Also profile subprocesses. Each subprocess gets its own profilerinstance and output file. Incompatible with
--live.
Mode options¶
- --mode<mode>¶
Sampling mode:
wall(default),cpu,gil, orexception.Thecpu,gil, andexceptionmodes are incompatible with--async-aware.
- --async-mode<mode>¶
Async profiling mode:
running(default) orall.Requires--async-aware.
Output options¶
- --pstats¶
Generate text statistics output. This is the default.
- --collapsed¶
Generate collapsed stack format for external flame graph tools.
- --flamegraph¶
Generate self-contained HTML flame graph.
- --gecko¶
Generate Gecko JSON format for Firefox Profiler.
- --heatmap¶
Generate HTML heatmap with line-level sample counts.
- -o<path>,--output<path>¶
Output file or directory path. Default behavior varies by format:
--pstatswrites to stdout,--flamegraphand--geckogeneratefiles likeflamegraph.PID.html, and--heatmapcreates a directorynamedheatmap_PID.
pstats display options¶
These options apply only to pstats format output.
- --sort<key>¶
Sort order:
nsamples,tottime,cumtime,sample-pct,cumul-pct,nsamples-cumul, orname. Default:nsamples.
- -l<count>,--limit<count>¶
Maximum number of entries to display. Default: 15.
- --no-summary¶
Omit the Legend and Summary of Interesting Functions sections from output.
Run command options¶
- -m,--module¶
Treat the target as a module name rather than a script path.
- --live¶
Start interactive terminal interface instead of batch profiling.
See also
profilingOverview of Python profiling tools and guidance on choosing a profiler.
profiling.tracingDeterministic tracing profiler for exact call counts and timing.
pstatsStatistics analysis for profile data.
- Firefox Profiler
Web-based profiler that accepts Gecko format output. See thedocumentation for usage details.
- FlameGraph
Tools for generating flame graphs from collapsed stack format.