
Holomics is an R Shiny application that enables users toperform single- and multi-omics analyses by providing a user-friendlyinterface to upload different omics datasets, select and run theimplemented algorithms and finally visualize the generated results.
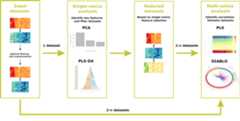
Holomics is primarily built on the R package mixOmics, whichoffers numerous algorithms for the integrative analysis of omicsdatasets. From this repertoire, the single-omics algorithms “PrincipalComponent Analysis” (PCA) and “Partial Least Squares DiscriminantAnalysis” (PLS-DA), the pairwise-omics analysis “sparse Partial LeastSquares” (sPLS) and the multi-omics framework DIABLO (“Data IntegrationAnalysis for Biomarker discovery using Latent variable approaches forOmics studies”) have been implemented inHolomics.
install.packages("Holomics")# Install devtools if it is not already installedinstall.packages("devtools")library(devtools)# Install Holomics package install_github("https://github.com/MolinLab/Holomics")You need to install the Bioconductor package separately.
if (!require("BiocManager", quietly = TRUE)) install.packages("BiocManager")BiocManager::install("mixOmics")BiocManager::install("BiocParallel")Either with
library(Holomics)run_app()or
Holomics::run_app()To use all the features offered, the following workflow should befollowed. First, datasets are uploaded, during which any necessarypre-filtering or transformation steps take place. Next, the user shouldproceed to the single-omics analysis, where key features are identifiedand the datasets are reduced accordingly. After completing thesingle-omics analyses, the user can apply multi-omics analyses toidentify correlations between two or more datasets. NOTE: Ifpre-filtered datasets (ideally generated earlier using Holomics) havealready been uploaded, it is possible to start directly with themulti-omics analysis.
For further information on how to use Holomics please have a look atourvignette.