targeted)
Various methods for targeted learning and semiparametric inferenceincluding augmented inverse probability weighted (AIPW) estimators formissing data and causal inference (Bang and Robins (2005)doi:10.1111/j.1541-0420.2005.00377.x), variableimportance and conditional average treatment effects (CATE) (van derLaan (2006)doi:10.2202/1557-4679.1008), estimators for riskdifferences and relative risks (Richardson et al. (2017)doi:10.1080/01621459.2016.1192546), assumption leaninference for generalized linear model parameters (Vansteelandt etal. (2022)doi:10.1111/rssb.12504).
You can install the released version of targeted fromCRAN with:
install.packages("targeted")And the development version fromGitHub with:
remotes::install_github("kkholst/targeted",ref="dev")Computations such as cross-validation are parallelized via the{future} package. To enable parallel computations andprogress-bars the following code can be executed
future::plan("multisession")progressr::handlers(global=TRUE)To illustrate some of the functionality of thetargetedpackage we simulate some data from the following model
library("targeted")simdata<-function(n, ...) { w1<-rnorm(n)# covariates w2<-rnorm(n)# ... a<-rbinom(n,1,plogis(-1+ w1))# treatment indicator y<-exp(- (w1-1)**2- (w2-1)**2)-# continuous response2*exp(- (w1+1)**2- (w2+1)**2)* a+# additional effect in treatedrnorm(n,sd=0.5**.5)data.frame(y, a, w1, w2)}set.seed(1)d<-simdata(5e3)head(d)#> y a w1 w2#> 1 -0.59239667 0 -0.6264538 -1.5163733#> 2 0.01794935 0 0.1836433 0.6291412#> 3 0.24968229 0 -0.8356286 -1.6781940#> 4 1.34434300 1 1.5952808 1.1797811#> 5 1.16367655 0 0.3295078 1.1176545#> 6 -0.94757031 0 -0.8204684 -1.2377359wnew<-seq(-3,3,length.out=200)dnew<-expand.grid(w1 = wnew,w2 = wnew,a =1)y<-with(dnew,exp(- (w1-1)**2- (w2-1)**2)-2*exp(- (w1+1)**2- (w2+1)**2)*a )image(wnew, wnew,matrix(y,ncol=length(wnew)),col=viridisLite::viridis(64),main=expression(paste("E(Y|",W[1],",",W[2],")")),xlab=expression(W[1]),ylab=expression(W[2]))
Methods for targeted and semiparametric inference rely on fittingnuisance models to observed data when estimating the target parameter ofinterest. The{targeted} package implements theR6 reference classlearnerto harmonize common statistical and machine learning models for theusage as nuisance models across the various implemented estimators, suchas thetargeted:cate function. Commonly used models areconstructed aslearner class objects through thelearner_* functions.
As an example, we can specify a linear regression model with aninteraction term between treatment and the two covariates
lr<-learner_glm(y~ (w1+ w2)*a,family = gaussian)lr#> ────────── learner object ──────────#> glm#>#> Estimate arguments: family=<function>#> Predict arguments:#> Formula: y ~ (w1 + w2) * a <environment: 0xaee788b30>To fit the model to the data we use theestimatemethod
lr$estimate(d)lr$fit#>#> Call: stats::glm(formula = formula, family = family, data = data)#>#> Coefficients:#> (Intercept) w1 w2 a w1:a w2:a#> 0.18808 0.13044 0.08253 -0.33517 0.15330 0.24068#>#> Degrees of Freedom: 4999 Total (i.e. Null); 4994 Residual#> Null Deviance: 3098#> Residual Deviance: 2741 AIC: 11200Predictions,\(E(Y\mid W_1, W_2)\),can be performed with thepredict method
head(d)|> lr$predict()#> 1 2 3 4 5 6#> -0.01878799 0.26395942 -0.05942914 0.68687155 0.32330487 -0.02109944pr<-matrix(lr$predict(dnew),ncol=length(wnew))image(wnew, wnew, pr,col=viridisLite::viridis(64),main=expression(paste("E(Y|",W[1],",",W[2],")")),xlab=expression(W[1]),ylab=expression(W[2]))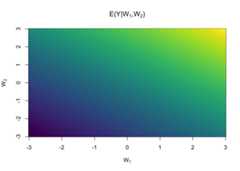
Similarly, a Random Forest can be specified with
lr_rf<-learner_grf(y~ w1+ w2+ a,num.trees =500)Lists of models can also be constructed for differenthyper-parameters with thelearner_expand_grid function.
To assess the model generalization error we can performcv method
mod<-list(glm = lr,rf = lr_rf)cv(mod,data = d,rep =2,nfolds =5)|>summary()#> , , mse#>#> mean sd min max#> glm 0.5498117 0.02987117 0.5085057 0.5969734#> rf 0.5070569 0.03177828 0.4597520 0.5534290#>#> , , mae#>#> mean sd min max#> glm 0.5907746 0.01516298 0.5686472 0.6148165#> rf 0.5684956 0.01659710 0.5453521 0.5953637An ensemble learner (super-learner) can easily be constructed fromlists oflearner objects
sl<-learner_sl(mod,nfolds =10)sl$estimate(d)sl#> ────────── learner object ──────────#> superlearner#> glm#> rf#>#> Estimate arguments: learners=<list>, nfolds=10, meta.learner=<function>, model.score=<function>#> Predict arguments:#> Formula: y ~ (w1 + w2) * a <environment: 0x15cf956c8>#> ─────────────────────────────────────#> score weight#> glm 0.5499084 0.03290729#> rf 0.5070931 0.96709271pr<-matrix(sl$predict(dnew),ncol=length(wnew))image(wnew, wnew, pr,col=viridisLite::viridis(64),main=expression(paste("E(Y|",W[1],",",W[2],")")),xlab=expression(W[1]),ylab=expression(W[2]))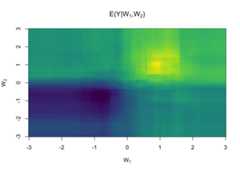
In the following we are interested in estimating the target parameter\(\psi_a(P) = E_P[Y(a)]\), where
then the target parameter can be identified from the observed datadistribution as\[E(E[Y|W,A=a]) =E(E[Y(a)|W]) = E[Y(a)]\] or\[E[YI(A=a)/P(A=a|W)] = E[Y(a)].\]
This suggests estimators based on outcome regression (
In practice, this requires plugin estimates of both the outcome model,\(Q(W,A) := E(Y\mid A, W)\), and of thetreatment propensity model\(\Pi_a(W) :=P(A=a\mid W)\). The corresponding estimator is consistent even ifjust one of the two nuisance models is correctly specified.
First we specify the propensity model
prmod<-learner_glm(a~ w1+ w2,family=binomial)We will reuse one of the outcome models from the previous section,and use thecate function to estimate the treatmenteffect
a<-cate(response.model = lr_rf,propensity.model = prmod,data = d,nfolds =5)a#> Estimate Std.Err 2.5% 97.5% P-value#> E[y(1)] -0.1700 0.02628 -0.2214840 -0.1185 9.939e-11#> E[y(0)] 0.1483 0.07595 -0.0005763 0.2971 5.089e-02#> ───────────#> (Intercept) -0.3183 0.07996 -0.4749849 -0.1615 6.892e-05In the output we get estimates of both the mean potential outcomesand the difference of those, the average treatment effect, given as theterm(Intercept).
summary(a)#> cate(response.model = lr_rf, propensity.model = prmod, data = d,#> nfolds = 5)#>#> Estimate Std.Err 2.5% 97.5% P-value#> E[y(1)] -0.1700 0.02628 -0.2214840 -0.1185 9.939e-11#> E[y(0)] 0.1483 0.07595 -0.0005763 0.2971 5.089e-02#> ───────────#> (Intercept) -0.3183 0.07996 -0.4749849 -0.1615 6.892e-05#>#> Average Treatment Effect:#> Estimate Std.Err 2.5% 97.5% P-value#> [E[y(1)]] - [E[y(0)]] -0.3183 0.08008 -0.4752 -0.1613 7.055e-05#>#> Null Hypothesis:#> [E[y(1)]] - [E[y(0)]] = 0Here we use thenfolds=5 argument to use 5-foldcross-fitting to guarantee that the estimates converges weaklyto a Gaussian distribution even though that the estimated influencefunction based on plugin estimates from the Random Forest does notnecessarily lie in a\(P\)-Donskerclass.
We use thedev branch for development and themain branch for stable releases. All releases followsemantic versioning, aretagged and notablechanges are reported inNEWS.md.
If you want to ask questions, require help or clarification, orreport a bug, we recommend to either contact a maintainer directly orthe following:
We will then take care of the issue as soon as possible.
targetedAll types of contributions are encouraged and valued. See theCONTRIBUTING.mdfor details about how to contribute code to this project.