The goal of omicsTools is to provide tools for processing andanalyzing omics data from genomics, transcriptomics, proteomics, andmetabolomics platforms. It provides functions for preprocessing,normalization, visualization, and statistical analysis, as well asmachine learning algorithms for predictive modeling. omicsTools is anessential tool for researchers working with high-throughput omics datain fields such as biology, bioinformatics, and medicine.
License: AGPL-3.0
if (!require("BiocManager",quietly =TRUE)) {install.packages("BiocManager") }if (!require("pvca",quietly =TRUE)) { BiocManager::install("pvca") }You can install the Stable version of omicsTools like so:
install.packages("omicsTools")#> Installing package into 'C:/Users/bach/AppData/Local/Temp/RtmpqQj0Pu/temp_libpath245079296a59'#> (as 'lib' is unspecified)#> package 'omicsTools' successfully unpacked and MD5 sums checked#>#> The downloaded binary packages are in#> C:\Users\bach\AppData\Local\Temp\RtmpIN7Pjh\downloaded_packagesTo get a bug fix, or use a feature from the development version, youcan install omicsTools from GitHub.
if (!require("devtools",quietly =TRUE))install.packages("devtools")#>#> Attaching package: 'devtools'#> The following object is masked from 'package:BiocManager':#>#> installdevtools::install_github("omicsTools")#> Using GitHub PAT from the git credential store.#> Downloading GitHub repo omicsTools@HEAD#> rlang (1.1.3 -> 1.1.4 ) [CRAN]#> cli (3.6.2 -> 3.6.3 ) [CRAN]#> digest (0.6.35 -> 0.6.36) [CRAN]#> fresh (0.2.0 -> 0.2.1 ) [CRAN]#> dbscan (1.1-12 -> 1.2-0 ) [CRAN]#> Installing 5 packages: rlang, cli, digest, fresh, dbscan#> Installing packages into 'C:/Users/bach/AppData/Local/Temp/RtmpqQj0Pu/temp_libpath245079296a59'#> (as 'lib' is unspecified)#> package 'rlang' successfully unpacked and MD5 sums checked#> package 'cli' successfully unpacked and MD5 sums checked#> package 'digest' successfully unpacked and MD5 sums checked#> package 'fresh' successfully unpacked and MD5 sums checked#> package 'dbscan' successfully unpacked and MD5 sums checked#>#> The downloaded binary packages are in#> C:\Users\bach\AppData\Local\Temp\RtmpIN7Pjh\downloaded_packages#> ── R CMD build ─────────────────────────────────────────────────────────────────#> checking for file 'C:\Users\bach\AppData\Local\Temp\RtmpIN7Pjh\remotes53285a4b5044\omicsTools-baa6316/DESCRIPTION' ... ✔ checking for file 'C:\Users\bach\AppData\Local\Temp\RtmpIN7Pjh\remotes53285a4b5044\omicsTools-baa6316/DESCRIPTION'#> ─ preparing 'omicsTools': (337ms)#> checking DESCRIPTION meta-information ... checking DESCRIPTION meta-information ... ✔ checking DESCRIPTION meta-information#> ─ checking for LF line-endings in source and make files and shell scripts#> ─ checking for empty or unneeded directories#> Omitted 'LazyData' from DESCRIPTION#> ─ building 'omicsTools_1.1.3.tar.gz'#>#>#> Installing package into 'C:/Users/bach/AppData/Local/Temp/RtmpqQj0Pu/temp_libpath245079296a59'#> (as 'lib' is unspecified)# Load the CSV datadata_file<-system.file("extdata","example1.csv",package ="omicsTools")data<- readr::read_csv(data_file)#> Rows: 85 Columns: 482#> ── Column specification ────────────────────────────────────────────────────────#> Delimiter: ","#> chr (1): Sample#> dbl (414): Urea_pos, Lipoamide_pos, AcetylAmino Sugars_pos, Glycerophosphoch...#> lgl (67): DBQ_pos.IS, Aminolevulinic Acid_pos, Leucine_pos, Homocystine_pos...#>#> ℹ Use `spec()` to retrieve the full column specification for this data.#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.# Apply the impute functionimputed_data<- omicsTools::handle_missing_values(data)#> Registered S3 method overwritten by 'GGally':#> method from#> +.gg ggplot2#> ℹ Starting missing value handling... 🚀#> ℹ Calculating missing value percentages... ⏳#> ✔ 446 features removed due to missing values exceeding the threshold. ✅#> ℹ Imputing missing values using method: mean 🔧#> ✔ Missing value handling completed! 🎉# Write the imputed data to a new CSV filereadr::write_csv(imputed_data,paste0(tempdir(),"/imputed_data.csv"))# Load the CSV datadata_file<-system.file("extdata","example2.csv",package ="omicsTools")data<- readr::read_csv(data_file)#> Rows: 63 Columns: 202#> ── Column specification ────────────────────────────────────────────────────────#> Delimiter: ","#> chr (1): Sample#> dbl (201): Urea_pos, Lipoamide_pos, Glycerophosphocholine_pos, Allanoate_pos...#>#> ℹ Use `spec()` to retrieve the full column specification for this data.#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.# Apply the normalize functionnormalized_data<- omicsTools::qc_normalize(data)# Write the normalized data to a new CSV filereadr::write_csv(normalized_data,paste0(tempdir(),"/normalized_data.csv"))# Using generated datalibrary(omicsTools)#>#> This is omicsTools version 1.1.3.#> omicsTools is free software and comes with ABSOLUTELY NO WARRANTY.#> Please use at your own risk.omics_data<-createOmicsData()#>#> ── Generating High-Dimensional Data with Anomalies ─────────────────────────────#> ℹ Introducing anomalies in specific features#> ✔ Data generation complete# Define custom thresholdscustom_thresholds<-define_thresholds(skewness =3,kurtosis =8)# Update the OmicsData object with custom thresholdsomics_data@thresholds<- custom_thresholds# Calculate measures for each featureomics_data<-calculate_measures(omics_data)#>#> ── Calculating Measures for Each Feature ───────────────────────────────────────#> Calculating [===>------------------------------------------------] 8% in 0sCalculating [===>------------------------------------------------] 9% in 0sCalculating [====>-----------------------------------------------] 9% in 0sCalculating [====>-----------------------------------------------] 10% in 0sCalculating [====>-----------------------------------------------] 11% in 0sCalculating [=====>----------------------------------------------] 11% in 0sCalculating [=====>----------------------------------------------] 12% in 0sCalculating [======>---------------------------------------------] 13% in 0sCalculating [======>---------------------------------------------] 14% in 0sCalculating [=======>--------------------------------------------] 14% in 0sCalculating [=======>--------------------------------------------] 15% in 0sCalculating [=======>--------------------------------------------] 16% in 0sCalculating [========>-------------------------------------------] 16% in 0sCalculating [========>-------------------------------------------] 17% in 0sCalculating [========>-------------------------------------------] 17% in 1sCalculating [========>-------------------------------------------] 18% in 1sCalculating [=========>------------------------------------------] 18% in 1sCalculating [=========>------------------------------------------] 19% in 1sCalculating [=========>------------------------------------------] 20% in 1sCalculating [==========>-----------------------------------------] 20% in 1sCalculating [==========>-----------------------------------------] 21% in 1sCalculating [==========>-----------------------------------------] 22% in 1sCalculating [===========>----------------------------------------] 22% in 1sCalculating [===========>----------------------------------------] 23% in 1sCalculating [===========>----------------------------------------] 24% in 1sCalculating [============>---------------------------------------] 24% in 1sCalculating [============>---------------------------------------] 25% in 1sCalculating [============>---------------------------------------] 26% in 1sCalculating [=============>--------------------------------------] 26% in 1sCalculating [=============>--------------------------------------] 27% in 1sCalculating [=============>--------------------------------------] 28% in 1sCalculating [==============>-------------------------------------] 28% in 1sCalculating [==============>-------------------------------------] 29% in 1sCalculating [==============>-------------------------------------] 30% in 1sCalculating [===============>------------------------------------] 30% in 1sCalculating [===============>------------------------------------] 31% in 1sCalculating [===============>------------------------------------] 32% in 1sCalculating [================>-----------------------------------] 32% in 1sCalculating [================>-----------------------------------] 33% in 1sCalculating [================>-----------------------------------] 34% in 1sCalculating [=================>----------------------------------] 34% in 1sCalculating [=================>----------------------------------] 35% in 1sCalculating [=================>----------------------------------] 36% in 1sCalculating [==================>---------------------------------] 36% in 1sCalculating [==================>---------------------------------] 37% in 1sCalculating [===================>--------------------------------] 38% in 1sCalculating [===================>--------------------------------] 39% in 1sCalculating [====================>-------------------------------] 39% in 1sCalculating [====================>-------------------------------] 40% in 1sCalculating [====================>-------------------------------] 41% in 1sCalculating [=====================>------------------------------] 41% in 1sCalculating [=====================>------------------------------] 42% in 1sCalculating [=====================>------------------------------] 43% in 1sCalculating [======================>-----------------------------] 43% in 1sCalculating [======================>-----------------------------] 44% in 1sCalculating [======================>-----------------------------] 45% in 1sCalculating [=======================>----------------------------] 45% in 1sCalculating [=======================>----------------------------] 46% in 1sCalculating [=======================>----------------------------] 47% in 1sCalculating [========================>---------------------------] 47% in 1sCalculating [========================>---------------------------] 48% in 1sCalculating [========================>---------------------------] 48% in 2sCalculating [========================>---------------------------] 49% in 2sCalculating [=========================>--------------------------] 49% in 2sCalculating [=========================>--------------------------] 50% in 2sCalculating [=========================>--------------------------] 51% in 2sCalculating [==========================>-------------------------] 51% in 2sCalculating [==========================>-------------------------] 52% in 2sCalculating [==========================>-------------------------] 53% in 2sCalculating [===========================>------------------------] 53% in 2sCalculating [===========================>------------------------] 54% in 2sCalculating [===========================>------------------------] 55% in 2sCalculating [============================>-----------------------] 55% in 2sCalculating [============================>-----------------------] 56% in 2sCalculating [============================>-----------------------] 57% in 2sCalculating [=============================>----------------------] 57% in 2sCalculating [=============================>----------------------] 58% in 2sCalculating [=============================>----------------------] 59% in 2sCalculating [==============================>---------------------] 59% in 2sCalculating [==============================>---------------------] 60% in 2sCalculating [==============================>---------------------] 61% in 2sCalculating [===============================>--------------------] 61% in 2sCalculating [===============================>--------------------] 62% in 2sCalculating [================================>-------------------] 63% in 2sCalculating [================================>-------------------] 64% in 2sCalculating [=================================>------------------] 64% in 2sCalculating [=================================>------------------] 65% in 2sCalculating [=================================>------------------] 66% in 2sCalculating [==================================>-----------------] 66% in 2sCalculating [==================================>-----------------] 67% in 2sCalculating [==================================>-----------------] 68% in 2sCalculating [===================================>----------------] 68% in 2sCalculating [===================================>----------------] 69% in 2sCalculating [===================================>----------------] 70% in 2sCalculating [====================================>---------------] 70% in 2sCalculating [====================================>---------------] 71% in 2sCalculating [====================================>---------------] 72% in 2sCalculating [=====================================>--------------] 72% in 2sCalculating [=====================================>--------------] 73% in 2sCalculating [=====================================>--------------] 74% in 2sCalculating [======================================>-------------] 74% in 2sCalculating [======================================>-------------] 75% in 2sCalculating [======================================>-------------] 76% in 2sCalculating [=======================================>------------] 76% in 2sCalculating [=======================================>------------] 77% in 2sCalculating [=======================================>------------] 78% in 2sCalculating [========================================>-----------] 78% in 2sCalculating [========================================>-----------] 79% in 2sCalculating [========================================>-----------] 79% in 3sCalculating [========================================>-----------] 80% in 3sCalculating [=========================================>----------] 80% in 3sCalculating [=========================================>----------] 81% in 3sCalculating [=========================================>----------] 82% in 3sCalculating [==========================================>---------] 82% in 3sCalculating [==========================================>---------] 83% in 3sCalculating [==========================================>---------] 84% in 3sCalculating [===========================================>--------] 84% in 3sCalculating [===========================================>--------] 85% in 3sCalculating [===========================================>--------] 86% in 3sCalculating [============================================>-------] 86% in 3sCalculating [============================================>-------] 87% in 3sCalculating [=============================================>------] 88% in 3sCalculating [=============================================>------] 89% in 3sCalculating [==============================================>-----] 89% in 3sCalculating [==============================================>-----] 90% in 3sCalculating [==============================================>-----] 91% in 3sCalculating [===============================================>----] 91% in 3sCalculating [===============================================>----] 92% in 3sCalculating [===============================================>----] 93% in 3sCalculating [================================================>---] 93% in 3sCalculating [================================================>---] 94% in 3sCalculating [================================================>---] 95% in 3sCalculating [=================================================>--] 95% in 3sCalculating [=================================================>--] 96% in 3sCalculating [=================================================>--] 97% in 3sCalculating [==================================================>-] 97% in 3sCalculating [==================================================>-] 98% in 3sCalculating [==================================================>-] 99% in 3sCalculating [===================================================>] 99% in 3sCalculating [===================================================>] 100% in 3sCalculating [====================================================] 100% in 3s#> ✔ Measures calculation complete# Flag anomaliesomics_data<-flag_anomalies(omics_data)#>#> ── Flagging Anomalies ──────────────────────────────────────────────────────────#> ✔ Anomaly flagging complete# Plotting distribution measuresplot_distribution_measures(omics_data)#>#> ── Plotting Distribution Measures ──────────────────────────────────────────────#> ✔ Distribution measures plot complete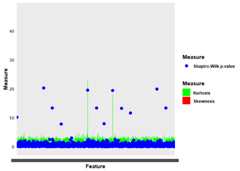
# Plotting sample measuresplot_sample_measures(omics_data)#>#> ── Plotting Sample Measures ────────────────────────────────────────────────────#> ✔ Sample measures plot complete
# Prepare data for UpSet plotupset_data<-prepare_upset_data(omics_data)#>#> ── Preparing Data for UpSet Plot ───────────────────────────────────────────────#> ✔ Data preparation for UpSet plot complete# Convert to binary matrix for UpSetRupset_matrix<-convert_to_binary_matrix(upset_data)#>#> ── Converting Data to Binary Matrix for UpSetR ─────────────────────────────────#> ✔ Data conversion to binary matrix complete# Ensure there are at least two columns for the UpSet plotensure_enough_sets_for_upset(upset_matrix)#>#> ── Checking Data for UpSet Plot ────────────────────────────────────────────────#> ✔ Enough sets for UpSet plot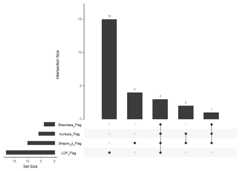
library(omicsTools)# Assuming user_data is a data frame where rows are samples and columns are featuresuser_data<-generate_data_with_anomalies(n_samples =100,n_features =200)# Replace with actual data loading if available#>#> ── Generating High-Dimensional Data with Anomalies ─────────────────────────────#> ℹ Introducing anomalies in specific features#> ✔ Data generation completeomics_data_user<-createOmicsData(data = user_data)# Define custom thresholdscustom_thresholds_user<-define_thresholds(skewness =4,kurtosis =6,shapiro_p =1e-5,cooks_distance =0.5,lof =10)# Update the OmicsData object with custom thresholdsomics_data_user@thresholds<- custom_thresholds_user# Calculate measures for each featureomics_data_user<-calculate_measures(omics_data_user)#>#> ── Calculating Measures for Each Feature ───────────────────────────────────────#> Calculating [=====================================>--------------] 74% in 0sCalculating [======================================>-------------] 74% in 0sCalculating [======================================>-------------] 75% in 0sCalculating [======================================>-------------] 76% in 0sCalculating [=======================================>------------] 76% in 0sCalculating [=======================================>------------] 77% in 0sCalculating [=======================================>------------] 78% in 0sCalculating [========================================>-----------] 78% in 0sCalculating [========================================>-----------] 79% in 0sCalculating [========================================>-----------] 80% in 0sCalculating [=========================================>----------] 80% in 0sCalculating [=========================================>----------] 81% in 0sCalculating [=========================================>----------] 82% in 0sCalculating [==========================================>---------] 82% in 0sCalculating [==========================================>---------] 83% in 0sCalculating [==========================================>---------] 84% in 0sCalculating [===========================================>--------] 84% in 0sCalculating [===========================================>--------] 85% in 0sCalculating [===========================================>--------] 86% in 0sCalculating [============================================>-------] 86% in 0sCalculating [============================================>-------] 87% in 0sCalculating [=============================================>------] 88% in 0sCalculating [=============================================>------] 89% in 0sCalculating [==============================================>-----] 90% in 0sCalculating [==============================================>-----] 91% in 0sCalculating [===============================================>----] 92% in 0sCalculating [===============================================>----] 93% in 0sCalculating [================================================>---] 94% in 0sCalculating [================================================>---] 95% in 0sCalculating [=================================================>--] 96% in 0sCalculating [=================================================>--] 97% in 0sCalculating [==================================================>-] 98% in 0sCalculating [==================================================>-] 99% in 0sCalculating [===================================================>] 100% in 0sCalculating [====================================================] 100% in 0s#> ✔ Measures calculation complete# Flag anomaliesomics_data_user<-flag_anomalies(omics_data_user)#>#> ── Flagging Anomalies ──────────────────────────────────────────────────────────#> ✔ Anomaly flagging complete# Plotting distribution measuresplot_distribution_measures(omics_data_user)#>#> ── Plotting Distribution Measures ──────────────────────────────────────────────#> ✔ Distribution measures plot complete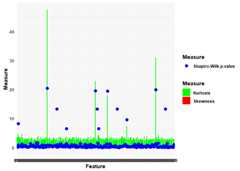
# Plotting sample measuresplot_sample_measures(omics_data_user)#>#> ── Plotting Sample Measures ────────────────────────────────────────────────────#> ✔ Sample measures plot complete
# Prepare data for UpSet plotupset_data_user<-prepare_upset_data(omics_data_user)#>#> ── Preparing Data for UpSet Plot ───────────────────────────────────────────────#> ✔ Data preparation for UpSet plot complete# Convert to binary matrix for UpSetRupset_matrix_user<-convert_to_binary_matrix(upset_data_user)#>#> ── Converting Data to Binary Matrix for UpSetR ─────────────────────────────────#> ✔ Data conversion to binary matrix complete# Ensure there are at least two columns for the UpSet plotensure_enough_sets_for_upset(upset_matrix_user)#>#> ── Checking Data for UpSet Plot ────────────────────────────────────────────────#> ✔ Enough sets for UpSet plot
Since this is a collaborative project, please adhere to the followingcode formatting conventions: * We use the tidyverse style guide (https://style.tidyverse.org/) * Please write roxygen2comments as full sentences, starting with a capital letter and endingwith a period. Brevity is preferred (e.g., “Calculates standarddeviation” is preferred over “This method calculates and returns astandard deviation of given set of numbers”).