Nikolas I. Krieger, M.S.;1 Adam T. Perzynski,Ph.D.;2 and Jarrod E. Dalton, Ph.D.1,3
1 Department of Quantitative Health Sciences, LernerResearch Institute, Cleveland Clinic, 9500 Euclid Avenue (JJN3),Cleveland, OH, 44195
2 Center for Healthcare Research and Policy, Case WesternReserve University at MetroHealth, 2500 MetroHealth Drive, Cleveland, OH44109
3 Cleveland Clinic Lerner College of Medicine, Case WesternReserve University
Acknowledgements:
The authors of this package acknowledge the support provided by membersof the Northeast Ohio Cohort for Atherosclerotic Risk Estimation(NEOCARE) investigative team: Claudia Coulton, Douglas Gunzler, DarcyFreedman, Neal Dawson, Michael Rothberg, David Zidar, David Kaelber,Douglas Einstadter, Alex Milinovich, Monica Webb Hooper, KristenHassmiller-Lich, Ye Tian (Devin), Kristen Berg, and Sandy Andrukat.
Funding:
This work was supported by The National Institute on Aging of theNational Institutes of Health under award number R01AG055480. Thecontent is solely the responsibility of the authors and does notnecessarily represent the official views of the National Institutes ofHealth.
You can installprojects with:
install.packages("projects")The goal of theprojects R package is to provide a setof tools that support an efficient project management workflow forstatisticians and data scientists who perform reproducible researchwithin team science environments. Theprojects package isbuilt upon some existing tools for reproducible research, particularlyRStudio, the R integrated development environment in which it dwells,and R Markdown, the file structure that allows users to assembledatasets, to perform analyses and to write manuscripts in a single file.Theprojects package is oriented towards efficient andreproducible academic research manuscript development and incorporatesprotocol and analysis templates based on widely-accepted reportingguidelines (viz., CONSORT and STROBE). When used on a shared file system(e.g., a server), theprojects package providesinfrastructure for collaborative research: multiple researchers can workon the same project and keep track of its progress without having torequest updates.
The primary features of the projects R package are the following:
At its outset, theprojects package creates a foldercalled/projects in a user-specified location. This directorywill contain all research projects created and maintained by the user.The/projects folder will also contain a relational database ofthe research projects and the persons who contribute to them. Thedatabase allows for users to input important metadata about the projectsand authors, including stage of research and contact information. Oncethis higher-level folder is created, users run R functions to createprojects, each of which is given its own folder. New project foldersautomatically include aptly named subfolders and templates in order toguide the project workflow (e.g., a “data” subfolder; a “datawork” RMarkdown template). Right away, users can begin working on the researchproject and edit the metadata of the project itself and its authors. Tolessen the burden of the mundane details of manuscript writing, theprojects package can output lines to the console that, whencopied into an R Markdown file, generates a title page with all relevantauthorship information of any given project. Finally, since users maycreate dozens of projects over time, users can run functions to organizetheir projects within grouping subfolders of the main/projectsfolder.
Reproducibility in research is the focus of the much debatedreplication crisis and is therefore an increasingly central goal of thecontemporary scientific process. In addition to a final report of studyresults, reproducible research processes include the entire workflowthat the researchers used to generate those results. Activelymaintaining and archiving this workflow is important to the evaluationand validation of the research. If other researchers can follow the sameworkflow to achieve the same results, it corroborates the results asscientific knowledge. When results are not produced by the sameworkflow, however, scientific knowledge is still advanced, as theworkflow is shown not to yield generalizable results. (Baker 2016)
There exist today widely available tools that aid with reproducibleresearch, such as R and other statistical programming languages, thatallow for precise documentation of some of the most detail-orientedportions of a project workflow. Researchers can distribute their codescripts alongside their results in order to communicate the integrity oftheir data processing and analysis. Unfortunately, statisticalprogramming languages per se only contribute to research reproducibilityinsofar as individual statistical programmers are able (1) to use thesetools effectively and (2) to integrate their own use of these tools withtheir collaborators’ work—which may not necessarily be oriented towardsreproducibility.
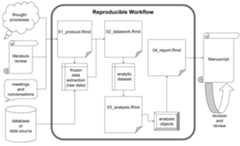
Although researchers of different disciplines may operate in nuancedways, there are aspects of the project workflow that are common to mostinvestigations. First, studies are conceptualized and designed accordingto a protocol that details the research questions and planned analyses.Data are collected, manipulated (or “tidied”) in order to make dataanalysis possible. The results of the analyses are compiled into areport, and ultimately an academic manuscript is drafted and submittedfor wider distribution.
When navigating this workflow, researchers strive for reproducibilitywherever possible, but especially during the intermediate, data-focusedphases of the workflow. Readers of the final manuscript should haveaccess to the study data in its most unrefined state possible: a frozendataset. A frozen dataset is almost invariably a digital file or set offiles that standard data analysis software can process. Whereas studydata may have been initially collected in a non-digital manner, a frozendata set represents the study data’s earliest state of simultaneousdigitization and consolidation. From this point forward through thereporting stage, total reproducibility is expected. Thanks to moderndata analysis via statistical programming languages, a reader should beable to exactly reproduce all data-derived results from the frozen dataset alone. With access to the exact scripts the researchers used toproduce their results, readers can scrutinize every function callperformed on the frozen data set and its descendants.
The middle stage of the assumed study workflow can be performed withnear perfect reproducibility, but the beginning and ending stages maynot. Researchers cannot document every thought process, literature probeand informal conversation that contributes to the development of theinitial study protocol, but they should strive to document it asmeticulously as possible. Databases tend to be dynamic such that a givenanalytic data set is merely a snapshot in time. As for the final stagesof project development, journals require that manuscripts adhere tospecific and unique stylistic guidelines and that they be digitallysubmitted with file types that are not independently conducive toreproducibility (e.g.,.pdf). For instance, even as RStudiosupports the creation of submission-ready documents directly from frozendatasets, the vast majority of project teams include experts who do notuse RStudio; therefore, the collaborative manuscript editing processultimately takes place in an environment (e.g., Microsoft Word) thatonly supports total reproducibility with extraordinary effort. In lightof these realities, researchers must do their best during manuscriptcreation, keeping the process in reproducible environments for as longas possible and otherwise documenting significant changes andalterations.
projects packageAll projects that the user creates with theprojectspackage—as well as its infrastructure—reside in a main folder called/projects. Users need not manually create this directory, andin fact they are encouraged not to manually manipulate any folders thattheprojects package involves. Instead, users run thefunctionsetup_projects(), providing the full file path ofthe directory in which the user wants the/projects folder toreside.
Data about authors, institutional and/or department affiliations andprojects are stored in.rds files within the main/project directory, so that the user only needs to enter thesedetails once (unless, for example, a co-author changes their name oraffiliations). These data are also used to assemble title pages ofreports, with automatically generated author lists and lists of authoraffiliations. We provide a complete example of this process below in theDemonstration section below.
The main metadata tables accessible to the user areprojects(),authors() andaffiliations(), via functions thusly named. Two additionaltables are internally created to keep track of associations betweenauthors and projects and between authors and affiliations (see theInternal Tables section).
id – an identification number, specifically an integer,unique among the other projects. This number can be used wheneverneeding to identify this project withinprojects packagefunctions.title – the title of the project. A nonambiguoussubstring oftitle (i.e., a substring that does not matchany other project) can be used whenever needing to identify this projectwithinprojects package functions.short_title – an optional unique nickname for theproject. A nonambiguous substring ofshort_title (i.e., asubstring that does not match any other project) can be used wheneverneeding to identify this project withinprojects packagefunctions. This is useful if users cannot remember the long, formalprojecttitle nor the projectid.current_owner – theid of the author whois responsible for taking action in order that work on the project mayproceed further.status – a short description of the status of theproject. For example, it may elaborate on the value ofcurrent_owner and/orstage.deadline_type – a simple description of the meaning ofthe date contained in the next field,deadline.deadline – a date indicating some kind of deadlinewhose meaning is described in the previous field,deadline_type.stage – one of seven predefined stages of projectdevelopment that the project is currently in:c("0: idea","1: design","2: data collection","3: analysis","4: manuscript","5: under review","6: accepted")path – the full file path where the project folder islocated.corresp_auth – theid of the author whoshould be contacted for any correspondence relating to the project. Thisauthor’s name will be especially marked on automatically generated titlepages for this project, and his or her contact information will beespecially displayed there as well in a “Corresponding Author”section.creator – theid of the author whoinitially created the project, or the value ofSys.info()["user"] if the author who rannew_project() did not enter a value.id – an identification number, specifically an integer,unique among the other authors. This number can be used whenever needingto identify this author withinprojects packagefunctions.given_names – the given name or names of the author. Anonambiguous substring ofgiven_names (i.e., a substringthat does not match any other author) can be used whenever needing toidentify this author withinprojects package functions.This is included in the automatically generated title pages of theprojects associated with this author.last_name – the last name or names of the author. Anonambiguous substring oflast_name (i.e., a substring thatdoes not match any other author) can be used whenever needing toidentify this author withinprojects package functions.This is included aftergiven_names in the automaticallygenerated title pages of the projects associated with this author.title – the job title of the author.degree – the abbreviation(s) of the author’s academicdegree(s). This is included afterlast_name in theautomatically generated title pages of the projects associated with thisauthor.email – the email address of the author. This isincluded in the “Corresponding Author” section of the automaticallygenerated title pages of projects whosecorresp_auth fieldcontains this author.phone – the phone number of the author. This isincluded in the “Corresponding Author” section of the automaticallygenerated title pages of projects whosecorresp_auth fieldcontains this author.id – an identification number, specifically an integer,unique among the other affiliations. This number can be used wheneverneeding to identify this affiliation withinprojectspackage functions.department_name – the department name of theaffiliation. A nonambiguous substring ofdepartment_name(i.e., a substring that does not match any other affiliation) can beused whenever needing to identify this affiliation withinprojects package functions. This is included in theaffiliations section of the automatically generated title page ofprojects associated with authors with this affiliation.institution_name – the name of the overall institutionof the affiliation. A nonambiguous substring ofinstitution_name (i.e., a substring that does not match anyother affiliation) can be used whenever needing to identify thisaffiliation withinprojects package functions. This isincluded afterdepartment_name in the affiliations sectionof the automatically generated title page of projects associated withauthors with this affiliation.address – the address of the affiliation. This isincluded afterinstitution_name in the affiliations sectionof the automatically generated title page of projects associated withauthors with this affiliation. It is also included in the “CorrespondingAuthor” section of the title page when a project’s corresponding authorhas this affiliation as his or her primary (i.e., first) affiliation(see theInternal Tables section).In keeping with relational database theory, there are two.rds files that keep track of the many-to-many relationshipsbetween projects and authors and between authors and affiliations. Eachhas two columns,id1 andid2, that contain theid numbers of these items. Each row of this table describesan association. Furthermore, theprojects package keepstrack of the order in which these associations appear so that theautomatically generated title pages list authors and affiliations in thecorrect order. Users are able to run functions to reorder theseassociations as needed.
Users create individual project folders with the functionnew_project(). By default, the name of each project folderis of the form pXXXX, whereXXXX is the project’sid padded with 0s on the left side. Its contents are copiedfrom a template project folder within the.templates directoryin the main/projects folder.
The default project folder template is structured as follows:
The included subfolders serve to organize the project, while the.Rmd files are templates that facilitate the user’s workflow.Thestyle.css andstyles.docx files are a cascadingstyle sheets (CSS) file and a Word style template, which allow forcustom styling of knitted HTML files and Word documents, respectively.Thecitations.bib file is an empty BibTeX file. The default01_protocol.Rmd and04_report.Rmd files alreadyreference the.bib and style files, easing the implementationof these features for the user.
The goal of theprojects package is to provide acomprehensive set of tools managing project files in a way that isself-contained in R and independent of the underlying operating system.On a daily basis, researchers make, move, copy, delete and archivefiles. Through theprojects package, researchers canperform all these actions in an organized manner with an automated filestructure. In fact, users are advised not to manipulate the/projects folder and its content with their operating system,so that the package does not lose track of these files. Multiuserapplication ofprojects requires a server or an otherwiseshared directory where multiple users can access the/projectsfolder. File-managing functions—along with all functions—aredemonstrated below in theDemonstration section.
Upon installation, theprojects package must be set upusingsetup_projects(). The user is to input the file pathof the directory wherein the/projects folder is to belocated.
library(projects)setup_projects("~")#> projects folder created at#> /tmp/RtmpH0SVxn/projects#>#> Add affiliations with new_affiliation(),#> then add authors with new_author(),#> then create projects with new_project()As the message suggests, it is in the user’s best interest to addaffiliations, followed by authors and projects.
new_affiliation(department_name ="Department of Physics",institution_name ="University of North Science",address ="314 Newton Blvd, Springfield CT 06003")#> New affiliation:#> # A tibble: 1 x 4#> id department_name institution_name address#> <int> <chr> <chr> <chr>#> 1 1 Department of Physi… University of North Sc… 314 Newton Blvd, Springfie…This affiliation has been successfully added to the “affiliations”table in theprojects relational database. Now to create afew more affiliations:
new_affiliation(department_name ="Impossibles Investigation Team",institution_name ="Creekshirebrook Academy of Thinks",address ="Let Gade 27182, 1566 Copenhagen, Denmark")#> New affiliation:#> # A tibble: 1 x 4#> id department_name institution_name address#> <int> <chr> <chr> <chr>#> 1 2 Impossibles Investigat… Creekshirebrook Academy… Let Gade 27182, 1566 C…new_affiliation(department_name ="Statistical Consulting Unit",institution_name ="Creekshirebrook Academy of Thinks",address ="196 Normal Ave, Columbus, OH ",id =50)#> New affiliation:#> # A tibble: 1 x 4#> id department_name institution_name address#> <int> <chr> <chr> <chr>#> 1 50 Statistical Consulting… Creekshirebrook Academy o… "196 Normal Ave, Col…Note that we chose a specificid number (50) for theaffiliation called the “Impossibles Investigation Team.”
Now we are ready to add authors to the “authors” table of theprojects database.
new_author(given_names ="Scott",last_name ="Bug",title ="Professor",affiliations =c(2,"Physics"),degree ="PhD",email ="scottbug@imPOSSible.net",phone ="965-555-5556")#> New author:#> # A tibble: 1 x 7#> id last_name given_names title degree email phone#> <int> <chr> <chr> <chr> <chr> <chr> <chr>#> 1 1 Bug Scott Professor PhD scottbug@impossible.… 965-555-55…#>#> New author's affiliations:#> # A tibble: 2 x 4#> affiliation_id department_name institution_name address#> <int> <chr> <chr> <chr>#> 1 2 Impossibles Investig… Creekshirebrook Acade… Let Gade 27182, 1…#> 2 1 Department of Physics University of North S… 314 Newton Blvd, …Notice that in creating associations between Scott Bug and hisaffiliations, we were able to enter both theid number ofone of them (2) and a substring of thedepartment_name ofthe other (“Physics”). Also notice that the email address was coerced tobe lowercase.
Now we’ll add more authors.
new_author(given_names ="Marie",last_name ="Curie",title ="Chemist",affiliations ="Unit",phone ="553-867-5309",id =86)#> New author:#> # A tibble: 1 x 7#> id last_name given_names title degree email phone#> <int> <chr> <chr> <chr> <chr> <chr> <chr>#> 1 86 Curie Marie Chemist <NA> <NA> 553-867-5309#>#> New author's affiliations:#> # A tibble: 1 x 4#> affiliation_id department_name institution_name address#> <int> <chr> <chr> <chr>#> 1 50 Statistical Consulti… Creekshirebrook Academ… "196 Normal Ave,…new_author(given_names ="George Washington",last_name ="Carver",title ="Astrophysicist",degree ="MA, MPhil, PhD",affiliations =c(1,2,50),id =1337)#> New author:#> # A tibble: 1 x 7#> id last_name given_names title degree email phone#> <int> <chr> <chr> <chr> <chr> <chr> <chr>#> 1 1337 Carver George Washington Astrophysicist MA, MPhil, PhD <NA> <NA>#>#> New author's affiliations:#> # A tibble: 3 x 4#> affiliation_id department_name institution_name address#> <int> <chr> <chr> <chr>#> 1 1 Department of Physics University of North … "314 Newton Blvd, …#> 2 2 Impossibles Investig… Creekshirebrook Acad… "Let Gade 27182, 1…#> 3 50 Statistical Consulti… Creekshirebrook Acad… "196 Normal Ave, C…new_author(last_name ="Archimedes",title ="Mathematician")#> New author:#> # A tibble: 1 x 7#> id last_name given_names title degree email phone#> <int> <chr> <chr> <chr> <chr> <chr> <chr>#> 1 2 Archimedes <NA> Mathematician <NA> <NA> <NA>#>#> New author's affiliations:#> None.new_author(last_name ="Wu",given_names ="Chien-Shiung",title ="Physicist",affiliations =c("of North","Statistical Consulting"),degree ="PhD",email ="wu@WU.wU")#> New author:#> # A tibble: 1 x 7#> id last_name given_names title degree email phone#> <int> <chr> <chr> <chr> <chr> <chr> <chr>#> 1 3 Wu Chien-Shiung Physicist PhD wu@wu.wu <NA>#>#> New author's affiliations:#> # A tibble: 2 x 4#> affiliation_id department_name institution_name address#> <int> <chr> <chr> <chr>#> 1 1 Department of Physi… University of North S… "314 Newton Blvd, …#> 2 50 Statistical Consult… Creekshirebrook Acade… "196 Normal Ave, C…Now that some authors and affiliations have been created, we can viewthese tables:
authors()#> # A tibble: 5 x 7#> id last_name given_names title degree email phone#> <int> <chr> <chr> <chr> <chr> <chr> <chr>#> 1 1 Bug Scott Professor PhD scottbug@impo… 965-555…#> 2 2 Archimedes <NA> Mathemati… <NA> <NA> <NA>#> 3 3 Wu Chien-Shiung Physicist PhD wu@wu.wu <NA>#> 4 86 Curie Marie Chemist <NA> <NA> 553-867…#> 5 1337 Carver George Washing… Astrophys… MA, MPhil… <NA> <NA>affiliations()#> # A tibble: 3 x 4#> id department_name institution_name address#> <int> <chr> <chr> <chr>#> 1 1 Department of Physics University of North Sci… "314 Newton Blvd, Spri…#> 2 2 Impossibles Investigat… Creekshirebrook Academy… "Let Gade 27182, 1566 …#> 3 50 Statistical Consulting… Creekshirebrook Academy… "196 Normal Ave, Colum…Now we will showcase project creation:
new_project(title ="Achieving Cold Fusion",short_title ="ACF",authors =c("Bug","Chien-Shiung",86,1337),current_owner ="Carver",corresp_auth ="Bug",stage ="1: design",deadline_type ="Pilot study",deadline ="2020-12-31")#>#> Project 1 has been created at#> /tmp/RtmpH0SVxn/projects/p0001#> # A tibble: 1 x 6#> id title stage status deadline_type deadline#> <int> <chr> <prjstg> <chr> <chr> <dttm>#> 1 1 Achieving Cold F… 1: design just crea… Pilot study 2020-12-31 00:00:00#>#> New project's authors:#> # A tibble: 4 x 7#> author_id last_name given_names title degree email phone#> <int> <chr> <chr> <chr> <chr> <chr> <chr>#> 1 1 Bug Scott Professor PhD scottbug@imp… 965-555…#> 2 3 Wu Chien-Shiung Physicist PhD wu@wu.wu <NA>#> 3 86 Curie Marie Chemist <NA> <NA> 553-867…#> 4 1337 Carver George Washin… Astrophys… MA, MPhi… <NA> <NA>#> # A tibble: 1 x 3#> current_owner corresp_auth creator#> <prjaut> <prjaut> <prjaut>#> 1 1337: Carver 1: Bug 0: kriegenNotice that since acreator was not specified, thisfield was populated with the value ofSys.info()["user"].
Notice also that the author order given to theauthorsargument innew_project() command has been preserved. Alsonotice that Scott Bug has been marked as the corresponding author, andhis contact information has been included. When knitted, this will be aproper title page.
Now a few more projects will be created:
new_project(title ="Weighing the Crown",short_title ="Eureka!",authors ="Archimedes",current_owner ="Archimedes",corresp_auth ="Archimedes",stage =4)#>#> Project 2 has been created at#> /tmp/RtmpH0SVxn/projects/p0002#> # A tibble: 1 x 6#> id title stage status deadline_type deadline#> <int> <chr> <prjstg> <chr> <chr> <dttm>#> 1 2 Weighing the … 4: manuscript just cre… <NA> NA#>#> New project's authors:#> # A tibble: 1 x 7#> author_id last_name given_names title degree email phone#> <int> <chr> <chr> <chr> <chr> <chr> <chr>#> 1 2 Archimedes <NA> Mathematician <NA> <NA> <NA>#> # A tibble: 1 x 3#> current_owner corresp_auth creator#> <prjaut> <prjaut> <prjaut>#> 1 2: Archimedes 2: Archimedes 0: kriegennew_project(title ="How I Learned to Stop Worrying and Love the Bomb",short_title ="Dr. Strangelove",authors =c("wu",1),creator ="wu",current_owner ="George",corresp_auth ="George",stage ="under review",deadline_type ="2nd revision",deadline ="2030-10-8",id =1945,status ="debating leadership changes",parent_directory ="top_secret",make_directories =TRUE)#>#> Project 1945 has been created at#> /tmp/RtmpH0SVxn/projects/top_secret/p1945#> # A tibble: 1 x 6#> id title stage status deadline_type deadline#> <int> <chr> <prjstg> <chr> <chr> <dttm>#> 1 1945 How I Learn… 5: under review debating… 2nd revision 2030-10-08 00:00:00#>#> New project's authors:#> # A tibble: 3 x 7#> author_id last_name given_names title degree email phone#> <int> <chr> <chr> <chr> <chr> <chr> <chr>#> 1 3 Wu Chien-Shiung Physicist PhD wu@wu.wu <NA>#> 2 1 Bug Scott Professor PhD scottbug@imp… 965-555…#> 3 1337 Carver George Washin… Astrophys… MA, MPhi… <NA> <NA>#> # A tibble: 1 x 3#> current_owner corresp_auth creator#> <prjaut> <prjaut> <prjaut>#> 1 1337: Carver 1337: Carver 3: Wunew_project(title ="Understanding Radon",short_title ="Rn86",authors =86,creator =86,corresp_auth =86,stage ="3",status ="Safety procedures")#>#> Project 3 has been created at#> /tmp/RtmpH0SVxn/projects/p0003#> # A tibble: 1 x 6#> id title stage status deadline_type deadline#> <int> <chr> <prjstg> <chr> <chr> <dttm>#> 1 3 Understandin… 3: analysis Safety proc… <NA> NA#>#> New project's authors:#> # A tibble: 1 x 7#> author_id last_name given_names title degree email phone#> <int> <chr> <chr> <chr> <chr> <chr> <chr>#> 1 86 Curie Marie Chemist <NA> <NA> 553-867-5309#> # A tibble: 1 x 3#> current_owner corresp_auth creator#> <prjaut> <prjaut> <prjaut>#> 1 86: Curie 86: Curie 86: CurieHere is the list of all projects that have been created:
projects()#> # A tibble: 4 x 5#> id title current_owner status stage#> <int> <chr> <prjaut> <chr> <prjstg>#> 1 1945 How I Learned to Stop Wor… 1337: Carver debating leade… 5: under review#> 2 2 Weighing the Crown 2: Archimedes just created 4: manuscript#> 3 3 Understanding Radon 86: Curie Safety procedu… 3: analysis#> 4 1 Achieving Cold Fusion 1337: Carver just created 1: designProjects, authors, and affiliations can all be edited with theirrespectiveedit_*() functions. For example, we can add toand remove affiliations from an author with:
edit_author(author ="Bug",affiliations =~+50- impossibles)When adding or removing affiliations/authors from an author/project,a one-sidedformula is used: it must begin with a tilde~, and elements are added with+ and removedwith-. Elements can be referred to by theirid numbers or their names, as described above.
A formula is also used in theauthors argument inedit_project():
edit_project("Cold",title ="Cold Fusion Is Actually Impossible",authors =~"archi",stage ="accepted")#>#> Header has changed. Reprint it with:#> header(1)Here, thetitle andstage of the projecthave also been edited.
Note that the default behavior when adding elements is to place thembefore the last author (unless there was only one author). This occursafter elements are removed, as specified by any minussigns (-) in theformula.
Thereorder_authors() function allows for the user tochange author order:
reorder_authors(project ="Cold Fusion","George","Bug",86)#>#> Header has changed. Reprint it with:#> header(1)Author order matters when obtaining title page YAML text to pasteinto a protocol or manuscript.Rmd file. Best practice is to doso late in the writing process, if, for example, the name of anaffiliation changes in the middle of a research project:
edit_affiliation(affiliation ="Impossibles",department_name ="Pseudoscience Debunking Unit")The text to be pasted in the YAML can be obtained for any givenproject usingheader():
header(project ="Cold")title:"Cold Fusion Is Actually Impossible"author:- George Washington Carver, MA, MPhil, PhD;^1,2,3^ Scott Bug, PhD;^1,3^\* Marie Curie;^3^ Chien-Shiung Wu, PhD;^1,3^ and Archimedes-^1^ Department of Physics, University of North Science,314 Newton Blvd, Springfield CT06003-^2^ Pseudoscience Debunking Unit, Creekshirebrook Academy of Thinks, Let Gade27182,1566 Copenhagen, Denmark-^3^ Statistical Consulting Unit, Creekshirebrook Academy of Thinks,196 Normal Ave, Columbus, OH- \* Corresponding author-314 Newton Blvd, Springfield CT06003-965-555-5556- scottbug@impossible.netIn order to organize projects, users can create subdirectories withinthe main/projects folder where individual project folders candwell. Among the examples above, this has already occurred with theproject with the nickname (i.e.,short_title)“Dr. Strangelove” because on its creation the argumentspath = top_secret andmake_directories = TRUEwere included. The latter argument must beTRUE if thedesired path does not already exist. Observe thepathcolumn among the existing projects (including thepathcolumn inprojects() output requires the argumentverbose = TRUE):
projects(verbose =TRUE)%>%select(id, short_title, path)#> # A tibble: 3 x 3#> id short_title path#> <int> <chr> <chr>#> 1 1945 Dr. Strangelove /tmp/RtmpH0SVxn/projects/top_secret/p1945#> 2 2 Eureka! /tmp/RtmpH0SVxn/projects/p0002#> 3 3 Rn86 /tmp/RtmpH0SVxn/projects/p0003Users can also create subdirectories with the functionnew_project_group():
new_project_group("Greek_studies/ancient_studies")#>#> The following directory was created:#> /tmp/RtmpH0SVxn/projects/Greek_studies/ancient_studiesIf a project has already been created, it can be movednot withedit_project() butmove_project(). Users can also copy projects usingcopy_project(); everything in the copy will be the sameexcept itsid, folder name (which, again, is based on itsid),path (which, again, is based on itsfolder name), and the name of its.Rproj file (which has thesame name as the folder name).
move_project("Crown",path ="Greek_studies/ancient_studies")#> # A tibble: 1 x 11#> id title short_title current_owner status deadline_type deadline#> <int> <chr> <chr> <prjaut> <chr> <chr> <dttm>#> 1 2 Weig… Eureka! 2: Archimedes just … <NA> NA#> # … with 4 more variables: stage <prjstg>, path <chr>, corresp_auth <prjaut>,#> # creator <prjaut>#>#> Project 2 moved so that its new path is#> /tmp/RtmpH0SVxn/projects/Greek_studies/ancient_studies/p0002copy_project(project_to_copy ="Radon",path ="dangerous_studies/radioactive_studies/radon_studies",make_directories =TRUE)#> # A tibble: 1 x 11#> id title short_title current_owner status deadline_type deadline#> <int> <chr> <chr> <prjaut> <chr> <chr> <dttm>#> 1 3 Unde… Rn86 86: Curie Safet… <NA> NA#> # … with 4 more variables: stage <prjstg>, path <chr>, corresp_auth <prjaut>,#> # creator <prjaut>#>#> Project 4 below is a copy of project 3 and is located at#> /tmp/RtmpH0SVxn/projects/dangerous_studies/radioactive_studies/radon_studies/p0004#> # A tibble: 1 x 11#> id title short_title current_owner status deadline_type deadline#> <int> <chr> <lgl> <prjaut> <chr> <chr> <dttm>#> 1 4 Unde… NA 86: Curie Safet… <NA> NA#> # … with 4 more variables: stage <prjstg>, path <chr>, corresp_auth <prjaut>,#> # creator <prjaut>#>#> The .Rproj file#> /tmp/RtmpH0SVxn/projects/dangerous_studies/radioactive_studies/radon_studies/p0004/p0003.Rproj#> was renamed to#> /tmp/RtmpH0SVxn/projects/dangerous_studies/radioactive_studies/radon_studies/p0004/p0004.Rproj#>#> Be sure to change all instances of "p0003" to "p0004" as desired#> (e.g., .bib files and references to them in YAML headers).projects(c("Crown","Radon"),verbose =TRUE)%>%select(id, title, path)#> # A tibble: 3 x 3#> id title path#> <int> <chr> <chr>#> 1 2 Weighing the C… /tmp/RtmpH0SVxn/projects/Greek_studies/ancient_studies/…#> 2 4 Understanding … /tmp/RtmpH0SVxn/projects/dangerous_studies/radioactive_…#> 3 3 Understanding … /tmp/RtmpH0SVxn/projects/p0003Projects can also be archived; they are moved into a subdirectorycalled/archive that is at the same level as the project folder(/pXXXX) before it was run. If this/archive folderdoes not exist, it will be created.
archive_project("Strangelove")#> # A tibble: 1 x 11#> id title short_title current_owner status deadline_type deadline#> <int> <chr> <chr> <prjaut> <chr> <chr> <dttm>#> 1 1945 How … Dr. Strang… 1337: Carver debat… 2nd revision 2030-10-08 00:00:00#> # … with 4 more variables: stage <prjstg>, path <chr>, corresp_auth <prjaut>,#> # creator <prjaut>#>#> The above project was archived and has the file path#> /tmp/RtmpH0SVxn/projects/top_secret/archive/p1945When a project is archived, it is no longer included inprojects() output unless the user setsarchived = TRUE.
projects(verbose =TRUE)%>%select(id, short_title, path)#> # A tibble: 3 x 3#> id short_title path#> <int> <chr> <chr>#> 1 2 Eureka! /tmp/RtmpH0SVxn/projects/Greek_studies/ancient_studies/p0002#> 2 4 <NA> /tmp/RtmpH0SVxn/projects/dangerous_studies/radioactive_stud…#> 3 3 Rn86 /tmp/RtmpH0SVxn/projects/p0003projects(verbose =TRUE,archived =TRUE)%>%select(id, short_title, path)#> # A tibble: 4 x 3#> id short_title path#> <int> <chr> <chr>#> 1 1945 Dr. Strangelo… /tmp/RtmpH0SVxn/projects/top_secret/archive/p1945#> 2 2 Eureka! /tmp/RtmpH0SVxn/projects/Greek_studies/ancient_studies/p…#> 3 4 <NA> /tmp/RtmpH0SVxn/projects/dangerous_studies/radioactive_s…#> 4 3 Rn86 /tmp/RtmpH0SVxn/projects/p0003Lastly, affiliations, authors and projects can be deleted with thedelete_*() functions. Deleting an author is complete: doingso removes the author from thecreator,current_owner andcorresp_auth fields of allprojects. Furthermore, deleting a project also deletes the entireproject folder. Use thedelete_*() functions withcaution.
delete_affiliation("north science")#> # A tibble: 1 x 4#> id department_name institution_name address#> <int> <chr> <chr> <chr>#> 1 1 Department of Physi… University of North Sc… 314 Newton Blvd, Springfie…#> # A tibble: 1 x 4#> id department_name institution_name address#> <int> <chr> <chr> <chr>#> 1 1 Department of Physi… University of North Sc… 314 Newton Blvd, Springfie…#> The above affiliation was deleted.delete_author(2)#> # A tibble: 1 x 7#> id last_name given_names title degree email phone#> <int> <chr> <chr> <chr> <chr> <chr> <chr>#> 1 2 Archimedes <NA> Mathematician <NA> <NA> <NA>#> # A tibble: 1 x 7#> id last_name given_names title degree email phone#> <int> <chr> <chr> <chr> <chr> <chr> <chr>#> 1 2 Archimedes <NA> Mathematician <NA> <NA> <NA>#> The above author was deleted.delete_project("Crown")#> # A tibble: 1 x 11#> id title short_title current_owner status deadline_type deadline#> <int> <chr> <chr> <prjaut> <chr> <chr> <dttm>#> 1 2 Weig… Eureka! NA just … <NA> NA#> # … with 4 more variables: stage <prjstg>, path <chr>, corresp_auth <prjaut>,#> # creator <prjaut>#> # A tibble: 1 x 11#> id title short_title current_owner status deadline_type deadline#> <int> <chr> <chr> <prjaut> <chr> <chr> <dttm>#> 1 2 Weig… Eureka! NA just … <NA> NA#> # … with 4 more variables: stage <prjstg>, path <chr>, corresp_auth <prjaut>,#> # creator <prjaut>#>#> The above project was deleted.Theprojects package provides a comprehensive set oftools for reproducible team science workflows. Efficiency in projectmanagement, including manuscript development, is facilitated by aninternal database that keeps record of project details as well as teammembers’ affiliations and contact information. For manuscripts, titlepages are automatically generated from this database, and a selection ofmanuscript outlines compliant with reporting guidelines are available inR Markdown format. We believe that theprojects package maybe useful for teams that manage multiple collaborative research projectsin various stages of development.
Baker, Monya. 2016. “1,500 Scientists Lift the Lid onReproducibility.”Nature News 533 (7604): 452.