Faster raster processing inR usingGRASS
fasterRaster is anR package designedspecifically to handle large-in-memory/large-on-disk spatial rasters andvectors.fasterRaster does this using Open SourceGeospatial’sGRASS
fasterRaster was created with five designprinciples:
fasterRaster complementsterra andsf, and is highly dependent on them!It is useful for analyzing large-in-memory/large-on-disk rasters andvectors that those packages struggle to handle. For medium- andsmall-size objects,terra andsf will almostalways be faster.terra, you basically know how to usefasterRaster! That’s because most of the functions have thesame name and almost the same arguments asterrafunctions.fasterRaster are the same as those from functions interra with the same name.GRASS requires users totrack things like “locations” or “projects”, “mapsets”, and “regions”for which there is no comparable analog in theterra orsf packages.fasterRaster handles these behindthe scenes so you don’t need to.rgrass package provides apowerful conduit through which you can runGRASS tools fromR. As such, it provides much more flexibility thanfasterRaster. However, to usergrass, you needto know whatGRASS tools you want to use and be familiarwithGRASS syntax.fasterRaster obviates thisstep but usesrgrass as a backend, allowing you to focus onR syntax and look up help for functions the normal way youdo inR. You don’t need to knowGRASS!fasterRaster makes heavy use of thergrasspackage by Roger Bivand and others, theterrapackage by Robert Hijmans, thesfpackage by Edzer Pebesma, Roger Bivand, and others, and of courseGRASS, so is greatlyindebted to all of these creators!
fasterRaster comes with four user-orientedvignettes, plus apkgdownsitewith full documentation:
oGettingstarted (also reproduced below)
oTypesofGRasters
oMakingfasterRaster faster
oAddons
oDocumentation
To installfasterRaster, please use:
install_packages('fasterRaster', dependencies = TRUE)
You can get the latest stable release using:
remotes::install_github('adamlilith/fasterRaster', dependencies = TRUE)
…and the development version from:
remotes::install_github('adamlilith/fasterRaster@intuitive_fasterRaster', dependencies = TRUE)
To usefasterRaster you must installGRASS version 8.3+ on your operatingsystem.You will need to use the stand-alone installer, not theOpen Source Geospatial (OS Geo) installer.
Optional: A few functions infasterRaster requireGRASS “addon”tools, which do not come bundled withGRASS. You do notneed to install these addons if you do not use functions that call them.A list of functions that require addons can be seen in the “addons”vignette (inR, usevignette("addons", package = "fasterRaster")). Thisvignette also explains how to install addons.
The example presented here is the same as that presented in the the“gettingstarted” vignette.
We’ll do a simple operation in which we:
Add a buffer to lines representing rivers, then
Calculate the distance to from each cell to the closest bufferand burn the distance values into a raster.
To do this, we’ll be using maps representing the middle of theeastern coast of Madagascar. We will also use theterra andsf packages.
library(terra) # GIS for rasters and vectorslibrary(sf) # GIS for vectorslibrary(fasterRaster)# Get example elevation raster and rivers vector:madElev <- fastData('madElev') # SpatRaster with elevationmadRivers <- fastData('madRivers') # sp vector with rivers# Plot inputs:plot(madElev)plot(st_geometry(madRivers), col = "lightblue", add = TRUE)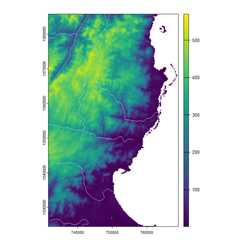
Before you use nearly any function in the package, you need to tellfasterRaster whereGRASS is installed on yoursystem. The installation folder will vary by operating system and maybeGRASS version, but will look something like this:
# Choose the appropriate one, and modify as needed:grassDir <- "C:/Program Files/GRASS GIS 8.4" # WindowsgrassDir <- "/Applications/GRASS-8.4.app/Contents/Resources" # Mac OSgrassDir <- "/usr/local/grass" # LinuxNow, use thefaster() function to tellfasterRaster whereGRASS is installed:
faster(grassDir = grassDir)Thefast() function is the key function for loading araster or vector intofasterRaster format. Rasters in thispackage are calledGRasters and vectorsGVectors (the “G” stands forGRASS). We willnow convert themadElev raster, which is aSpatRaster from theterra package, into aGRaster.
elev <- fast(madElev)elevYou will see theGRasters metadata:
class : GRastertopology : 2D dimensions : 1024, 626, NA, 1 (nrow, ncol, ndepth, nlyr)resolution : 59.85157, 59.85157, NA (x, y, z)extent : 731581.552, 769048.635, 1024437.272, 1085725.279 (xmin, xmax, ymin, ymax)coord ref. : Tananarive (Paris) / Laborde Grid name(s) : madElev datatype : integer min. value : 1 max. value : 570Next, we’ll do the same for the rivers vector. In this case, thevector is ansf object from thesf package,but we could also use aSpatVector from theterra package.
rivers <- fast(madRivers)riversclass : GVectorgeometry : 2D lines dimensions : 11, 11, 5 (geometries, sub-geometries, columns)extent : 731627.1, 762990.132, 1024541.235, 1085580.454 (xmin, xmax, ymin, ymax)coord ref. : Tananarive (Paris) / Laborde Grid names : F_CODE_DES HYC_DESCRI NAM ISO NAME_0 type : <chr> <chr> <chr> <chr> <chr> values : River/Stream Perennial/Permanent MANANARA MDG Madagascar River/Stream Perennial/Permanent MANANARA MDG Madagascar River/Stream Perennial/Permanent UNK MDG Madagascar ...and 8 more rowsNow, let’s add a 1000-m buffer to the rivers usingbuffer(). As much as possible,fasterRasterfunctions have the same names and same arguments as their counterpartsin theterra package to help users who are familiar withthat package.
Note, though, that the output fromfasterRaster is notnecessarily guaranteed to be the same as output from the respectivefunctionsterra. This is because there are differentmethods to do the same thing, and the developers ofGRASSmay have chosen different methods than the developers of other GISpackages.
# width in meters because CRS is projectedriver_buffers <- buffer(rivers, width = 1000)Now, let’s calculate the distances between the buffered areas and allcells on the raster map usingdistance().
dist_to_rivers_meters <- distance(elev, river_buffers)Finally, let’s plot the output.
plot(dist_to_rivers_meters)plot(river_buffers, border = 'white', add = TRUE)plot(rivers, col = "lightblue", add = TRUE)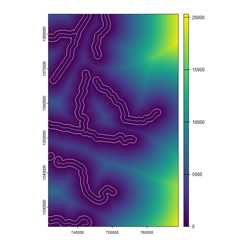
And that’s how it’s done! You can do almost anything infasterRaster you can do withterra. Theexamples above do not show the advantage offasterRasterbecause the they do not use in large-in-memory/large-on-disk spatialdatasets. For very large datasets,fasterRaster can be muchfaster! For example, for a large raster (many cells), thedistance() function interra can take manydays to run and even crashR, whereas infasterRaster, it could take just a few minutes orhours.
GRasters andGVectors from aGRASS sessionYou can convert aGRaster to aSpatRasterraster usingrast():
terra_elev <- rast(elev)
To convert aGVector to theterra package’sSpatVector, usevect():
terra_rivers <- vect(rivers)You can usewriteRaster() andwriteVector()to savefasterRaster rasters and vectors directly to disk.This willalways be faster than usingrast() orvect() and then saving.
elev_temp_file <- tempfile(fileext = ".tif") # save as GeoTIFFwriteRaster(elev, elev_temp_file)vect_temp_shp <- tempfile(fileext = ".shp") # save as shapefilevect_temp_gpkg <- tempfile(fileext = ".gpkg") # save as GeoPackagewriteVector(rivers, vect_temp_shp)writeVector(rivers, vect_temp_gpkg)fasterRaster versions will look something like8.3.1.2, or more generally,M1.M2.S1.S2. Here,M1.M2 will mirror the version ofGRASS forwhichfasterRaster was built and tested. For example,fasterRaster version 8.4.x.x will work usingGRASS 8.4 (and version 8.3). The values inS1.S2 refer to “major” and “minor” versions offasterRaster. That is, a change in the value ofS1 (e.g., fromx.x.1.0 tox.x.2.0) indicates changes that potentially break oldercode developed with a prior version offasterRaster. Achange inS2 refers to a bug fix, additional functionalityin an existing function, or the addition of an entirely newfunction.
Note that theM1.M2 andS1.S2 incrementindependently. For example, if the version changes from8.3.1.5 to8.4.1.5, then the new version hasbeen tested onGRASS 8.4, but code developed with version8.3.1.x offasterRaster should still work.
NOTE: WhilefasterRaster is still inbeta/alpha release, the version will look something like8.3.0.7XXX, following Hadley Wickham’s guidelines forversioning under development.
terrapackage and Edzer Pebesma’ssf packageare good places to start if you are not familiar with doing GIS inR.rgrasspackage allows users to call anyGRASS function with all ofits functionality, which in some cases is far beyond what is allowed byfasterRaster.GRASSfunctions used in this package and more.GRASS inR orR inGRASS will help you to become a power-user ofGRASS inR.A publication is forthcoming! In the meantime, please see andcite:
fasterRaster: GIS inRusingGRASS for large vectors and rasters. EarthArXivpreprint doi: <href =“https://doi.org/10.31223/X52R0M”>10.31223/X52R0MAdam